Abstract
Human embryonic stem cell–derived neural progenitors (NP) present an important tool for understanding human development and disease. Optimal utilization of NP cells, however, requires an enhanced ability to monitor these cells in vitro and in vivo. Here we report production of the first genetically modified self-renewing human embryonic stem cell–derived NP cells that express fluorescent proteins under constitutive as well as lineage-specific promoters, enabling tracking and monitoring of cell fate. Nucleofection, transfection, and lentiviral transduction were compared for optimal gene delivery to NP cells. Transduction was most efficient in terms of transgene expression (37%), cell viability (39%), and long-term reporter expression (>3 months). Further, the constitutive gene promoters, cytomegalovirus, elongation factor 1α, and ubiquitin-C, exhibited comparable silencing (20–30%) in NP cells over a 2-month period, suggesting their suitability for long-term reporter expression studies. Transduced NP cells maintained their progenitor state and differentiation potential, as demonstrated by expression of endogenous NP markers and neuronal markers after differentiation. We also detected reporter expression in astrocytes generated from NP cells transduced with an astrocyte-specific gene promoter, glial fibrillary acidic protein, demonstrating the usefulness of this approach. The genetically manipulated NP cells described here offer great potential for live cell–tracking experiments, and a similar approach can as well be used for expression of proteins other than reporters.
Introduction
Amajor advance in human neurobiology was the generation of a unique self-renewing population of lineage-restricted neural progenitor (NP) cell line from human embryonic stem cells (hESC).1 These cells can be differentiated to produce various types of human neurons, astrocytes, and oligodendrocytes, and share molecular characteristics with neuroepithelial cells present in the developing embryo.1,2 Use of hESC-derived NP cells offer several significant advantages. Majority of neural stem cells isolated from fetal or adult tissues lack naïve unprimed molecular state of NP cells derived from an embryonic stem cell origin, making the latter far more suitable for studying early development and disease. Being a self-renewing population, these NP cells provide a convenient starting point for derivation of neural cells.1 Moreover, being lineage restricted, they eliminate the variability often associated with derivation of neural cells directly starting from hESC. Thus, they provide a powerful and hitherto unavailable resource for studying human neural development, differentiation, and related neurological conditions.
Optimal utilization of these NP cells and their derivatives requires thorough characterization both in vitro and in animal models. For example, to test whether these cells will migrate, differentiate, and functionally integrate into the recipient's nervous system requires an appropriate system to follow migration and cell fate3,4 of these NP cells. Expression of fluorescent markers under constitutive or tissue-specific promoters will generate a cell population that can be genetically identified in mixed population culture, and in vivo, using less invasive techniques than currently possible.5 Genetically manipulated NP cells can also provide a source for neural cells expressing other proteins of interest, including those required to generate appropriate in vitro models for neurological disorders. Therefore, it is necessary to explore reliable techniques to stably express exogenous genes in NP cells. Here, for the first time, we describe genetic manipulation of our self-renewing NP cell population derived from hESC. The methods described could potentially serve as a general guideline for genetic alteration of NP cells.
Previous reports have described genetic manipulation at the level of ES cells, with subsequent differentiation to neural cells,6 or have used NPs isolated from fetal sources and modified them genetically.7,8 For example, fetal rat NP cells were modified for spinal cord or brain transplantation utilizing a lentiviral delivery system and the cytomegalovirus (CMV) promoter driving expression of green fluorescent protein or neurotrophic factors (brain-derived neurotrophic factor, ciliary neurotrophic factor, glial cell derived neurotrophic factor, neurotrophin-3 and its mutated forms).7 A similar approach has been tested in a spinal cord model utilizing a retroviral delivery system.8 None produced a genetically modified self-renewing NP population.
Genetic manipulation of hESC by both transfection and transduction has been reported.9 Transfection in hESC was beset with poor rate of stable integration.10–12 Moreover, when successfully integrated, transgene expression, especially that of fluorescent proteins, was often not maintained for prolonged periods in transfected hESC.13 Transduction, on the other hand, generated stable hESC lines that retained fluorescent protein expression upon differentiation to neural cells.6,14 However, none of the previously described processes produced self-renewing NP cells of hESC origin that can repeatedly be used as a starting point for production of neural cells.
Our objective was to generate hESC-derived NP cells that permanently express fluorescent reporter proteins under control of each of three ubiquitous and a tissue-specific promoter. In addition, we tested integration efficiency, viability, and expression of exogenous genes, and differentiation potential of the modified NP lines. Three different NP cell lines derived from the same hESC line were delivered with a constitutive mouse ubiquitin-C (Ubc) promoter driving enhanced green fluorescent protein (EGFP) (Ubc-EGFP) expression. To determine an optimal method for gene delivery, three systems (nucleofection, transfection, and lentiviral transduction) were compared in each NP line. Lentiviral transduction demonstrated optimal insertion efficiency and cell viability among the methods evaluated. Apart from delivery methods, three constitutive promoters, CMV, elongation factor 1α (EF1α), and Ubc, were compared for silencing with time in continued culture. All three promoters proved suitable for long-term tracking experiments, as 70–80% of the transduced cells maintained EGFP expression over an extended period of culture. We also used the more traditional approach and produced an EGFP+ stable hESC line. NP cells were subsequently generated from these hESCs and evaluated for continued reporter expression.
To our knowledge, this is the first report describing genetic manipulation of any hESC-derived self-renewing neural lineage-restricted progenitor population. We demonstrate the production of hESC-derived genetically marked NP cells and their derivatives (e.g., neuronal and astrocyte phenotypes) that can potentially be monitored in live culture and in vivo.
Materials and Methods
hESC culture
All cells were cultured in a 5% CO2 incubator at 37°C. Reagents are from Gibco–Invitrogen, Carlsbad, CA, unless mentioned otherwise. WA09 hESC with normal karyotype were cultured on ICR mouse (Harlan, Prattville, AL) embryonic fibroblast feeders inactivated by mitomycin-C (Sigma-Aldrich, St. Louis, MO). The cells were cultured in ES cell medium, consisting of Dulbecco's modified Eagle's medium/F12 supplemented with 20% knockout serum replacement, 2 mM L-glutamine, 0.1 mM nonessential amino acids, 50 units/mL penicillin/50 μg/mL streptomycin, 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 4 ng/mL fibroblast growth factor-2 (FGF2) (Sigma-Aldrich). Cells were passaged as previously described by our laboratory.15
Human NP and fetal cortex cell culture
NP cells were derived from WA09 hESC as described previously.1 Cells were propagated and maintained in NP proliferation medium consisting of neurobasal medium supplemented with 2 mM L-glutamine, 1 × penicillin/streptomycin, 1 × B27, 20 ng/mL of FGF2 (Sigma-Aldrich), and 10 ng/mL leukemia inhibitory factor (Chemicon–Millipore, Temecula, CA). Cells were passaged by mechanical dissociation via trituration and replating on polyornithine-coated (20 μg/mL) and laminin-coated (5 μg/mL) plates.
Human cerebral cortex cells from 16- to 20-week-old brain tissue (Cambrex–Lonza, Walkersville, MD; #PT-2599, HHNP) were adapted to monolayer culture (manuscript in preparation) in NP proliferation medium. These cells were used for validation of viral vectors.
Human astrocytoma and rat astrocyte cell culture
Human astrocytoma cells (ATCC, Manassas, VA; #CRL-1718) were maintained in tissue culture according to supplier's protocol, in RPMI-1640 medium (ATCC; # 30–2001) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). They were gradually adapted to NP differentiation medium (NP proliferation medium with no FGF2). Astrocyte growth medium was completely replaced by serum-free NP differentiation medium over a 1-week period. For production of Astrocytoma-conditioned medium (ACM), NP differentiation medium, exposed to astrocytoma culture for 24 h, was harvested, filtered (0.2 μM; Millipore, Carrigtwohill, Co. Cork, Ireland), and used for differentiation of NP cells to glial fibrillary acidic protein (GFAP+) cells.
Rat astrocytes were harvested from postnatal day 1 Sprague-Dawley pups and propagated as described previously.16 All procedures involving animals were approved by the Institutional Animal Care and Use Committee at University of Georgia. Cells were grown in Dulbecco's modified Eagle's medium/F12 with 10% fetal bovine serum and used for validation of viral vector.
NP differentiation to GFAP+ cell lineage
To enrich GFAP+ cells in culture, NP cells plated on polyornithine- and laminin-coated dishes were cultured with freshly harvested ACM mixed with NP differentiation medium (1:1) for 24 days. Medium was replaced every third day.
pFUGW GFAP-EGFP and GFAP-tdTomatoRed transfer vector construction
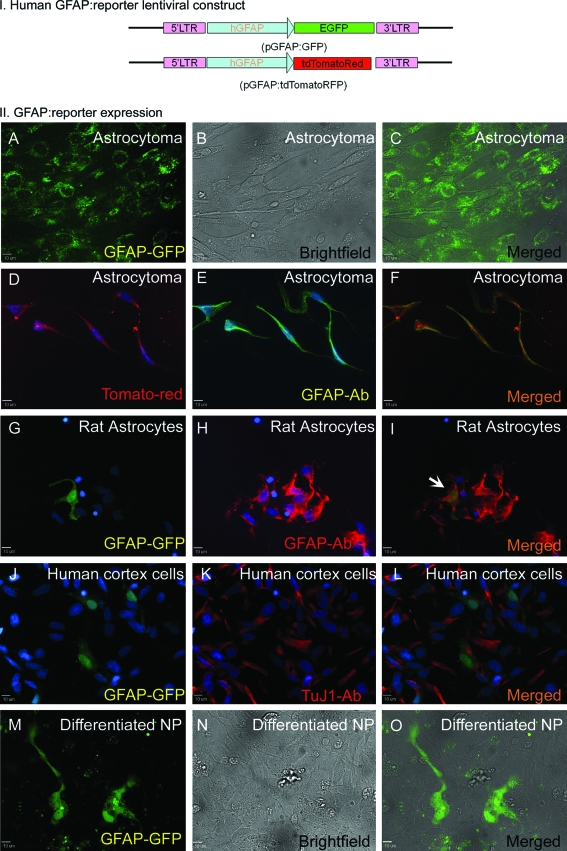
pFUGW lentiviral transfer vector was a gift from Dr. James Lah at Emory University. A BglII–SalI fragment containing the hGFAP promoter was excised from pGfa2-cLac plasmid17 and inserted into the corresponding cloning site of vector DsRed Express-1 (Clontech, Mountain View, CA). Subsequently, the promoter was moved from DsRed Express-1 to pFUGW using AfeI and BamHI, yielding pFUGW GFAP-EGFP. To generate a GFAP-tdTomatoRed vector, EGFP was replaced by tandem-dimer Tomato-Red fluorescent protein (tdTomatoRed, GenBank Id: 55420-623) construct. For this, the BamHI–NotI-digested fragment containing tdTomatoRed sequence from pRSET-beta plasmid (a gift from Dr. Roger Tsien) was inserted into the shuttle pZsGreen1N1 (Clontech) plasmid. The fragment was recovered with AflII digestion (blunted after digestion) followed by BamHI digestion. Fragment was inserted into pFUGW GFAP-EGFP digested with EcoRI (subsequently blunted) followed by BamHI to produce pFUGW GFAP-tdTomato-Red. EGFP and tdTomatoRed sequences were confirmed by sequencing using primer located on the GFAP promoter sequence18 (Table 1).
Table 1.
List of Primers Used for Reverse-Transcription Polymerase Chain Reaction and Sequencing
| Name | Size (bp) | Sequence |
|---|---|---|
| EGFP | 734 | Forward: CACATGAAGCAGCACGACTT |
| Reverse: TGCTCAGGTAGTGGTTGTCG | ||
| PAX6 | 363 | Forward: CCGGCAGAAGATTGTAGAGC |
| Reverse: CTAGCCAGGTTGCGAAGAAC | ||
| NESTIN | 313 | Forward: CAGGAGAAACAGGGCCTACA |
| Reverse: TAAGAAAGGCTGGCACAGGT | ||
| NCAM1 | 751 | Forward: GCCCCTAGGTCTGTCGCTCA |
| Reverse: CCTGGCCTGGATGGTAGGTG | ||
| GAPDH | 983 | Sequence not available (Clontech Cat. # 639005) |
Primer for sequencing of plasmid containing GFAP promoter: CAGAGCAGGTTGGAGAGGAGA.
EGFP, enhanced green fluorescent protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein.
Lentiviral production and transduction of NP cells
CMV-EGFP and EF1α-EGFP lentiviruses were provided by Dr. John Wakefield (Open Biosystems). Other lentiviral particles were produced and titered in the laboratory of Dr. James Lah (Emory University). Vector particles were produced in HEK293T cells by transient cotransfection with the transfer vector(s), the HIV-1 packaging vector ΔR8.9, and the Vesicular Somatitis Virus-G (VSV-G) envelope glycoprotein. Medium was replaced the morning posttransduction. Virus-containing supernatant was removed after 28 h, filtered (0.45 μM), concentrated (centrifugation at 23,000 rpm for 2 h at 4°C), and frozen at −80°C. Viral titers were calculated using serial dilution.
For transduction, NP cells were cultured in 35 mm dishes. At ∼60% confluency they were exposed to the proliferation medium containing viral particles with a multiplicity of infection (MOI) of 5 (experiment) or 0 (negative control). Cells were centrifuged (250 g for 5 min at 23°C) and incubated overnight. Cultures were washed with phosphate buffered saline (PBS+/+ with Ca2+ and Mg2+), and fresh NP proliferation medium was added after 24 h.
Transfection
NP cells were transfected using ExGen500 (MBI Fermentas, Glen Burnie, MD) reagent following a protocol adapted for hESC transfection.19 About 1 × 106 NP cells were plated overnight in a 35 mm tissue culture dish. Immediately before transfection, old medium was replaced with 1 mL fresh medium. Two micrograms of DNA was diluted in 100 μL of 150 mM NaCl and 3.3 μL of ExGen500, vortexed (10 s), spun briefly, and incubated at room temperature for 10 min. Postincubation, the DNA–reagent solution was added to the culture, gently mixed, and centrifuged (280 g for 5 min). After incubation for 30 min, transfection solution was removed, and cells were washed with PBS+/+ and incubated with fresh NP proliferation medium.
Nucleofection
One million cells were resuspended in 100 μL of Nucleofector Solution Kit V (Amaxa, Gaithersburg, MD) at room temperature. Plasmid DNA (5 μg), diluted in TE buffer, was transferred into the nucleofection cuvette. The cell suspension was added and nucleofection was performed as per the manufacturer's settings (electrical setting B-16). Immediately after pulsing, the cells were transferred to prewarmed NP proliferation medium and plated on polyornithine- and laminin-coated six-well plates.
For stable transfection of hESC and NP cells by nucleofection, ApaLI (New England Biolabs, Ipswich, MA)–linearized pZsGreen1N1 plasmid (Clontech) was used. G418 selection (200 μg/mL) was started 72 h postnucleofection and continued for 2 weeks.
Immunocytochemistry
Immunostaining was performed as described previously.2 Briefly, cells were grown on permanox (Nalge Nunc, Naperville, IL) four-chamber well slides, fixed with 4% paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature, and stained with antibodies against SOX2 (1:200; R&D Systems, Minneapolis, MN), MUSASHI1 (1:100; Neuromics), NESTIN (1:400; Neuromics, Edina, MN), TUJ1 (1:500; Neuromics), tyrosine hydroxylase (TH) (1:50; Neuromics), gamma aminobutyric acid (GABA) (1:2000; Sigma), and GFAP (1:500; Sigma-Aldrich) and appropriate fluorescent secondary antibodies (1:1000; Molecular Probes–Invitrogen, Eugene, OR). All wells were counter-stained with DAPI (4,6′-diamidino-2-phenylindole). Pictures were acquired using an Olympus with Disc-Spinning Unit and Slide Book Software (Intelligent Imaging Innovations).
Reverse-transcription polymerase chain reaction
RNA was isolated from NP cells using RNeasy plus kit (Qiagen, Valencia, CA).2 One microgram of RNA was reverse transcribed using Advantage reverse-transcription polymerase chain reaction (RT-PCR) kit (Clontech). PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI). Primer sequences are provided in Table 1.
Flow cytometry and cell sorting (fluorescence-activated cell sorting)
Transduced NP cells were washed twice with PBS+/+, harvested by mechanical dissociation, and centrifuged at 1000 g at 23°C for 4 min. Pellet was gently resuspended in 0.5 mL of fresh prewarmed NP proliferation medium and stored on ice. Flow cytometry was performed using fluorescence-activated cell sorting (FACS) Calibur system (BD Biosciences, San Jose, CA) and FlowJo analysis software (Tree Star, Ashland, OR). Propidium iodide (PI, 20 μg/mL) was used to count dead cells. Forward- and side-scatter plots were used to exclude dead cells and debris from the histogram analysis. Cells were sorted on a MoFlo (Beckman Coulter, Miami, FL) using a 100 μm nozzle. GFP was excited at 488 and the fluorescence was collected through a 530/40 BP filter. Postsort, cells were grown on polyornithine- and laminin-coated plates in NP proliferation medium.
Statistical analysis
Analysis of variance (ANOVA) and comparison of means (Tukey's test) were performed using general linear models (GLM) procedure by SAS 8.01 (SAS Institute).
Results
Stably transfected hESC undergo silencing during neural differentiation
Significant efforts have been made to improve genetic modification of hESC for long-term reporter expression that will be maintained in its differentiated derivatives. Efforts include improved gene delivery schemes and use of various promoters that may be less prone to silencing. In our hESC line we used the human cytomegalovirus immediate-early (CMV) promoter, a popular candidate for ubiquitous reporter expression,13 to drive EGFP expression (pZsGreen1N1; Clontech). In transient transfection experiments, delivery efficiency was higher with nucleofection (20–25% EGFP+ cells) than with transfection using ExGen500 (∼5%). Therefore, for stable transfection, the linearized construct was delivered using nucleofection, generating ∼10 stable clones per million transfected cells (Fig. 1A–B). Stable clones were selected using a neomycin resistance gene (G418 selection) driven by the SV40 promoter. One in five G418 resistant stable hESC clones generated by nucleofection expressed EGFP. These clones were continuously cultured, and could be frozen–thawed and subcultured (tested for 10 passages) in the presence of G418 without affecting either colony morphology or EGFP expression.
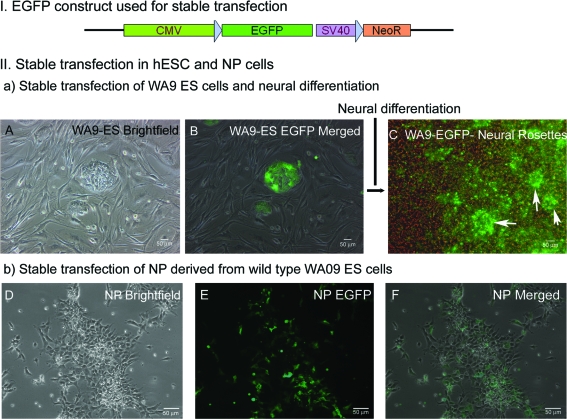
FIG. 1.
Stable transfection of human embryonic stem cells (hESCs) and hESC-derived neural progenitor (NP) cells. I. Schematic diagram of the linear construct used in transfection experiment. II. Stable hESC clones expressing EGFP generated using nucleofection (Amaxa). (A) Bright field and (B) overlay of bright-field with EGFP. An EGFP-expressing hESC clone was differentiated toward the neural lineage, forming early neural rosette structures (at 10–12 days) expressing EGFP (arrow heads in C). NP cells were derived from hESC, and then stably transfected with the EGFP construct: bright-field (D), epifluorescence (E), and merged fields of stably transfected NP cells (F). However, continued EGFP expression in long-term culture was not achieved with either approach. EGFP, enhanced green fluorescent protein. Color images available online at www.liebertonline.com/ten.
Ten to 12 days of neural differentiation from these hESC produced early neural rosette structures. EGFP expression similar to that in the parental hESC clone was maintained throughout this early period of differentiation (Fig. 1C). However, EGFP expression could not be detected after this initial period. hESC clones also lost EGFP expression after 2 months of propagation, even though neomycin resistance was maintained. These EGFPlow/G418+ hESC clones (Fig. 1A–C) did not generate EGFP+ neural rosettes. Thus, even though we could produce stably transfected hESC, reporter expression was lost during neural differentiation.
Transfection of NP derived from hESC did not produce stable lines
To fully realize the many advantages of genetically modified NP cells, and to circumvent the problem of silencing of reporter genes in hESC during neural differentiation, we next attempted to stably transfect NP cells derived from wild-type hESC.
NP cells were derived from WA09 hESC using published protocol1 and were transfected using the same linear construct and techniques described in the previous section. In NP cells, nucleofection produced more (15–20%; Fig. 1D–F) EGFP expressing cells than ExGen500 (5%) (not shown) 24 h posttransfection. After 2 weeks of G418 selection, NP cells transfected with ExGen500 lost neomycin resistance, while a few of the NP cells that underwent nucleofection survived neomycin selection (<1 in 5 million). These G418-resistant NP cells could not be propagated and also did not show EGFP expression. These results suggest that similar to the hESC, stable integration and expression of heterologous DNA in NP is not easily achieved by these two methods.
Lentiviral transduction is the most efficient system for gene delivery in NP cells
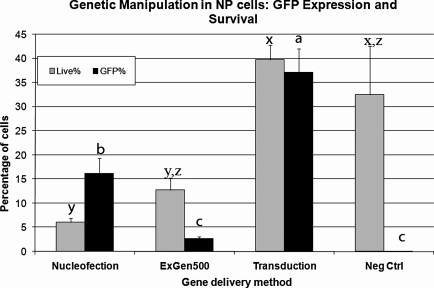
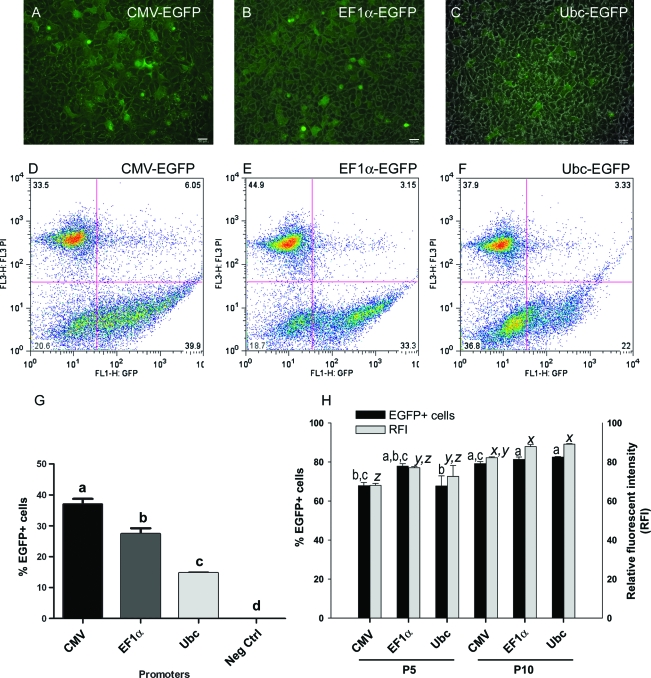
To compare various methods for gene delivery in NP cells, a construct containing EGFP driven by the mouse Ubc promoter (pFUGW vector) was introduced into NP cells using electroporation transfection with a nucleofection kit (Amaxa), transfection with ExGen500 (MBI Fermentas), and lentiviral transduction (MOI of 5) (Fig. 2). Gene delivery efficiency for each method was determined by measuring the percentage of cells expressing EGFP and viability after 48 h. ExGen500 transfection produced the least number of EGFP expressing cells (<3%), followed by 15% in nucleofected cells (Fig. 2). Lentiviral transduction resulted in the maximum number of EGFP+ cells (35–40%). Overall, cell viability was highly compromised after genetic manipulation, with only 6% cells surviving after nucleofection, followed by 12.7% cells for ExGen500 transfection and a significantly higher proportion of 37% (p < 0.05) surviving after lentiviral transduction (Fig. 2). The above results for viability and EGFP expression were similar for three different lines of NP cells all derived from the same WA09 hESC. In terms of both cell viability and transgene delivery, lentiviral transduction appeared to be the best method for genetic manipulation of hESC-derived NP cells.
FIG. 2.
Comparison of efficiency and viability for three different gene delivery methods in NP Cells. EGFP under the control of Ubiquitin-C (Ubc) promoter was delivered to three different NP cell lines. Three different delivery methods were used for each cell line: Nucleofection, transfection with ExGen500, and lentiviral transduction. Mean viability (superscripts x, y, and z) and EGFP expression (superscripts a, b, and c) were determined by flow cytometry. Nonmodified wild-type NP cells were used as negative control for this experiment. No significant differences in viability were seen between Nucleofection and ExGen500 (y), negative control and Exgen500 (z), and between transduction and negative control (x). Different superscripts indicate a statistical difference at p < 0.05 significance level.
Lentivirally transduced NP cells express NP markers and retain ability to differentiate
Genetic manipulation is known to affect the differentiation potential of some cell types, including embryonic stem cells,20 and molecular properties and differentiation potential require reevaluation after transduction.6,21
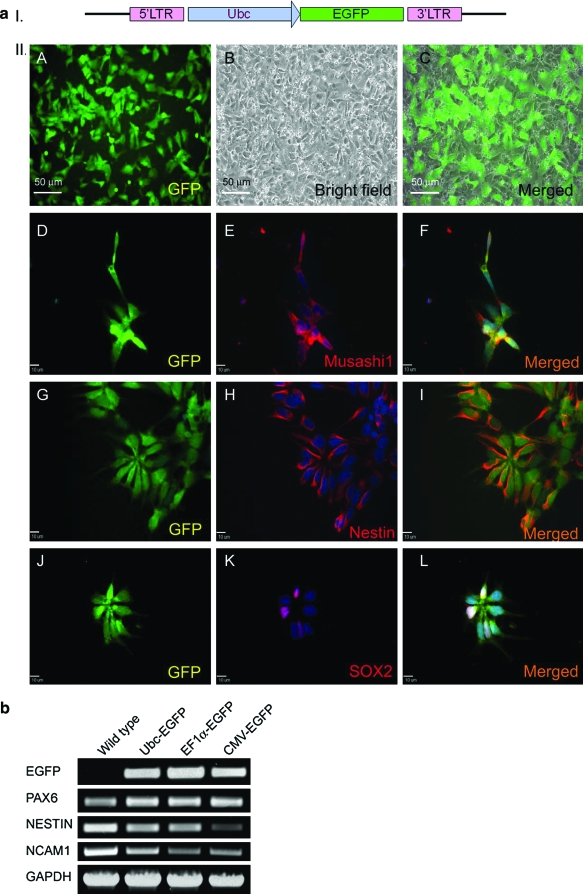
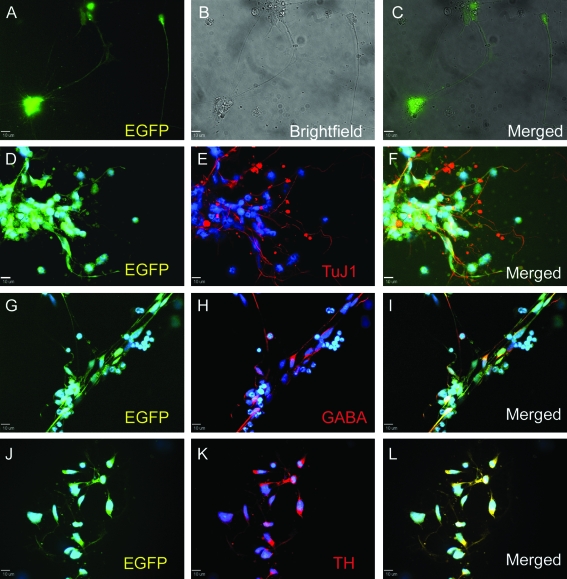
To assess if lentiviral transduction had any negative effect on the differentiation potential of hESC-derived NP cells, transduced NP cells were differentiated into neurons for 14 days. Both transduced NP cells and neurons derived from them were tested for expression of known neural and neuronal markers (Figs. 3 and 4). Immunostaining of EGFP+ NP cells showed expression of MUSASHI1 (RNA-binding protein), NESTIN (intermediate filament protein), and SOX2 (transcription factor) (Fig. 3a). RT-PCR also detected transcripts for EGFP, NES (nestin), and other NP associated genes such as the transcription factor PAX6 and a neural cell adhesion molecule NCAM1 (Fig. 3b). Upon random differentiation, transduced NP cells produced neurons that retained EGFP expression at levels similar to the NP cells (data not shown for EF1α-EGFP and CMV-EGFP). In addition, these neurons expressed TUBB3 (beta3 tubulin, TUJ1), GABA, and TH (Fig. 4). This indicates that transduced NP cells still express NP markers (MUSASHI1, NES, SOX2, PAX6, and NCAM1), and can be further differentiated into neurons expressing neuronal markers (TUJ1, GABA, and TH).
FIG. 3.
Marker expression in transduced NP cells. Lentivirally transduced NP cells retain the expression of NP markers. (a) Shown are both live (A–C) and immunostained images (D–L) of lentivirally transduced NP cells. Ubc-EGFP–transduced NP cells expressed EGFP (A, D, G and J). Panels corresponding to each EGFP field are bright field (B) or immunostained (red) (E, H, K) images of NP markers, MUSASHI1 (E), NESTIN (H), and SOX2 (K). The rightmost column (C, F, I, L) shows the merged images of the EGFP field with either bright field or immunostained images presented in first two columns (A, B, D, E, G, H, J, K). Nuclei were stained with DAPI (blue). (b) Reverse-transcription polymerase chain reaction performed on NP cells transduced with Ubc-EGFP, EF1α-EGFP, and CMV-EGFP. Even after prolonged propagation (>4 months) the NP cells maintained expression of NP markers PAX6, NESTIN, and NCAM1. Nontransduced NP cells serve as negative control. EF1α, elongation factor 1α; CMV, cytomegalovirus. Color images available online at www.liebertonline.com/ten.
FIG. 4.
Marker expression in neurons differentiated from transduced NP cells. Transduced NP cells differentiate to produce EGFP-positive neurons. NP cells transduced with Ubc-EGFP construct (shown in Fig. 3) generated neuronal cells after 2 weeks of differentiation. Live EGFP (A), bright field (B), and merged (C). Differentiated cells were immunostained for neuronal markers TUJ1 (neuronal beta III tubulin) (E, F), GABA (gamma amino butyric acid, an inhibitory neuronal marker) (H, I), and TH (tyrosine hydroxylase, an excitatory neuronal marker) (K, L). Colocalization with EGFP expression (D, G, J) is shown in F, I, and L. Nuclei were stained with DAPI (in blue). Color images available online at www.liebertonline.com/ten.
Glial-like cells derived from NP cells express fluorescent reporter proteins under the glial-specific promoter, GFAP
Isolation and tracking of specific cell phenotypes derived from hESC sources are of significant interest to the research community. To address this need we examined the ability of cells derived from NP cells to express fluorescent reporter proteins under the glial-specific promoter, GFAP. The constitutive Ubc promoter from the pFUGW lentiviral transfer plasmid was excised and replaced with hGFAP promoter17,18 to drive the expression of either EGFP, or tomato-Red fluorescent reporter proteins, yielding GFAP-EGFP and GFAP-tdTomatoRed lentiviral plasmids, respectively. Human astrocytoma, rat astrocytes, and astrocytes in human fetal cortical cell culture express endogenous GFAP and were used for validation of the reporter constructs. Separate transduction experiments with GFAP-EGFP and GFAP-tdTomatoRed lentiviral constructs resulted in the expression of fluorescent protein (Fig. 5A–F for astrocytoma, G–I for rat astrocytes) that colocalized with the GFAP antibody upon immunostaining (Fig. 5D–I). We also transduced cortex cells from human fetal brain tissue and immunostained with neuronal marker TUJ1 (Fig. 5J–L). EGFP expression was not found in TUJ1+ cells, indicating the specificity of the GFAP promoter. Generation of glial-like cells from hESC-derived NP cells involved differentiation of NP cells in the presence of ACM differentiation medium for 24 days. Three days posttransduction, no more than 5% of the ACM-differentiated cells expressed EGFP (Fig. 5M–O), indicating endogenous GFAP promoter activity, whereas NP cells in cultures not conducive to astrocyte differentiation did not express EGFP. Undifferentiated NP cells were also transduced and served as negative control. This suggests that lentiviral system can be used for genetically expressing exogenous genes in hESC-derived NP cells or in further differentiated lineages.
FIG. 5.
Transduction can track astrocyte-like cells expressing glial fibrillary acidic protein (GFAP) in vitro. I. EGFP or tdTomato Red genes under control of the glial-specific GFAP promoter. II. Human astrocytoma (ATCC; #CRL-1718) cells transduced with the GFAP-reporter lentiviral construct (A–F). EGFP (A), bright field (B), and merge (C). Transduction of human astrocytoma cells with Tomato-Red fluorescent reporter expression in human astrocytoma cells (D), and endogenous GFAP protein expression, determined by immunostaining (green in E) colocalize (merged in F). Note that morphology of astrocytoma cells differ depending on culture density (A–F). Rat astrocytes were transduced with the GFAP-EGFP construct (G–I). Endogenous GFAP expression (red in H) and EGFP reporter expression (green in G) are shown. Reporter expression colocalized with endogenous GFAP (merge in I, arrow head). To further demonstrate specificity of the GFAP promoter, human fetal cortex cells were transduced with the GFAP-EGFP construct and immunostained with neuronal marker TUJ1. No TUJ1-expressing cells (red in K and L) showed EGFP expression (green in J). Nuclei were stained with DAPI (blue). NP cells were differentiated for 24 days in Astrocyte-conditioned differentiation medium and transduced with the GFAP-EGFP lentiviral construct. Forty-eight hours posttransduction, several cells with astrocyte-like morphology (bright field, N) expressed EGFP (M), and merged image (O) suggests that about 5% total cells expressed EGFP. Color images available online at www.liebertonline.com/ten.
Comparison of silencing of constitutive promoters in long-term culture of lentivirally transduced NP cells
Lentivirally transduced NP cells showed variation in EGFP expression under all promoters we examined (Fig. 6). We compared three different constitutive promoters, CMV, EF1α, and Ubc. Forty-eight hours after gene delivery (MOI of 10), we quantified EGFP+ cells and viability by flow cytometry (Fig. 6A–G). Transduction with the CMV promoter resulted in a 37% EGFP+ population (Fig. 6G). This was significantly (p < 0.05) higher than the EGFP+ populations produced by transduction with the other mammalian promoters EF1α (27%) and Ubc (15%) (Fig. 6D–G). A significantly higher level (p < 0.05) of EGFP+/PI+ cells were detected (average 3.5% false-positive EGFP population) with the CMV promoter than with EF1α (1.4%) and Ubc (1.6%) (Fig. 6D–F). EGFP signals were more readily detected with CMV-based transduction, suggesting that the CMV promoter is more efficient in driving transcription of EGFP than the other two promoters (Fig. 6A–C).
FIG. 6.
Comparison of transgene expression and silencing for three ubiquitous promoters. NP cells were transduced for constitutive EGFP expression driven by CMV, EF1α, or UbC promoters. Merged, bright field, and epifluorescent images of transduced NP cells (A–C). Corresponding flow cytometry analysis quantifying percentage of EGFP+ cells before sorting (D–F). Mean and standard errors of EGFP+ cells before sorting, n = 3 (G). Transduced cells were propagated postsort, and EGFP+ cells were quantified at passages 5 and 10 (H). Relative fluorescent intensity was measured (as median channel numbers) and analyzed. Significant differences (p < 0.05) (for EGFP+ cells superscripts a, b, and c, and for relative fluorescent intensity superscripts x, y, and z) are indicated by different superscripts on bar diagram and presence of common superscripts indicates lack of significant differences between measures. Color images available online at www.liebertonline.com/ten.
Transduced cells were further sorted by FACS to obtain an enriched EGFP+ NP population (designated passage P0) and replated for subculture. One month later, at P5, percentage of EGFP+ cells was quantified by flow cytometry analysis. This was repeated at P10 approximately 2 months after the initial FACS. The overall numbers of EGFP+ cells (p < 0.0004) differed significantly between the two time points. Irrespective of the promoters used, at both P5 and P10, the percentages of EGFP+ cells were less than at P0 (100% after sorting). The number of EGFP+ cells remaining at P5 was 68% for CMV and Ubc promoters and 78% for the EF1α promoter (average from three replicates) (Fig. 6H). However, at P10, cell populations for all three promoters showed about 81% EGFP+ cells. Thus, the difference in number of EGFP+ cells remaining at P5 and at P10 was statistically significant (p < 0.05) for both CMV and Ubc promoters but not for the CMV promoter. We further quantified median relative fluorescent intensity (RFI) in transduced and nontransduced NP cells (negative control). The RFI median values ranged between 68 and ∼89 channel numbers, with the Ubc promoter producing the highest channel number at P10 (RFI 89.1) and the CMV promoter producing the lowest channel number at P5 (RFI 68.1). Thus, promoter has an effect on RFI values in NP cells (p < 0.001). Although there was an increase in RFI value for all three promoters when cells were propagated from P5 to P10 (p < 0.001), the promoter-specific differences in brightness observed initially were lost, and at both P5 and P10, the average RFI median values became the same for the three promoters. Thus, for long-term (P10 or beyond) maintenance of EGFP expression, all three promoters are efficient and do not differ in activity in transduced NP cells.
Discussion
Genetically manipulated human NP cell lines of hESC origin provide an important resource for furthering our understanding of neural development and disease. Our results demonstrate efficient genetic modification by lentiviral transduction, resulting in long-term constitutive and lineage-specific reporter expression without altering NP characteristics. Further, we demonstrated enrichment of transgene expressing cells via cell sorting (FACS) based on reporter expression. We also established the suitability of both viral and eukaryotic promoters for extended expression of exogenous genes in NP cells.
Previously, attempts have been made to generate neural tissues from hESC that express reporter proteins.22,23 However, to the best of our knowledge, we are the first to report a hESC-derived self-renewing cell population that was modified with a permanent reporter system for continuous cell-tracking capabilities. These genetically marked NP cells, which then serve as a source of genetically modified neurons and glia of hESC origin, have significant advantages: (a) less variability compared to derivation of neural cells each time from hESC; (b) during differentiation, transgene expression is sustained better by NP cells; (c) during terminal differentiation, nonneural cells are not generated from NP cells. This approach will overcome limitations of monitoring gene expression in live cells both in culture and in vivo with less invasive techniques. This system presents an opportunity to study neural development and subsequent differentiation in vitro to enhance the understanding of neurodegenerative diseases. Further, NP cells transduced with suitable transgene could potentially serve as vehicle for delivery of therapeutic protein(s) in future transplantation experiments.
Previously, stable hESC clones have been generated via transfection.10,11,24,25 Among various methods employed, nucleofection has been used most successfully to generate stable hESC clones.6,26,27 With nucleofection, Siemen et al. demonstrated an efficiency of 66% transient transgene expression and obtained one stable EGFP+ clone in every nine drug-resistant clones.27 Using the same technique we obtained EGFP-expressing hESC clones (Fig. 1) at almost twice the efficiency (one for every five drug-resistant clones). However, the same method failed to generate stably transfected NP populations. The absence of stable G418-resistant/EGFP+ NP cell lines with nucleofection was possibly due to poor clonal propagation28 or suboptimal gene delivery with nucleofection, leading us to explore other methods for transgenesis of NP cells.
Three gene delivery systems, nucleofection, transfection, and transduction, were compared in different NP lines derived from hESC. To avoid the possibility of silencing observed with viral promoters like CMV, the eukaryotic Ubc promoter (mouse) was used to drive constitutive expression of EGFP for these experiments. In a previous study, Ubc promoter was used successfully to generate stable hESC clones.13 An early transgenic hESC clone was generated using the transfection reagent.11 Since then, a number of laboratories have also used VSV-G pseudotyped lentiviral transduction to infect a variety of hESC lines.6,29–34 When VSV-G pseudotyped lentivirus were used for transduction of hESC grown on feeder cells, the feeder mouse embryonic fibroblast cells were also transduced, reducing efficiency of hESC transduction.29 Our feeder-free NP culture system1 circumvents this problem.
Both nucleofection and ExGen500 had minimal effect on hESC viability (survival > 70%),27,35 but severely reduced viability of NP cell lines, suggesting NP-specific cytotoxicity. NP cell survival was not affected by lentiviral transduction (Fig. 2). It is important to note that nonmanipulated control NP cells also showed reduced viability in our flow cytometry results. This is possibly due to sample preparation stress, and this reduction in viability should occur in all test samples and should not affect the comparison. Apart from exhibiting the highest survival, lentiviral transduction with mouse Ubc-EGFP virus was also the most efficient (∼37%) in WA09 hESC-derived NP cells (Fig. 2). These results also suggest that efficiency of transgenesis varied greatly with the delivery systems and not on the cell line NP being manipulated.
Maintenance of NP properties and differentiation potential is a critical consideration in choosing a method for genetically modifying the NP cells. Our results demonstrate that lentivirally transduced NP cells do express NP markers (Fig. 3)1,2 and maintain their ability to differentiate as indicated by the expression of TUJ1, GABA, and TH (Fig. 4) while also expressing EGFP expression over 30 passages. Marker expression in transduced cells (Figs. 3 and 4) was similar to nontransduced NP cells. For example, SOX2 is expressed in about 25% of both transduced and wild-type NP cells (results not shown). Previous efforts at genetic manipulation were directed either at demonstrating that modified hESC could retain reporter expression in differentiated neural cells,6 or to show hESC-derived neurons could be marked with lineage-specific reporter.23 Unlike the previous studies the hESC-derived transgenic NP cells described here continue to proliferate in culture as undifferentiated but lineage-restricted progenitors containing stably integrated reporter expression constructs. We further explored the adaptability of our system to monitor presence of glial-like cells in differentiating NP cell cultures with GFAP reporter constructs (Fig. 5). This human GFAP promoter sequence is well characterized and has been widely used36–38 for lineage-specific reporter expression.17 Specificity was further verified by transduction of astrocytes from multiple sources (human astrocytoma, rat astrocytes, and human fetal brain cells). Even though the differentiation protocol for NP cells used in our study generated only a limited glial population, those astrocytes could be detected 24 h after lentiviral transduction, suggesting the efficacy of the system for monitoring cell fate. However, silencing of this GFAP promoter in long-term culture remains to be studied. Hypothetically, a GFAP reporter construct could be used to isolate and enrich for unique cell phenotypes via FACS as was accomplished here using constitutive promoters. Utility of the GFAP reporter construct will hinge on whether silencing of the reporter gene can be averted. This will be tested once we determine culture conditions conducive to deriving a higher percentage of astrocytes in the initial cultures (>5%). Without being able to isolate a larger number of GFAP+ starting cells, studies on gene silencing over extended time will be problematic. Lentiviral gene delivery was optimal for transgenesis of hESC-derived NP cells when viability, efficiency, and long-term reporter expression were considered. Moreover, NP properties and differentiation potential were not compromised after transduction. The NP cell lines used were all derived from the WA09 hESC line. Outcomes of similar experiment in other hESC-derived NP cell lines remain to be tested.
Important applications of NP cell transgene expression include animal transplantation studies and ex vivo cell-based assays where cell tracking capabilities are imperative for the constant monitoring migration, expansion, and differentiation. Sustained transgene expression is crucial for such applications. Viral promoters are noted for their inability to sustain transgene expression in hESC,13 and the problem is likely to be enhanced in cells that divide rapidly.39 Intriguingly, it was shown recently that expression of transgenes delivered by lentiviral vectors was suppressed in a promoter-dependent manner specifically in hESC, but not in mouse ESC.40 In the absence of data from other laboratories on transgenesis in NP cells, we discuss our results in the context of what is known for hESC. In the present study the percentages of EGFP+ NP cells obtained with CMV and EF1α promoters (37% and 27%, respectively) were higher than that obtained in hESC with the same promoters (1.1% and 18.6%, respectively) but with a threefold lower MOI.40 However, the lower MOI alone does not explain this difference. In another study, the EF1α promoter, used at a much higher MOI than what we used for our NP cells, did not increase transduction efficiency beyond 26% in hESC.6 This suggests that the differences in transgene expression efficiency can be attributed to a combination of cell property, promoter, and the amounts of transducing units used.
To enrich EGFP+ NP population in vitro, we sorted and maintained these cells over an extended period in culture (more than 2 months), and evaluated reporter expression at passage 5 and 10 by flow cytometry. In contrast to results in hESC described by Xia et al.,40 we did not observe any preferential silencing among the three promoters (CMV, Ubc, and EF1α) driving EGFP expression. After 2 months in culture, 70–80% of the total cells were still EGFP+. Considering temporal changes in EGFP expression, EF1α varied the least and maintained the highest fluorescence intensity over time (Fig. 6H). Silencing of lentivirally delivered transgene was minimal (20–30%) in hESC-derived NP cells. A further reduction in silencing may be achieved by the addition of an internal ribosome entry site element linked to drug resistance genes. This strategy has successfully reduced silencing of EGFP expression in differentiating hESC.6 Alternatively, homologous recombination technology can be used to knock-in transgene at a specific genomic site for long-term reporter activity.41 It is expected that all three promoters, driving EGFP in NP cells, could aid in vivo transplantation studies because they retained reporter expression longer than required for a typical transplantation study.42–44 It should be noted that transduced NP cells described here were not subjected to clonal analysis, and EGFP+ neurons were not tested for electrical activities or transplanted in animal models.
NP cells derived from hESC are nontransformed, neural lineage restricted, and yet highly proliferative and easy to propagate, and hence are a unique source for human neural cells for basic and therapeutic applications. Transgenic NP cells are a convenient and renewable starting point for generation of transgenic human neural cells. Sustained transgene expression in NP cells add great value to efforts at tracking migration, expansion, and differentiation of these cells in real time, both in culture and in vivo. Transgenic NP cells can also be potentially used to express proteins of interest in vivo or to express short-hairpin RNA molecules for targeting gene expression in disease models.45,46
In conclusion, NP cells derived from hESC can be genetically modified by nucleofection, transfection, and lentiviral transduction. For long-term transgene expression, transduction was found to be the most suitable method. Transgene expression could be achieved with number of constitutive (CMV, EF1α, and Ubc) and lineage-specific (GFAP) promoters. Lentiviral-based transduction exhibited a marginal silencing (20–30%) in NP cells over a 2-month period, proving its suitability for longer-term transgene expression. The genetically manipulated NP cells described here offer great potential for live cell–tracking experiments, and a similar approach can as well be used for expression of proteins other than reporters.
Acknowledgments
We would like to thank Dr. Michael Brenner (University of Alabama, Birmingham, AL) for the Gfa2 promoter, Dr. Roger Y. Tsien (University of California, San Diego, CA) for the tdTomatoRed plasmid, Dr. John K. Wakefield (Open Biosystems) for CMV-EGFP and EF1α-EGFP viruses, and Dr. James J. Lah (Emory University, GA) for the Ubc-EGFP virus and packaging of the GFAP reporter constructs. We also thank Julie Nelson for help with Flow Cytometry, and Dr. Franklin West for useful comments on the manuscript. Funding was provided by the GRA Eminent Endowment, National Institutes of Health (R41-NS053272-01), and Department of Defense Award (N00014-08–1-0989).
Disclosure Statement
S.L.S. is a founder of Aruna Biomedical, Inc., and declares that this article could be considered a competing interest.
References
- 1.Shin S. Mitalipova M. Noggle S. Tibbitts D. Venable A. Rao R. Stice S.L. Long-term proliferation of human embryonic stem cell-derived neuroepithelial cells using defined adherent culture conditions. Stem Cells. 2006;24:125. doi: 10.1634/stemcells.2004-0150. [DOI] [PubMed] [Google Scholar]
- 2.Dhara S.K. Hasneen K. Machacek D.W. Boyd N.L. Rao R.R. Stice S.L. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation. 2008;76:454. doi: 10.1111/j.1432-0436.2007.00256.x. [DOI] [PubMed] [Google Scholar]
- 3.Schaefer A.W. Juliano S.L. Migration of transplanted neural progenitor cells in a ferret model of cortical dysplasia. Exp Neurol. 2008;210:67. doi: 10.1016/j.expneurol.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Guzman R. Bliss T. De Los Angeles A. Moseley M. Palmer T. Steinberg G. Neural progenitor cells transplanted into the uninjured brain undergo targeted migration after stroke onset. J Neurosci Res. 2008;86:873. doi: 10.1002/jnr.21542. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y. Shah K. Messerli S.M. Snyder E. Breakefield X. Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14:1247. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 6.Koch P. Siemen H. Biegler A. Itskovitz-Eldor J. Brustle O. Transduction of human embryonic stem cells by ecotropic retroviral vectors. Nucleic Acids Res. 2006;34:e120. doi: 10.1093/nar/gkl674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blits B. Kitay B.M. Farahvar A. Caperton C.V. Dietrich W.D. Bunge M.B. Lentiviral vector-mediated transduction of neural progenitor cells before implantation into injured spinal cord and brain to detect their migration, deliver neurotrophic factors and repair tissue. Restor Neurol Neurosci. 2005;23:313. [PubMed] [Google Scholar]
- 8.Ohori Y. Yamamoto S. Nagao M. Sugimori M. Yamamoto N. Nakamura K. Nakafuku M. Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J Neurosci. 2006;26:11948. doi: 10.1523/JNEUROSCI.3127-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates F. Daley G.Q. Progress and prospects: gene transfer into embryonic stem cells. Gene Ther. 2006;13:1431. doi: 10.1038/sj.gt.3302854. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y.P. Dovzhenko O.V. Garthwaite M.A. Dambaeva S.V. Durning M. Pollastrini L.M. Golos T.G. Maintenance of pluripotency in human embryonic stem cells stably over-expressing enhanced green fluorescent protein. Stem Cells Dev. 2004;13:636. doi: 10.1089/scd.2004.13.636. [DOI] [PubMed] [Google Scholar]
- 11.Eiges R. Schuldiner M. Drukker M. Yanuka O. Itskovitz-Eldor J. Benvenisty N. Establishment of human embryonic stem cell-transfected clones carrying a marker for undifferentiated cells. Curr Biol. 2001;11:514. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 12.Schuldiner M. Itskovitz-Eldor J. Benvenisty N. Selective ablation of human embryonic stem cells expressing a “suicide” gene. Stem Cells. 2003;21:257. doi: 10.1634/stemcells.21-3-257. [DOI] [PubMed] [Google Scholar]
- 13.Liew C.G. Draper J.S. Walsh J. Moore H. Andrews P.W. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 14.Nolden L. Edenhofer F. Haupt S. Koch P. Wunderlich F.T. Siemen H. Brustle O. Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat Methods. 2006;3:461. doi: 10.1038/nmeth884. [DOI] [PubMed] [Google Scholar]
- 15.Mitalipova M. Calhoun J. Shin S. Wininger D. Schulz T. Noggle S. Venable A. Lyons I. Robins A. Stice S.L. Human embryonic stem cell lines derived from discarded embryos. Stem Cells. 2003;21:521. doi: 10.1634/stemcells.21-5-521. [DOI] [PubMed] [Google Scholar]
- 16.Irons H.R. Cullen D.K. Shapiro N.P. Lambert N.A. Lee R.H. Laplaca M.C. Three-dimensional neural constructs: a novel platform for neurophysiological investigation. J Neural Eng. 2008;5:333. doi: 10.1088/1741-2560/5/3/006. [DOI] [PubMed] [Google Scholar]
- 17.Brenner M. Structure and transcriptional regulation of the GFAP gene. Brain Pathol. 1994;4:245. doi: 10.1111/j.1750-3639.1994.tb00840.x. [DOI] [PubMed] [Google Scholar]
- 18.Besnard F. Brenner M. Nakatani Y. Chao R. Purohit H.J. Freese E. Multiple interacting sites regulate astrocyte-specific transcription of the human gene for glial fibrillary acidic protein. J Biol Chem. 1991;266:18877. [PubMed] [Google Scholar]
- 19.Drukker M. Dhara S.K. Benvenisty N. Genetic engineering of human embryonic stem cells. In: Odorico J.S., editor; Zhang S-C., editor; Pedersen R.A., editor. Human Embryonic Stem Cells. Oxford: Garland Science/BIOS Scientific Publishers; 2005. pp. 215–230. [Google Scholar]
- 20.Wobus A.M. Boheler K.R. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol Rev. 2005;85:635. doi: 10.1152/physrev.00054.2003. [DOI] [PubMed] [Google Scholar]
- 21.Cao F. Lin S. Xie X. Ray P. Patel M. Zhang X. Drukker M. Dylla S.J. Connolly A.J. Chen X. Weissman I.L. Gambhir S.S. Wu J.C. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladewig J. Koch P. Endl E. Meiners B. Opitz T. Couillard-Despres S. Aigner L. Brustle O. Lineage selection of functional and cryopreservable human embryonic stem cell-derived neurons. Stem Cells. 2008;26:1705. doi: 10.1634/stemcells.2008-0007. [DOI] [PubMed] [Google Scholar]
- 23.Singh Roy N. Nakano T. Xuing L. Kang J. Nedergaard M. Goldman S.A. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196:224. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Gerrard L. Zhao D. Clark A.J. Cui W. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005;23:124. doi: 10.1634/stemcells.2004-0102. [DOI] [PubMed] [Google Scholar]
- 25.Dhara S.K. Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakshmipathy U. Pelacho B. Sudo K. Linehan J.L. Coucouvanis E. Kaufman D.S. Verfaillie C.M. Efficient transfection of embryonic and adult stem cells. Stem Cells. 2004;22:531. doi: 10.1634/stemcells.22-4-531. [DOI] [PubMed] [Google Scholar]
- 27.Siemen H. Nix M. Endl E. Koch P. Itskovitz-Eldor J. Brustle O. Nucleofection of human embryonic stem cells. Stem Cells Dev. 2005;14:378. doi: 10.1089/scd.2005.14.378. [DOI] [PubMed] [Google Scholar]
- 28.Elkabetz Y. Panagiotakos G. Al Shamy G. Socci N.D. Tabar V. Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang J.E. Shaw K. Yu X.J. Petersen D. Pepper K. Lutzko C. Kohn D.B. Specific and stable gene transfer to human embryonic stem cells using pseudotyped lentiviral vectors. Stem Cells Dev. 2006;15:109. doi: 10.1089/scd.2006.15.109. [DOI] [PubMed] [Google Scholar]
- 30.Gropp M. Itsykson P. Singer O. Ben-Hur T. Reinhartz E. Galun E. Reubinoff B.E. Stable genetic modification of human embryonic stem cells by lentiviral vectors. Mol Ther. 2003;7:281. doi: 10.1016/s1525-0016(02)00047-3. [DOI] [PubMed] [Google Scholar]
- 31.Ma Y. Ramezani A. Lewis R. Hawley R.G. Thomson J.A. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 2003;21:111. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Dor I. Itsykson P. Goldenberg D. Galun E. Reubinoff B.E. Lentiviral vectors harboring a dual-gene system allow high and homogeneous transgene expression in selected polyclonal human embryonic stem cells. Mol Ther. 2006;14:255. doi: 10.1016/j.ymthe.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer A. Ikawa M. Dayn Y. Verma I.M. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc Natl Acad Sci USA. 2002;99:2140. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue T. Cho H.C. Akar F.G. Tsang S.Y. Jones S.P. Marban E. Tomaselli G.F. Li R.A. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation. 2005;111:11. doi: 10.1161/01.CIR.0000151313.18547.A2. [DOI] [PubMed] [Google Scholar]
- 35.Eiges R. Genetic manipulation of human embryonic stem cells by transfection. Methods Mol Biol. 2006;331:221. doi: 10.1385/1-59745-046-4:221. [DOI] [PubMed] [Google Scholar]
- 36.Casper K.B. Jones K. McCarthy K.D. Characterization of astrocyte-specific conditional knockouts. Genesis. 2007;45:292. doi: 10.1002/dvg.20287. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y. Messing A. Su M. Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y. Su M. Messing A. Brenner M. Astrocyte heterogeneity revealed by expression of a GFAP-LacZ transgene. Glia. 2006;53:677. doi: 10.1002/glia.20320. [DOI] [PubMed] [Google Scholar]
- 39.Bartlett D.W. Davis M.E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia X. Zhang Y. Zieth C.R. Zhang S.C. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez S.L. Russell D.W. Hurlin P.J. Development of human gene reporter cell lines using rAAV mediated homologous recombination. Biol Proced Online. 2007;9:84. doi: 10.1251/bpo136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leor J. Gerecht S. Cohen S. Miller L. Holbova R. Ziskind A. Shachar M. Feinberg M.S. Guetta E. Itskovitz-Eldor J. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93:1278. doi: 10.1136/hrt.2006.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keirstead H.S. Nistor G. Bernal G. Totoiu M. Cloutier F. Sharp K. Steward O. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25:4694. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aharonowiz M. Einstein O. Fainstein N. Lassmann H. Reubinoff B. Ben-Hur T. Neuroprotective effect of transplanted human embryonic stem cell-derived neural precursors in an animal model of multiple sclerosis. PLoS ONE. 2008;3:e3145. doi: 10.1371/journal.pone.0003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An D.S. Xie Y. Mao S.H. Morizono K. Kung S.K. Chen I.S. Efficient lentiviral vectors for short hairpin RNA delivery into human cells. Hum Gene Ther. 2003;14:1207. doi: 10.1089/104303403322168037. [DOI] [PubMed] [Google Scholar]
- 46.Sapru M.K. Yates J.W. Hogan S. Jiang L. Halter J. Bohn M.C. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp Neurol. 2006;198:382. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]