Abstract
The crystal structure of the hypothetical protein TA1238 from Thermoplasma acidophilum was solved with multiple-wavelength anomalous diffraction and refined at 2.0 Å resolution. The molecule consists of a typical four-helix antiparallel bundle with overhand connection. However, its oligomerization into a trimer leads to a coiled ‘super-helix’ which is novel for such bundles. Its central feature, a six-stranded coiled coil, is also novel for proteins. TA1238 does not have significant sequence relatives in databases, but shows strong structural homologues with some proteins in the Protein Data Bank. The function could not be inferred from the sequence but the structure, with some rearrangement, bears some resemblance to the active site region of cobalamin adenosyltransferase (TA1434). Specifically, TA1238 retains Arg104, which is structurally equivalent to functionally critical Arg119 of TA1434. For such conformational change, the overhand connection of TA1238 might need to be involved in a gating mechanism that might be modulated by ligands and/or by interactions with the physiological partners. This allowed us to hypothesize that TA1238 could be involved in cobalamin biosyntheses.
Keywords: cobalamin biosynthesis, crystal structure, four-helix bundle, gating mechanism, MAD phasing, overhand connection, six-stranded coiled coil
Introduction
One of the major benefits of the Protein Structure Initiative (http://www.nigms.nih.gov/psi/) is the opportunity to infer the molecular and cellular functions of novel proteins by comparisons with the known structures. To this end, extensive effort has gone into developing and enhancing widely accessible databases and bioinformatics tools (e.g., www.ncbi.nlm.nih.gov, http://ca.expasy.org, www.ebi.ac.uk, http://scop.berkeley.edu) which can often be utilized successfully for identifying the function of unknown proteins (e.g., [1, 2]). It is to be expected that as the number of solved protein structures increases, the success rate for their functional annotation should also increase, in part because of enrichment of the databases with new information and in part due to improvement in the bioinformatics tools themselves.
Here we report the crystal structure of the TA1238 gene (gi16082243) product from Thermoplasma acidophilum, solved and refined at 2.0 Å resolution. The TA1238 protein displays remarkably high structural homology with a number of proteins with wide range of functions, while it has no sequence similarity with them. Automated analysis of patterns of sequence and structural conservation did not produce reliable indications of the protein's function. We carried out automated searches with ProFunc [3] as well. ProFunc contains bioinformatics tools such as InterPro [4] to scan for sequence motifs; BLAST search vs. PDB entries; matching folds detected by SSM; residue conservation analysis; gene neighbors; search for DNA-binding motifs (helix-turn-helix); NEST analysis to identify structural motifs that are often found in functionally important regions of protein structures [5–7] and 3D functional template searches [8, 9]. None of these searches yielded strong functional clues.
Detailed comparison with cobalamin adenosyltransferase led us to hypothesize that TA1238 is involved in cobalamin biosynthesis. This biological function should be tested.
Materials and methods
Cloning, purification, and crystallization
The gene (GenBank Accession No. gi16082243) encoding protein TA1238 was cloned from T. acidophilum genomic DNA and its gene product was expressed, selenomethionine(Se–Met)-labeled, and purified from a bacterial system using the NiNTA affinity procedure as described elsewhere for Methanococcus thermoautotrophicum proteins [10]. In brief, TA1238 recombinants were expressed in the Escherichia coli strain BL21 Gold in 1 l Luria-Bertani medium supplemented with 50 μg/ml kanamycin and 50 μg/ml ampicillin. The culture was incubated at 37 °C with shaking until it reached an optical density of 0.7 at 600 nm. The culture was induced with 0.4 mM isoprolylthiogalactoside (IPTG) for 3 h at 37 °C, then allowed to grow overnight with shaking at 24 °C. Cells were harvested by centrifugation, disrupted by sonication, and the insoluble cellular material was removed by centrifugation. TA1238 was purified using Ni-NTA affinity chromatography with the N-terminal hexahistidine affinity tag. Purified TA1238 was concentrated and quantified at 280 nm.
Crystallization experiments were conducted using the hanging-drop vapor diffusion method at room temperature. The purified protein was crystallized with the hexahistidine tag still attached. The final crystallization conditions were 16% polyethylene glycol 3350 (PEG3350) as the precipitant, 0.3 M ammonium chloride, 0.1 M sodium acetate at pH 4.6, and 12% glycerol. For X-ray diffraction studies, crystals were flash frozen by plunging from the crystallization drops directly into liquid nitrogen.
Crystallographic data collection
A three-wavelength anomalous diffraction experiment was carried out on the 19ID beamline of the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory). First, the fluorescence spectrum in the 25-eV range immediately before and after the Se absorption edge was recorded from the test crystal of Se–Met-substituted TA1238 protein. The CHOOCH program [11] was used to calculate f′ values from measured values of f″, and to plot them vs. energy. Diffraction data were collected from the fresh crystal at the Se absorption peak, at the rising inflection point, and at remote energy (13 keV) above the absorption edge. An additional data set was collected from a native protein crystal on the same beamline. This data set was collected in two steps – (50–2.66 Å) and (50–2.0 Å) resolution swipes. An approximately 40 times higher dose rate was used for the higher resolution swipe, resulting in some overloads at low resolution and better measured higher resolution reflections. All data were measured with a custom-built 3 × 3 tiled CCD detector [12]. The d*TREK program suite [13] was used for data collection and visualization. All data were integrated and scaled with the program package HKL2000 [14]. Some of the basic statistics of data collection and processing are given in Table 1.
Table 1.
Basic data collection and processing statistics for TA1238 crystals.
| Number of amino acid residues/methionines | 152/4 | ||
|---|---|---|---|
| Number of molecules in the asymmetric unit | 1 | ||
| Crystal lattice | |||
| Native | I23 | a = b = c = 101.25 Å | α = β = γ = 90° |
| Se–Met | I23 | a = b = c = 101.42 Å | α = β = γ = 90° |
| Crystal 1 (Se–Met) |
Crystal 2 (native) |
||||
|---|---|---|---|---|---|
| Infl. Point |
Peak |
Remote |
Low res. |
High res. |
|
| Energy (keV) | 12.6660a | 12.6678a | 13.0000 | 12.6463 | 12.6463 |
| Wavelength (Å) | 0.97887 | 0.97875 | 0.95372 | 0.98040 | 0.98040 |
| Resolution (Å) | 50–2.5 | 50–2.5 | 50–2.5 | 50–2.66 | 50–2.0 |
| No. of observations | 266013 | 268388 | 217084 | 35204 | 83217 |
| Uniqueb reflections | 11656 | 11655 | 11700 | 5017 | 11640 |
| Completeness (%)c | 99.99 (99.99) | 99.99(99.99) | 99.99 (99.99) | 98.4 (99.2) | 98.3 (99.99) |
| I/σ (I)c | 50.2 (7.6) | 50.2 (8.2) | 45.1 (7.1) | 46.9 (8.4) | 49.8 (4.6) |
| Rsymc | 0.076 (0.50) | 0.074 (0.47) | 0.074 (0.48) | 0.040 (0.215) | 0.033 (0.39) |
| Rmerge between low- and high-resolution passes on native rystals | 0.011 | ||||
Energy deviation from usual values of Se absorption edge reflects calibration of the monochromator at the time of the experiment.
Bijvoet pairs for scaling the MAD data sets were kept separately.
The numbers in parentheses correspond to the last resolution shell.
Results and discussion
Structure determination and refinement
MAD phasing of TA1238 data up to 2.5 Å resolution was carried out with the CNS program suite [15]. Little less than half of the model was built automatically with the ARP/wARP program [16] and the model was completed interactively with QUANTA (Accelrys, Inc.). The model was then refined against later-obtained 2.0 Å native data with one global cycle of CNS consisting of rigid body, simulated annealing, B-factor, and positional refinements. All subsequent refinement was performed with REFMAC5 [17], as implemented in CCP4 [18]. Phasing and refinement parameters are shown in Table 2. The coordinates of TA1238 have been deposited into the Protein Data Bank [19] with the entry code 1NIG.
Table 2.
Phasing and refinement statistics for TA1238 structure.
| Energies | Resolution (Å)a | Phasing Number of reflectionsa | Phasing powera | FOMa |
|---|---|---|---|---|
| Infl. point | 40–2.5 (2.81–2.7) | 11161 (1147) | 5.17 (1.80) | 0.50 (0.37) |
| Peak | 40–2.5 (2.81–2.7) | 11154 (1158) | 5.85 (2.08) | 0.49 (0.31) |
| Remote | 40–2.5 (2.81–2.7) | 11101 (1127) | 4.88 (1.64) | 0.49 (0.29) |
| Overall | 41.4–2.5 (2.81–2.7) | 11431 (1244) | 0.81 (0.52) |
| Refinement | ||
|---|---|---|
| Resolution (Å) | 70–2.0b | 2.05–2.0 |
| Number of reflections | 10810 | 735 |
| R factor % | 21.3 | 25.4 |
| R free % | 25.6 | 33.5 |
| Correlation | 94.9 | |
| Correlation (free) | 91.6 | |
| Number of all atoms | 1309 | |
| Number of solvent atoms | 41 | |
| Mean B factor | 32.85 | |
| Deviations from ideal |
Refined |
Target |
| Covalent bonds | 0.024 | 0.021 |
| Bond angles | 1.88 | 1.950 |
| Planarity | 0.016 | 0.02 |
| Chiral centers | 0.127 | 0.20 |
| Torsion angles | 7.96 | 5.0 |
| VDW contacts | 0.227 | 0.20 |
Parentheses show the statistics in the highest-resolution shell.
Low resolution limit is as assigned by the CCP4 suite.
Spatial model of TA1238
In the asymmetric unit of TA1238 crystals, 148 of 152 amino acid residues of the protein monomer and 41 solvent molecules were located. Residues Glu149, Val150, Arg151, and Leu152, as well as the hexahistidine tag, could not be fitted unambiguously and are missing from the model. For the same reason, the side chains of Met1, Asp2, Pro64, Glu65, Asp63, Lys67, Asp70, Phe71, Lys74, Phe78, Lys89, Glu121, and Lys128 were modeled only partially.
Residues Val19, Ser39, Lys43, Arg45, Arg48, Glu49, Ile50, Lys87, Glu88, Asn91, Asn92, Gly93, Asn94, Ser105, Arg116, Glu119, Glu130, and Leu135 have been modeled in more than one conformation. Of these, Val19, Ser39, Arg45, Asn92, Gly93, and Ser105 can be reliably modeled as distinct rotamers. The disorder of Lys43, Arg48, Glu49, Ile50, Lys87, Glu88, Asn91, Asn94, Arg116, Glu119, Glu130, and Leu135 is more complicated. These side chains do not have discrete electron densities, most likely because of disorder at more than one χ angle and/or several closely related values of these angles. Segment 91–94, connecting the helices α4 and α5, has two conformations in its backbone as well.
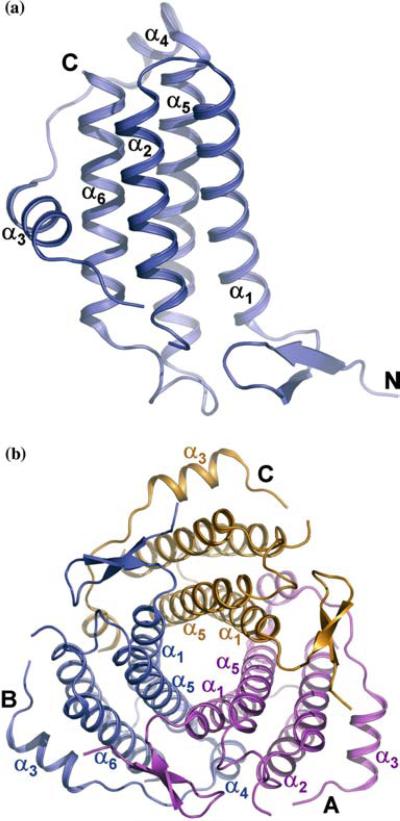
The TA1238 molecule consists of six α helices, α1 (residues 18–40), α2 (43–60), α3 (69–79), α4 (85–92), α5 (94–119), and α6 (128–148) (Figure 1a). At its N-terminus, there are also two short β strands, β1 (residues 4–7) and β2 (residues 12–15), forming a β hairpin. The four long helices – α1, α2, α5 and α6 – form a typical left-handed [19], anti-parallel α helical bundle with α1↑α2↓α6↑α5↓ topology. Helices α2 and α5 are linked with a long (residues 62–93) overhand connection [20], common for this class of helical bundles. It contains two shorter helices, α3 and α4, and wraps around the bundle. The non-helical parts of this connection are rather flexible, as indicated by alternative conformations and disorder of several residues.
Figure 1.
Cartoon representation of TA1238 monomer and trimer. (a) Helices α1, α2, α5, and α6 form a typical four-helix bundle, while α3 and α4 are part of a long overhand connection between α2 and α5. Breaks in the chain between helices α2 and α3 are due to several missing residues caused by their disorder. Four residues are missing at the C-terminus as well. (b) Monomers and labels in the TA1238 trimer are color coded. For clarity, only the labels of the secondary structure elements that are discussed in the text are shown. Orientations in a and b are related by a rotation of about 45° around the horizontal axis in the plane of the paper. (Figure was generated with PyMol, www.delanoscientific.com.)
The monomer of TA1238 belongs to the so-called square bundles, in which all helix-helix interaction angles are parallel [21]. They are more stable as stand-alone structural formations than other types of bundles [22] because they allow larger hydrophobic cores to be buried. Furthermore, in antiparallel packing of square bundles, favorable dipole–dipole interactions occur between antiparallel pairs [20, 22, 23]. The TA1238 monomer is tightly packed with strictly hydrophobic internal residues.
The distance between the centerlines of α1 and α5 is about 7.5 Å, which is considerably shorter than the observed mean distance of 9.6 Å between the closest adjacent helix pairs in other four-helix bundles [24]. To enable such a close approach, all side chains except Ala and Gly are pointing away from the interface between α1 and α5. This facilitates formation of a tightly packed trimer with a core of three α1/α5 helix pairs (Figure 1b). To our best knowledge, such a six-stranded coiled coil has not been observed previously. The α1 helix of each monomer is interacting with the α5 of the same monomer and with the α5 and α4 of the other. Due to the close approach of α1 and α5, the core of the trimer is compact, allowing additional side-chain interactions between the monomers.
Further stabilizing interactions also exist on the trimer surface. For example, the shortest helix, α4, from the overhand connection of one monomer (monomer B in Figure 1b), is interacting with helices α1 and α2 of monomer A. Furthermore, the β hairpin at the N-terminus of monomer A is interacting with helices α5 and α6 and with the overhand connection of monomer B. All of these interactions are replicated by the molecular triad.
About 30% of the molecular surface of each monomer is buried within the oligomerization interface. This super-coiled trimer can be referred to as a ‘parallel trimer’ because all three molecules have the same orientation relative to the trimer axis, with their N-termini on one end of the coil and their C-termini on the other.
Structural and sequence homologues
A sequence homology search with NCBI BLAST [25] against a nonredundant protein database resulted in only two hits, both from other archaea: 66% sequence identity with uncharacterized conserved protein (gi13541189) from Thermoplasma volcanium and 33% identity with hypothetical protein (gi22405829) from Ferroplasma acidarmanus. All other matches were with small segments of much larger proteins with widely varying functions.
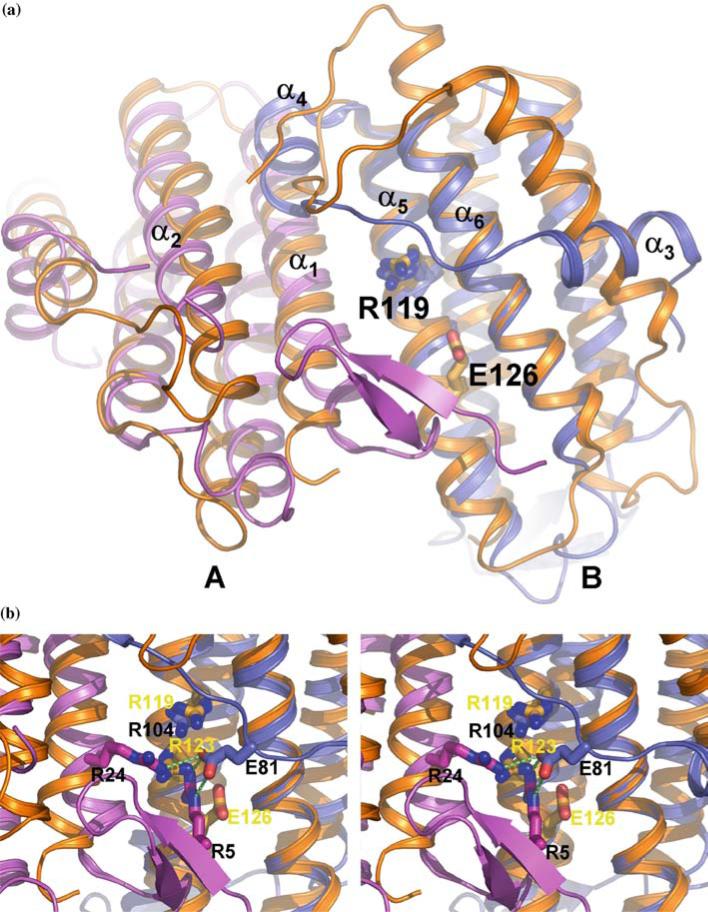
Despite a very low level of sequence conservation, structural comparisons carried out using the secondary-structure-matching (SSM) algorithm (www.ebi.ac.uk/msd-srv/ssm) produced a large number of hits. Perhaps this is not surprising, since helical bundles are well represented in intra- and inter-molecular interactions. The strongest similarity was found with the structures of cobalamin adenosyltransferases (Figure 2), TA1434 from T. acidophilum [26] (PDB entry 1NOG) and YvqK protein from Bacillus subtilis (PDB entry 1RTY) with RMS deviations of 2.42 Å for 110 aligned Cα atoms and 2.39 Å for 113 Cα atoms, respectively. 1NOG and 1RTY are virtually identical to each other, with RMSD of 1.13 Å for 146 aligned Cα atoms.
Figure 2.
Comparison of the TA1238 and TA1434 structures. (a) Two monomers of TA1238 (A, pink; B, teal) and TA1434 (orange) are shown superimposed; for clarity, the third monomer of each trimer is omitted. Arg119 and Glu126, the active site residues of TA1434, are represented by the stick models of their side chains. Arg104 of TA1238 is equivalent to Arg119 and is also shown but not labeled. Helices α5 and α6 align very well, while there is some shift in helices α1 and α2 (in TA1238 numbering) caused by the β hairpin of one monomer bumping into the N-terminal end of the other. The shifts are better seen in monomer A. The biggest differences between the two structures are in the overhand connection between helices α2 and α5. Instead of one long helix in TA1434 parallel to the four-helix bundle, there are two shorter helices in TA1238, α3 and α4. Helix α3 is almost perpendicular to its counterpart from TA1434. The rest of this segment also has different conformations in the two structures. The N-terminus in TA1434 is disordered and not seen on electron density maps, which indirectly points to a lack of β strands. Also, the configuration of this segment in TA1434 apparently does not lead to bumping of the α helices as in TA1238. (b) Close up of the presumed active site. Black labels correspond to TA1238 and yellow to TA1434. Hydrogen bonds between Glu81 from overhand connection and two arginines in the active site are shown in green dashed lines. (Figure was generated with PyMol, www.delanoscientific.com.).
Because of such remarkable structural similarity, including their oligomerization, it might be tempting to suggest a functional relation between TA1238 and cobalamin adenosyltransferase. On the other hand, the sequence identity of TA1238 with these two proteins is only 13 and 19%, respectively, with another 29 and 23% of residues displaying similarity. Moreover, the conserved residues do not form specific patterns of conservation and are distributed along the whole sequence. Furthermore, many of these residues are not structurally conserved. Therefore, one could argue that TA1238 and cobalamin adenosyltransferase, while close in the fold, are not functionally related, and that their sequences merely support their overwhelmingly helical architectures.
Functional implications of structural similarity with cobalamin adenosyltransferase
More detailed inspection of the structural similarities and differences between TA1238 and TA1434 may shed light on the function of TA1238. Both proteins comprise four-helix bundles with one long overhand connection and a small extension at the N-terminus. They both form tight and very similar oligomers (Figure 2). There are several notable differences between the structures of TA1238 and TA1434. The N-terminus is not visible in TA1434, so it is not clear whether there is a β hairpin similar to that in TA1238. However, the absence of this part of the model is most likely caused by its disorder, indicating a lack of organization into a secondary structure. Also, in TA1238, the β hairpin causes relative shifts of α helices (Figure 2). Lack of such tell-tail signs in TA1434 suggests the absence of the hairpin or its different conformation.
The overhand connection is the biggest difference between the structures of TA1238 and TA1434. In the former, it consists of 32 residues and contains two short α helices, α3 and α4. The longer of these, α3, is essentially perpendicular to the main bundle. In TA1434, this connection is longer, 38 residues, most of which form a single long α helix almost parallel to the bundle (Figure 2).
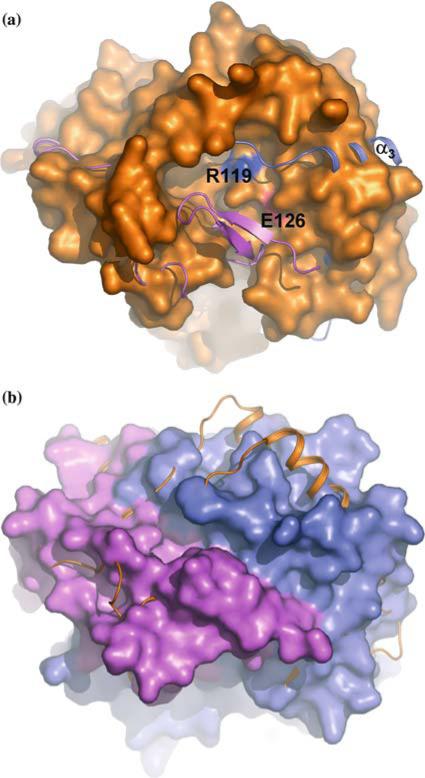
In TA1434, there is a large cavity close to the middle of the super-coiled bundle (Figure 3b). Two helices from different monomers (α1 and α5 in the TA1238 nomenclature) form the bottom of the cavity. Helices α2 from one monomer and α6 from the other create its two sides, and the C-terminal segment of the overhand connection contributes to the third side. The exact make up of the remaining forth side is not clear because the N-terminus of TA1434 is disordered and absent from the model. The equivalent region in TA1238 is partially occupied with the N-terminal β hairpin (Figure 2a). In TA1238, this cavity is mostly filled by the β hairpin of one monomer and by the extended loop of the overhand connection of the other, leaving a narrow channel in its place (Figure 3b).
Figure 3.
Surface depressions of the TA1238 and TA1434 trimers. The orientation and color scheme are the same as in Figure 2. (a) The surface of the TA1434 trimer is shown together with the cartoon of TA1238. The third monomer is visible fading away at the bottom. Active site residues Arg119 and Glu124 of TA1434 are at the bottom of the well-defined depression in the interface between the two monomers and are seen as blue and red patches. In TA1238, corresponding residues are isolated from the external solvent by the segment of the overhand connection of one monomer (blue) and by the β hairpin of the other (pink). (b) The surface of the TA1238 trimer is shown together with the cartoon of TA1434. The wide open depression is blocked by the overhand connection (see also Figure 2) and is transformed into a very narrow cleft. (Figure was generated with PyMol, www.delanoscientific.com.)
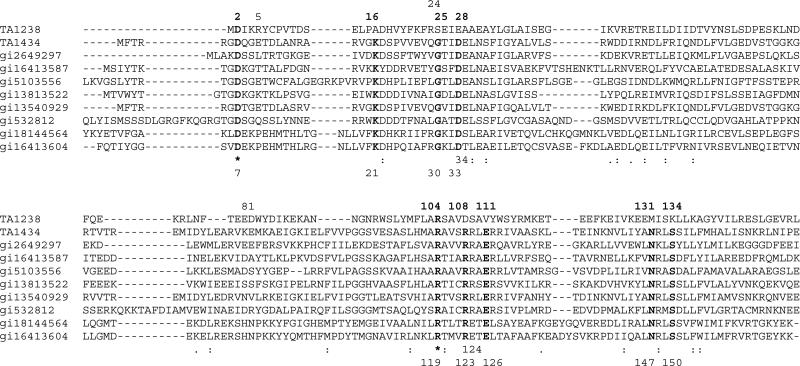
In Pfam01923, to which cobalamin adenosyltransferases belong, there are nine invariant residues (Figure 4). In TA1434 numbering, these are Asp7, Lys21, Gly30, Asp33, Arg119, Arg123, Glu126, Asn147, and Ser150. It was shown that Arg119 and Glu126, along with the conserved but not invariant Arg124, are essential for the enzymatic activity of TA1434 [26]. The invariant residues Asn147, and Ser150 interact with Glu126, stabilizing its side chain, and perhaps are also involved in catalysis. Furthermore, Asp33, donated by another monomer, stabilizes Arg119. Arg124 is in the interface between the two monomers and its function in TA1434 is most likely to stabilize the active site. This is supported by the observation that Arg124 is always paired with Glu34 and when the former changes to Glu, the latter also changes to serine or threonine, allowing the formation of hydrogen bonds between the side chains. It is diffcult to understand the roles of Asp7 and Lys21, since they are missing from the model of TA1434. The role of Arg123, which is in the active site of TA1434, has not been studied.
Figure 4.
Sequence alignment of Pfam01923 proteins and TA1238. The proteins are from the following organisms: TA1238 from Thermoplasma acidophilum, TA1434 from Thermoplasma acidophilum, gi2649297 from Archaeoglobus fulgidus, gi16413587 from Listeria innocua, gi5103556 from Aeropyrum pernix, gi13813522 from Sulfolobus solfataricus, gi13540929 from Thermoplasma volcanium, gi532812 from Caenorhabditis elegans, gi18144564 from Clostridium perfringens, and gi16413604 from Listeria innocua. First, Pfam proteins were aligned and then TA1238 sequence was added based on structural alignment of TA1238 and TA1434. Bold letters indicate the residues that are invariant throughout the family, excluding TA1238. Stars show the only two residues that are also invariant in TA1238. Numbers at the top of the alignment are those from the TA1238 sequence and at the bottom – from TA1434. Note residues 34 and 124 in Pfam and 5, 24, and 81 in TA1238, whose possible roles are also discussed in the text.
Despite very strong structural similarity, conservation of these residues is dramatically different in TA1238. The only residues that remained invariant in TA1238 are Asp2 (Asp7 in TA1434 numbering) and Arg104 (Arg119 in TA1434). Interaction of Asp33 and Arg119 was replaced by that of Glu28 and Arg104 in TA1238. The catalytically critical Glu126 in TA1434 was replaced by a valine residue in TA1238 and surrounding residues are also mostly hydrophobic. Thus, from the functionally important residues, only Arg104 is structurally conserved in TA1238 (Figure 2). It is noteworthy that in TA1238, Arg5 from monomer C and Arg24 from monomer A occupy the same general position as Arg123 in TA1434, which is conserved in Pfam01923 but is replaced by Asp108 in our structure. These arginines form hydrogen bonds with the side chain of Glu81 from monomer A (Figure 2b). The absence of the catalytic glutamate, equivalent to the one in TA1434 and conserved in Pfam, casts doubt on the function of TA1238 as cobalamin adenosyltransferase. However, the architecture of the surface depression is nearly identical in the two proteins (except for the overhand connection), suggesting that TA1238 still might be involved in cobalamin biosynthesis in some capacity and bind similar ligands. Moreover, the Arg/Glu content of this site is similar in the two proteins: Arg119, Arg123, Arg124, and Glu126 in TA1434, and Arg104, Arg5, Arg24, and Glu81 in TA1238. Maintenance of similar residues with different configuration may indicate the possible binding by TA1238 of the compounds from the cobalamin biosynthesis pathway. The ‘problematic’ overhand connection which blocks the entrance to this site in the TA1238 structure could be involved in a gating mechanism, modulated by such factors as the ligand binding itself and/or protein–protein interactions. This segment is characterized with considerable flexibility, which might allow the conformation changes to expose the active site.
In the current model of TA1238, helix α3 is flanked by two phenylalanines, Phe71 and Phe78, both of which are fully extended into the solvent and are disordered. Also, in the middle of the overhand connection, Trp83 is also extended into the solvent. This somewhat unusual exposure of strongly hydrophobic residues to the solvent might point to the potential of this segment to be a protein–protein interaction site. It might also point to the intrinsic propensity of this segment for conformational rearrangement in order to internalize the hydrophobic residues. In either case, the potential active site depression could be exposed for ligand binding. Furthermore, the stabilizing interactions of Glu81 from overhand connection with Arg5 and Arg24 (Figure 2b) would certainly be affected by ligand binding, thus serving as one of the triggers for a gating mechanism. If the conserved Arg1104 can be presumed responsible for specific binding of ligand(s), then the absence of the catalytic Glu might point to the function of TA1238 as transporter protein rather than enzyme.
It is also possible that at the temperature of ~95 °C and pH range of 2–4, which are prevalent in the natural environment of T. acidophilum, the active site of the protein is more exposed. Indeed, extended polypeptide chains were shown to increase their α helical contents at lower pH [27, 28]. If the existing α3 helix were to lengthen, it would most likely align with α2 and α6 because such helix–helix interactions would be energetically favorable. It would require considerable rotation of the α3 helix, thus opening the access to the conserved Arg104.
Even though it is outside the scope of this work within structural genomics, this hypothesis could probably be tested by studying the interactions of TA1238 with various relevant ligands from the cobalamin biosynthesis pathway; by measuring its helical content at different pH values; and by potential protein–protein interactions with the members of the cobalamin biosynthesis pathway.
Other structural homologues
The TA1238 protein is structurally also similar to rubrerythrins (PDB codes 1JYB, 1DVB, 1B71, and 1LKM) and ferritins (PDB codes 1HRS, 1KRQ, and 1EUM). These also form four-helix bundles, but other structural details are significantly different than in TA1238; for example, the four-helix bundles are longer. Additional structural elements are larger than the N-terminal β hairpin of TA1238 and are on the C-terminus of these proteins; their overhand connections are mostly without any a helices and are on the opposite side of the bundle as compared with TA1238. The latter also lacks the characteristic ‘buckle’ in the middle of the bundle where the metal ion is bound in rubrerythrins.
Some similarity was also found between TA1238 and interferons (PDB codes 1B5L, 1AU1), but this is likely due to the prevalently α helical structures, which form five-helix bundles. Another dramatic difference is that TA1238 and cobalamine adenosyltransferases form a novel super-coiled bundle that is functionally necessary. To our best knowledge, this type of oligomerization of four-helix bundles has not been previously observed.
Acknowledgements
We wish to thank all members of the Structural Biology Center at Argonne National Laboratory and the Structural Proteomics In Toronto (SpiT) group for their help in conducting experiments. This work was supported by a Grant from the National Institutes of Health, National Institute of General Medical Sciences to A.J. (GM62414); by the US Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-Eng-38; and by the Ontario Research and Development Challenge Fund.
References
- 1.Kim SH, Shin DH, Choi IG, Schulze-Gahmen U, Chen S, Kim R. J. Struct. Funct. Genomics. 2003;4(2–3):129–135. doi: 10.1023/a:1026200610644. [DOI] [PubMed] [Google Scholar]
- 2.Sanishvili R, Yakunin AF, Laskowski RA, Skarina T, Evdokimova E, Doherty-Kirby A, Lajoie GA, Thornton JM, Arrowsmith CH, Savchenko A, Joachimiak A, Edwards AM. J. Biol. Chem. 2003;278(28):26039–26045. doi: 10.1074/jbc.M303867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laskowski RA, Watson JD, Thornton JM. J. Struct. Funct. Genomics. 2003;4:167–177. doi: 10.1023/a:1026127927612. [DOI] [PubMed] [Google Scholar]
- 4.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley RR, Courcelle E, Das U, Durbin R, Falquet L, Fleischmann W, Griffiths-Jones S, Haft D, Harte N, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Lonsdale D, Silventoinen V, Orchard SE, Pagni M, Peyruc D, Ponting CP, Selengut JD, Servant F, Sigrist CJ, Vaughan R, Zdobnov EM. Nucleic Acids Res. 2003;31(1):315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal D, Suhnel J, Weiss MS. Angew Chem. Int. Ed. Engl. 2002;41(24):4663–4665. doi: 10.1002/anie.200290009. [DOI] [PubMed] [Google Scholar]
- 6.Watson JD, Milner-White EJ. J. Mol. Biol. 2002;315(2):183–191. doi: 10.1006/jmbi.2001.5228. [DOI] [PubMed] [Google Scholar]
- 7.Watson JD, Milner-White EJ. J. Mol. Biol. 2002;315(2):171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- 8.Wallace AC, Borkakoti N, Thornton JM. Protein Sci. 1997;6(11):2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker JA, Thornton JM. Bioinformatics. 2003;19(13):1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- 10.Christendat D, Saridakis V, Dharamsi A, Bochkarev A, Pai EF, Arrowsmith CH, Edwards AM. J. Biol. Chem. 2000;275(32):24608–24612. doi: 10.1074/jbc.C000238200. [DOI] [PubMed] [Google Scholar]
- 11.Evans G, Pettifer RF. J. Appl. Crystallogr. 2001;34:82–86. [Google Scholar]
- 12.Westbrook EM, Naday I. Methods Enzymol. 1997;276:244–268. [PubMed] [Google Scholar]
- 13.Pflugrath JW. Acta. Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, GrosseKuunstleve RW, Kuszewski J, Nilges M, Pannu N, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project Acta Crystal-logr. 1994;D50:760–763. [Google Scholar]
- 20.Presnell SR, Cohen FE. Proc. Natl. Acad. Sci. USA. 1989;86(17):6592–6596. doi: 10.1073/pnas.86.17.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crick FHC. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 22.Harris NL, Presnell SR, Cohen FE. J. Mol. Biol. 1994;236(5):1356–1368. doi: 10.1016/0022-2836(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 23.Hol WG, van Duijnen PT, Berendsen HJ. Nature. 1978;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Weber PC, Salemme FR. Nature. 1980;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]
- 26.Saridakis V, Yakunin A, Xu X, Anandakumar P, Pennycooke M, Gu J, Cheung F, Lew JM, Sanishvili R, Joachimiak A, Arrowsmith CH, Christendat D, Edwards AM. J. Biol. Chem. 2004;279:23646–23653. doi: 10.1074/jbc.M401395200. [DOI] [PubMed] [Google Scholar]
- 27.Hua Y, Raleigh DP. J. Mol. Biol. 1998;278(4):871–878. doi: 10.1006/jmbi.1998.1736. [DOI] [PubMed] [Google Scholar]
- 28.Dubovskii PV, Li H, Takahashi S, Arseniev AS, Akasaka K. Protein Sci. 2000;9(4):786–798. doi: 10.1110/ps.9.4.786. [DOI] [PMC free article] [PubMed] [Google Scholar]