Abstract
The spatial organization of cells of different phenotypes is an important and often defining determinant of tissue function. In tissue engineering, which attempts to rebuild functional tissues from cellular and synthetic components, spatial patterning of cells onto biomaterials is likely to be equally important. We have printed combinatorial arrays of extracellular matrix (ECM) and screened them for attachment by HepG2 hepatocytes, LX-2 hepatic stellate cells, primary portal fibroblasts, and bovine aortic endothelial cells—cells selected as representative phenotypes found in adult liver. Differential cell attachment to the underlying matrix proteins allowed us to establish two-dimensional co-cultures of HepG2 with these non-parenchymal cell types. These general approaches were then translated to tissue engineering scaffolds where deposition of ECM proteins onto electrospun polylactide meshes resulted in patterned HepG2 cultures. We observed that the spatial organization of fibronectin deposits influenced HepG2 attachment and the establishment of co-cultures on our arrays. These micropatterned co-culture systems should serve as valuable tools for studying the soluble and insoluble signals involved in liver development, function, and disease.
Introduction
Long-term and stable hepatocyte cultures are valuable systems for studying liver structure, function, and disease, as well as for developing therapeutic applications. Some long-term culture methods for hepatocytes have used special extracellular matrix (ECM) materials such as Matrigel™, a matrix derived from Engelbreth-Holm-Swarm (EHS) tumors grown in mice.1 Hepatocytes sandwiched between two EHS gels have been shown to exhibit a morphology and an intracellular organization that closely mimics the liver, where these cells are bound by ECM at each of their opposite basolateral domains.2 In addition, EHS matrix has been shown to induce expression of gap junctions and epidermal growth factor (EGF) receptors that are otherwise absent in hepatocytes cultured between collagen I gels.2,3 Sandwich cultures mimic the environment seen by hepatocytes in vivo, which may contribute to the success of this method. However, this culture format has been difficult to scale up because of nutrient transport limitations.
As an alternative to sandwich cultures, co-cultures of hepatocytes with non-parenchymal cell types can also stabilize hepatocyte viability and liver-specific function.4 Photolithography techniques have been developed to micropattern two distinct cell populations in spatially controlled configurations.4–9 This technique offers the possibility of presenting different adhesive ligands to different cell types within a co-culture and, by dictating the size and geometry of the cell patterns, permits control of cell-cell interactions. For example, co-cultures of hepatocytes and NIH/3T3 fibroblasts have been established by first culturing hepatocytes on immobilized collagen I islands and then allowing NIH/3T3 fibroblasts to adhere to serum proteins adsorbed to the surrounding spaces.4,5 The degree of homo- and heterotypic cell–cell interactions were manipulated by altering the size of the collagen I islands and varying the area available for fibroblast adhesion.7 This approach was used to systematically investigate the hepatocelluar response to changes in fibroblast number and variations in the magnitude of the heterotypic cell-cell interface. Co-culture systems retain all of the liver-specific functions observed in the sandwich cultures, and stable function can be maintained for 3 weeks or longer depending on the cell types co-cultured with hepatocytes.3,10 Hepatocytes in co-cultures also exhibit a morphology and intracellular organization reminiscent of hepatocytes in vivo.
Technologies for patterning multiple cell types onto artificial surfaces are highly desirable for advancing the development of cell-based sensors and drug screening platforms and for fabrication of tissue-engineering architectures. In a novel cell patterning approach, Hui et al. used a micromachined silicon substrate with moving parts to examine spatial and temporal influences on the communication between hepatocytes and stromal cells11 or NIH/3T3 fibroblasts.12 Although cell patterning using lithography and microcontact printing can position cells precisely and with high resolution, these methods require specialized equipment, and the cell culture surfaces are often modified chemically in a manner that may restrict their application in tissue engineering.13,14 As an alternative, inkjet printing uses an inexpensive desktop printer to deposit biological macromolecules onto substrates15,16 and has been used to pattern co-cultures of neurons and glia cells,17 but this technique produces lower-resolution patterns than lithographic approaches. Another innovative patterning technique uses complimentary base-pairing between DNA strands functionalized onto the surface of living cells and their cognate sequences printed onto glass slides to immobilize adherent and non-adherent cell types.18 This methodology allows cells to be patterned independently of their natural surface properties but requires using complex metabolic oligosaccharide engineering and azide chemistry.19
Here, we employed protein printing to define conditions that modulate attachment of different cell types found in the adult liver. Our combinatorial ECM array was modeled after a protein array described by Flaim et al., but we expanded their approach to screen twice as many ECM combinations and include several more cell types from the liver.20 The ECM array was used to screen for attachment by HepG2 hepatocytes, LX-2 hepatic stellate cells (HSCs), primary portal fibroblasts (PFs), and bovine aortic endothelial cells (BAECs). HSCs produce a number of ECM components (types I, III, IV, and VI collagen and fibronectin, laminin, and proteoglycans) and express several ECM-degrading enzymes implicated in initiating hepatocyte differentiation and revascularization during liver regeneration.21 Because hepatic stellate cells (HSCs) play an important role in rebuilding the architecture of the liver after injury or disease, we established co-culture systems with hepatocytes and LX-2 HSCs. The liver is also a highly vascularized organ, and we used our protein arrays to form co-cultures of hepatocytes with BAECs. PFs are a novel cell type whose role in regulating the proliferation of bile duct cells has been described only recently.22 This is the first report of screening primary PFs for attachment to multiple combinations of ECM proteins. Our approaches and findings on the two-dimensional (2D) arrays were then translated to tissue engineering scaffolds, where deposition of ECM proteins onto electrospun polylactide meshes resulted in patterned HepG2 cultures. Together, this information may allow production of more-relevant hepatocyte co-culture systems using micropatterning techniques.
Materials and Methods
ECM microarray fabrication
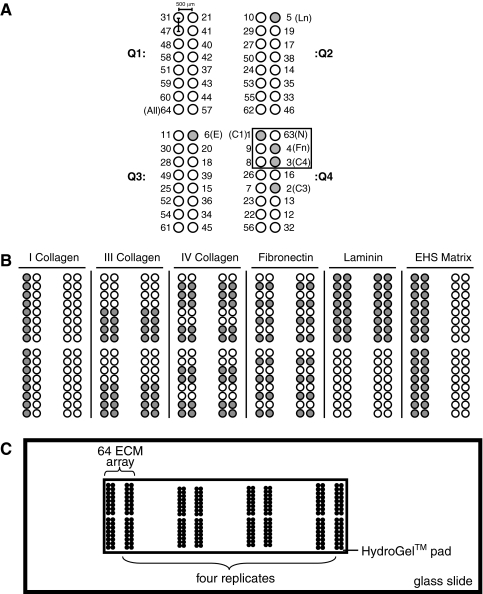
ECM protein microarrays were printed on HydroGel™ (Perkin Elmer, Waltham, MA) microarray slides consisting of a commercially fabricated acrylamide gel pad (40 mm × 12 mm) mounted on glass (Fig. 1C). All 64 unique combinations of six ECM proteins were formulated using a single-protein printing buffer described previously.20 This print buffer was shown to produce high-quality protein microarrays by decreasing the aggregation of laminin and collagen and increasing the amount of ECM material deposited at each spot.20 Stock solutions of rat collagen I (C1, BD Biosciences, San Jose, CA), bovine collagen III (C3, BD Biosciences), mouse collagen IV (C4, BD Biosciences), human fibronectin (Fn, Sigma, St Louis, MO) mouse laminin (Ln, Sigma), and Matrigel™ (EHS, BD Biosciences) were prepared in print buffer at a concentration of 500 μg/mL, and the 64 different ECM mixtures were combined in a 384-well plate. The range of protein concentrations for any mixture varied from 80 μg/mL to 500 μg/mL; in all cases, the total protein concentration was maintained constant. Hydrogel slides were prepared for printing according to the manufacturer's suggestions, and the arrays were printed in a 4°C cold room at the Yale Center for Genomics and Proteomics (YCGP). Four replicates of each protein mixture were deposited on the Hydrogel surface using a Virtek ChipWriter Pro (BioRad, Hercules, CA) robotic microarrayer equipped with Stealth SMP 2.0 split pins (TeleChem International, Sunnyvale, CA). After printing, slides were incubated at room temperature in a chamber with 65% to 75% relative humidity for 16 h, washed extensively in phosphate buffered saline supplemented with 0.5% Tween-20 (PBST), and dried. The printed ECM microarrays were stored in a desiccator at 4°C for up to 1 month.
FIG. 1.
Extracellular matrix protein microarray. (A) Diagram showing composition and position of all 64 unique compositions of collagen I, III, and IV; fibronectin (Fn); laminin (Ln); and Engelbreth-Holm-Swarm matrix (E). Number adjacent to each spot corresponds to the identity (ID) of the mixture in Table 1. Position of the pure Fn spot is indicated, and the box shows its nearest extracellular matrix (ECM) neighbors. (B) Diagram showing pattern of protein deposition for each of the six ECM proteins used to print the microarray. (C) Proteins were deposited in four replicates onto HydroGel microarray slides using a Virtek ChipWriter Pro robotic microarrayer.
Table 1.
Components of Extracellular Matrix (ECM) Combinatorial Array
| ID | Collagen I (C1) | Collagen III (C3) | Collagen IV (C4) | Fibronectin (Fn) | Laminin (Ln) | EHS Matrix (E) | No. of ECM proteins |
|---|---|---|---|---|---|---|---|
| 1 | Y | N | N | N | N | N | 1 |
| 2 | N | Y | N | N | N | N | |
| 3 | N | N | Y | N | N | N | |
| 4 | N | N | N | Y | N | N | |
| 5 | N | N | N | N | Y | N | |
| 6 |
N |
N |
N |
N |
N |
Y |
|
| 7 | Y | Y | N | N | N | N | 2 |
| 8 | Y | N | Y | N | N | N | |
| 9 | Y | N | N | Y | N | N | |
| 10 | Y | N | N | N | Y | N | |
| 11 | Y | N | N | N | N | Y | |
| 12 | N | Y | Y | N | N | N | |
| 13 | N | Y | N | Y | N | N | |
| 14 | N | Y | N | N | Y | N | |
| 15 | N | Y | N | N | N | Y | |
| 16 | N | N | Y | Y | N | N | |
| 17 | N | N | Y | N | Y | N | |
| 18 | N | N | Y | N | N | Y | |
| 19 | N | N | N | Y | Y | N | |
| 20 | N | N | N | Y | N | Y | |
| 21 |
N |
N |
N |
N |
Y |
Y |
|
| 22 | Y | Y | Y | N | N | N | 3 |
| 23 | Y | Y | N | Y | N | N | |
| 24 | Y | Y | N | N | Y | N | |
| 25 | Y | Y | N | N | N | Y | |
| 26 | Y | N | Y | Y | N | N | |
| 27 | Y | N | Y | N | Y | N | |
| 28 | Y | N | Y | N | N | Y | |
| 29 | Y | N | N | Y | Y | N | |
| 30 | Y | N | N | Y | N | Y | |
| 31 | Y | N | N | N | Y | Y | |
| 32 | N | Y | Y | Y | N | N | |
| 33 | N | Y | Y | N | Y | N | |
| 34 | N | Y | Y | N | N | Y | |
| 35 | N | Y | N | Y | Y | N | |
| 36 | N | Y | N | Y | N | Y | |
| 37 | N | Y | N | N | Y | Y | |
| 38 | N | N | Y | Y | Y | N | |
| 39 | N | N | Y | Y | N | Y | |
| 40 | N | N | Y | N | Y | Y | |
| 41 |
N |
N |
N |
Y |
Y |
Y |
|
| 42 | N | N | Y | Y | Y | Y | 4 |
| 43 | N | Y | N | Y | Y | Y | |
| 44 | N | Y | Y | N | Y | Y | |
| 45 | N | Y | Y | Y | N | Y | |
| 46 | N | Y | Y | Y | Y | N | |
| 47 | Y | N | N | Y | Y | Y | |
| 48 | Y | N | Y | N | Y | Y | |
| 49 | Y | N | Y | Y | N | Y | |
| 50 | Y | N | Y | Y | Y | N | |
| 51 | Y | Y | N | N | Y | Y | |
| 52 | Y | Y | N | Y | N | Y | |
| 53 | Y | Y | N | Y | Y | N | |
| 54 | Y | Y | Y | N | N | Y | |
| 55 | Y | Y | Y | N | Y | N | |
| 56 |
Y |
Y |
Y |
Y |
N |
N |
|
| 57 | N | Y | Y | Y | Y | Y | 5 |
| 58 | Y | N | Y | Y | Y | Y | |
| 59 | Y | Y | N | Y | Y | Y | |
| 60 | Y | Y | Y | N | Y | Y | |
| 61 | Y | Y | Y | Y | N | Y | |
| 62 |
Y |
Y |
Y |
Y |
Y |
N |
|
| 63 |
N |
N |
N |
N |
N |
N |
0 |
| 64 | Y | Y | Y | Y | Y | Y | 6 |
Indirect immunofluorescence
Protein immobilization on the microarray substrate was evaluated using indirect immunofluorescence. In brief, ECM microarray slides were first blocked with PBST containing 1% (w/v) bovine serum albumin for 1 h at room temperature. The slides were then washed with PBST, probed for 1 h at room temperature with a primary rabbit anti-rat antibody against collagen I (1:40, Chemicon, Temecula, CA), and washed extensively with PBST before probing with a secondary AlexaFluor 633 goat anti-rabbit polyclonal antibody (1:200, Molecular Probes, Eugene, OR) for 1 h at room temperature. Slides were scanned for the presence and spatial distribution of fluorescence using a GenePix Professional 4200A scanner (Molecular Devices) at the YCGP.
Cell culture
Microarrays were cultured with HepG2 hepatocytes, LX-2 HSCs, PFs, and BAECs. HepG2 hepatocytes were maintained in Eagle's minimum essential medium (E-MEM, American Type Culture Collection, Grand Island, NY) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, Grand Island, NY) and 1% (v/v) penicillin/streptomycin (Sigma). LX-2 HSCs were maintained in Dulbecco's modified Eagle medium/nutrient mix F-12 (DMEM/F-12, Gibco, Eugene, OR) supplemented with 1% (v/v) each FBS, penicillin/streptomycin, and L-glutamine (Gibco). Primary portal fibroblasts were maintained in DMEM/F-12 supplemented with 10% (v/v) FBS, 3% (v/v) penicillin/streptomycin, 0.2% (v/v) gentamicin (Gibco), and 0.4% (v/v) Fungizone (Gibco). BAECs were maintained in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin.
Printed microarray slides were sterilized by exposure to ultraviolet light in a laminar flow hood for 20 min and then rinsed twice with sterile culture medium before cell seeding. Slides were placed inside sterile cell culture dishes (P-100, BD Biosciences) and a suspension of approximately 3 × 105 cells/mL was applied in a volume sufficient to cover the printed slide area. The slides were incubated for 2 h to allow cell attachment, and then cells were cultured from 1 to 7 d with daily exchanges of medium.
Screening for cell attachment and viability
Cells cultured on microarrays were rinsed and fixed with 4% (w/v) paraformaldehyde and permeabilized with ice-cold acetone. Cell attachment on each ECM island was assessed using CyQuantNF® Cell Proliferation Assay Kit (Invitrogen, Norwood, MA), dried, and then scanned on a GenePix 4200A fluorescent scanner (Molecular Devices, Sunnyvale, CA). Cell viability was determined using the Live/Dead Assay Viability/Cytotoxicity Kit (Invitrogen). Calcein-AM (live) and ethidium homodimer-1 (dead) dyes were applied to unfixed cells as suggested by the manufacturer. Microarrays cultured with cells and probed with the live/dead stains were then fixed with 4% (w/v) paraformaldehyde, dried, and scanned. Trypan blue (Gibco) exclusion was also used to detect dead cells on the cultured microarrays. In this case, slides were rinsed in PBS, incubated briefly in trypan blue, rinsed, and then visualized using a Nikon TS100 microscope mounted with an Olympus digital camera.
Establishing micropatterned co-cultures
Co-cultures of HepG2 hepatocytes with other cell types were established using a sequential seeding protocol. In brief, approximately 3 × 105 cells/mL hepatocytes were seeded onto the ECM arrays in serum-free medium for 2 h at 37°C. Unattached HepG2 hepatocytes were gently aspirated, and the second cell type was seeded in the appropriate serum-containing medium. The second cell type was seeded at the same cell density for 2 h, followed by a fresh exchange of medium and overnight culture. In all co-cultures, HepG2 hepatocytes were prestained with CellTracker Orange CMTMR (HepG2-CMTMR; Invitrogen) to differentiate them from the second cell type.
Patterning co-cultures on 3D meshes of polylactic acid
Three-dimensional polylactic acid (PLA) meshes were formed by electrospinning. In brief, a 13% (w/v) solution of PLA in a 3:1 chloform:acetone mixture was ejected from a syringe at a flow rate of 15 μL/min toward a stationary copper plate. The needle of the syringe and the copper plate were attached to the positive and negative termini of a high-voltage generator, respectively. A distance of 16 cm was set between the needle and the collection plate. Using an 18-gauge needle fixed to a support and a high-voltage power supply to produce an electric field of 20 kV, the voltage gradient formed between the syringe and the copper plate pulled the polymer to form PLA fibers that were drawn to the plate, creating a non-woven mesh. The resulting scaffold thickness was measured using calipers. The PLA scaffolds were imaged using scanning electron microscopy (SEM), and fiber diameter was determined by image analysis of SEM micrographs using ImageJ (Rasband, 1997–2005). Mechanical strength of the PLA scaffolds was measured using a 5848 MicroTester (Instron). Scaffold samples measuring 1 cm × 4 cm were placed in the clamps of the Instron and tested at a rate of 10 mm/min using a 5 N load cell. The scaffolds were mounted onto glass microscope slides for protein printing and cell culture as described above.
Results
Printing and characterization of ECM protein microarray
A Virtek ChipWriter Pro (BioRad) robotic microarrayer equipped with Stealth SMP 2.0 split pins (Telechem) was used to print all combinations of rat C1, bovine C3, mouse C4, human Fn, mouse Ln, and mouse EHS matrix (Fig. 1). The 64 combinations were printed in four quadrants, each containing a 2 × 8 array (Fig. 1A). The protein mixtures were grouped into regions on the array based on their specific ECM composition (Fig. 1B). For example, mixtures containing C1 were located in the first column of quadrants 1 and 4, whereas mixtures containing C1 were located in quadrants 1 and 2. Each quadrant was printed on the HydroGel™ microarray pad such that an individual mixture was represented in four replicates (Fig. 1C).
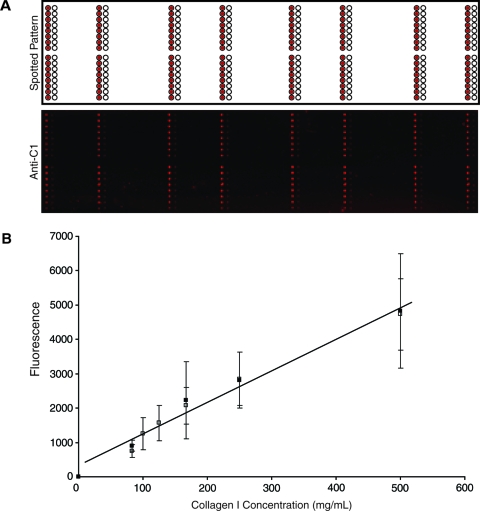
The individual pins each deposited approximately 1 to 2 nL of material and generated spots with a diameter of approximately 300 μm and a center-to-center spacing of approximately 500 μm. Indirect immunofluorescence was used to characterize the location and integrity of immobilized ECM material after printing. Antigenic recognition of immobilized C1 resulted in fluorescence at the expected locations (Fig. 2A). We also characterized the quality and quantity of C1 remaining immobilized on the microarray substrate after incubation in culture medium. We compared the fluorescence intensity of immobilized C1 directly after printing and after a 24-h incubation at 37°C in culture medium. Indirect immunofluorescence showed that C1 remained localized to the regions of deposition, and magnification of individual islands showed that C1 distribution was homogenous over the entire printed area before and after incubation. Furthermore, C1 concentration after incubation was similar to the values measured directly after printing (Fig. 2B). Thus, we did not detect any change in the quality or quantity of the immobilized C1 under the conditions that we used for cell seeding.
FIG. 2.
Characterization of extracellular matrix (ECM) microarray. (A) Specified position and immunofluorescent image of an ECM microarray probed with a primary antibody against collagen I and using a secondary antibody conjugated to Alexafluor 633. Immunofluorescent image was obtained using a GenePix 4200A scanner. (B) Correlation of fluorescence intensity and rat collagen I concentration after protein printing (□) and following incubation (▪) for 24 h at 37°C in cell culture medium. Color images available online at www.liebertonline.com/ten.
Attachment and viability of HepG2 hepatocytes on the ECM microarray
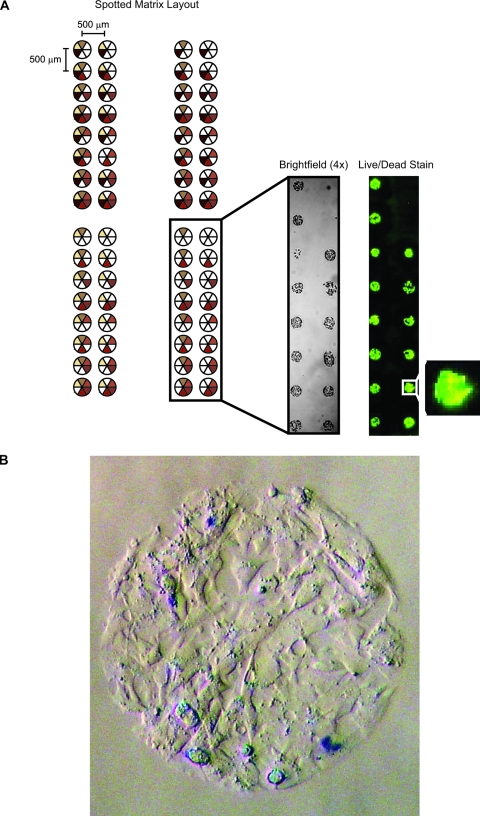
The ECM microarrays were seeded with HepG2 hepatocytes, and the cells were allowed to attach for 2 h and then cultured overnight after a fresh exchange of medium. Cultured ECM arrays were washed with several exchanges of PBS and fixed for imaging. HepG2 attachment occurred only at sites of protein deposition and did not show any attachment to the substrate areas lacking ECM proteins (Fig. 3A, brightfield). The cells were confined to the areas of protein deposition and grew to confluency across this region. Cells spread over the printed area to occupy an island with an average diameter of approximately 300 μm. Cell attachment was consistent over the entire printed area, and HepG2 islands were established reproducibly on replicate ECM material. Viability of cells cultured on the microarrays was assessed using the Live/Dead assay (Fig. 4A, fluorescence (Invitrogen) and trypan blue exclusion (Fig. 4B). Both assays showed that a significant fraction of the HepG2 cultured on the ECM microarrays was viable after 24 h.
FIG. 3.
Viability of cultured cells on extracellular matrix (ECM) protein arrays. (A) Live/dead staining reveals viability of HepG2 cells on ECM combinatorial array. Co-localization of green (live) and red (dead) fluorescence results in a yellow signal. (B) Brightfield image (20×) of a single HepG2 island after exposure to Trypan blue. Brightfield images were obtained using an Olympus digital camera mounted on a Nikon TS100 microscope. Fluorescent images were obtained using a GenePix 4200A scanner. Color images available online at www.liebertonline.com/ten.
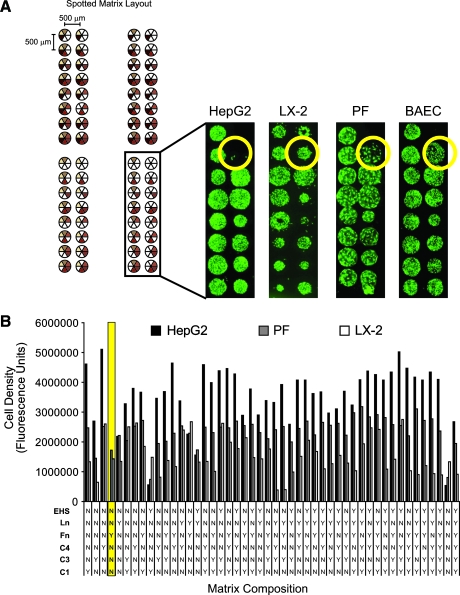
FIG. 4.
Cell attachment to extracellular matrix (ECM) protein microarrays. (A) Representative images of HepG2 hepatocytes, LX-2 hepatic stellate cells (HSCs), portal fibroblasts, and bovine aortic endothelial cells cultured on the combinatorial ECM protein arrays. Cell cultures were established overnight, washed, fixed, and then permeabilized and stained using CyQuantNF. Cells were imaged on a GenePix 4200A fluorescent laser scanner. Yellow circle indicates position of fibronectin island. (B) The amount of cells occupying each ECM island was estimated by measuring the total cellular fluorescence and plotted as a function of the underlying ECM material. Cultured cells do not show patterns of differential attachment but HepG2 cells are observed to attach less to fibronectin spots than LX-2 HSCs or portal fibroblasts. Color images available online at www.liebertonline.com/ten.
Influence of ECM composition on the attachment of various cell types
ECM protein microarrays were used to identify matrix compositions that would mediate specific attachment of hepatocytes or non-parenchymal cell types. HepG2 hepatocytes, LX-2 HSCs, primary PFs, and BAECs were screened for attachment to all possible combinations of the six ECM proteins. We used a nuclear stain to measure the number of cells occupying each ECM island (Fig. 4A). We observed that LX-2 HSCs, PFs, and BAECs attached equally to the different ECM mixtures and not to areas lacking ECM proteins. The spatial distribution of the cells cultured on any individual island probably reflects the quality of protein deposition rather than any preferential attachment to the underlying matrix proteins. Comparing values of total cellular fluorescence did not identify a unique adhesive substrate for any of the cell types cultured on the ECM microarrays (Fig. 4B). Although the scanned fluorescence was not normalized for the DNA content of each cell type, brightfield and fluorescent images suggested only nominal differences in cell type–specific attachment to the ECM mixtures printed on the arrays. In general, all cell types attached and remained established on each ECM substrate for at least 24 h. However, in contrast to the other cell types, we consistently observed that HepG2 hepatocytes did not attach to Fn islands (Fig. 4A, yellow circle).
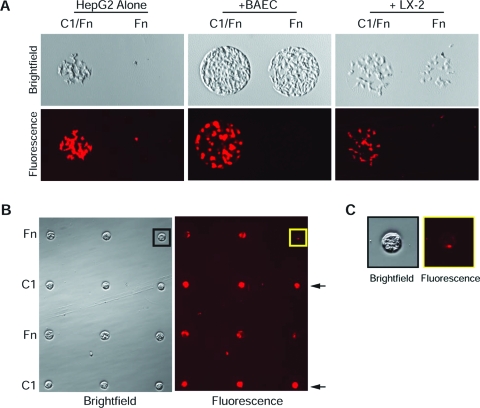
We took advantage of this selective non-attachment of HepG2 hepatocytes to Fn and employed a sequential seeding protocol to establish co-cultures of HepG2 hepatocytes on the ECM protein arrays. Seeding HepG2-CMTMR first onto the ECM microarrays resulted in their attachment to all ECM compositions on the array except to the Fn island (Fig. 5A, left panels). A second seeding onto the same arrays with BAECs (Fig. 5A, middle panels) or LX-2 HSCs (Fig. 5A, right panels) led to co-cultures with the HepG2 hepatocytes. We printed a simple 4 × 4 grid pattern composed of alternating rows of Fn and C1 spots and used the sequential seeding protocol to establish a co-culture of BAECs with HepG2-CMTMR (Fig. 5B). Brightfield images of the Fn/C1 4 × 4 arrays cultured with BAECs and HepG2-CMTMR show cells occupying and spread across the areas where ECM proteins were deposited. Fluorescent images of the same arrays show intense red fluorescence for rows printed with C1. The spatial distribution of fluorescence at these positions correspond to the spatial distribution of cells seen under brightfield. This result suggests that HepG2-CMTMR were the predominant cell type attached on the C1 rows. We also observed some red fluorescence in rows printed with Fn, which indicates the presence of HepG2-CMTMR at these positions. However, the spatial distribution of fluorescence at the Fn positions did not correspond to the respective cell islands seen under brightfield (Fig. 5C), and small fraction of the cells on these spots were fluorescent. We expect that the rows printed with Fn contain HepG2-CMTMRs and BAECs but are predominantly occupied by BAECs.
FIG. 5.
Extracellular matrix (ECM) arrays support co-cultures of HepG2 cells with bovine aortic endothelial cells (BAECs) or LX-2 hepatic stellate cells (HSCs). (A) ECM microarrays support co-cultures of HepG2 hepatocytes were prestained with CellTracker Orange CMTMR (HepG2-CMTMRs) with BAECs or LX-2 HSCs. Sequential seeding with BAECs (middle panel) or LX-2 HSCs (right panel) onto the 64 ECM combinatorial protein arrays produced co-cultures with HepG2 cells, which do not attach to fibronectin (Fn) spots (left panel). (B) ECM arrays composed of alternating rows of Fn and rat collagen I were sequentially seeded with BAECs and HepG2-CMTMRs to establish co-cultures patterns. Brightfield images show the pattern and boundary of the cell cultures in the 4 × 4 array. Fluorescent images show that rows of rat collagen I (black arrows) are occupied by HepG2-CMTMRs and that rows of Fn are cultured with both cell types. (C) Magnified images of boxed regions from (B). Brightfield (black box) and fluorescence (yellow box) image showing a Fn island occupied mainly by BAECs. Brightfield images were obtained using an Olympus digital camera mounted on a Nikon TS100 microscope. Fluorescent images were acquired using an Olympus IX71 inverted fluorescent microscope. Color images available online at www.liebertonline.com/ten.
ECM deposition and cell culture on 3D electrospun biodegradable scaffolds
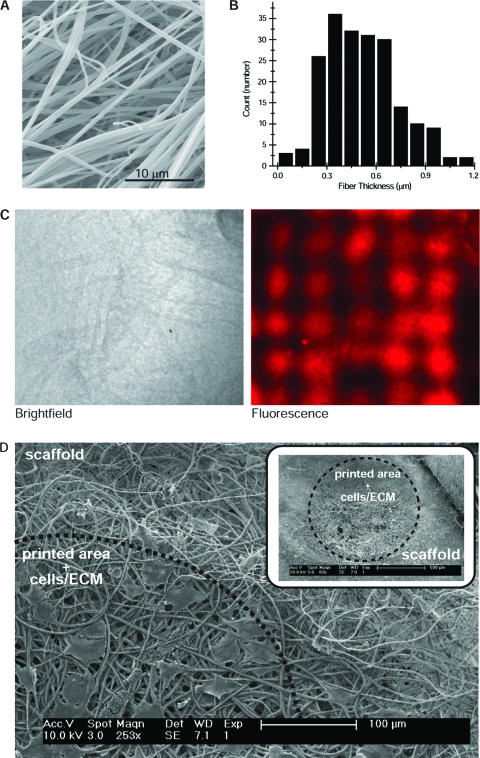
We fabricated 3D scaffolds by electrospinning PLA, a biodegradable polyester. Fibers were collected until a uniform mesh formed with a thickness of approximately 100 μm. The electrospinning device produced a flat, porous, nonwoven matrix with random fiber alignment (Fig. 6A). We found that a 13% (w/v) polymer solution concentration formed reproducible PLA fibers free of defects. Fibers spun at a flow rate of 15 μL/min and 20 kV produced the smallest average fiber diameter (480 ± 150 nm) (Fig. 6B). The minimum and maximum fiber diameters measured were 100 nm and 1.2 μm, respectively.
FIG. 6.
HepG2 hepatocytes cultured on electrospun biodegradable scaffolds. (A) Scanning electron microscopy (SEM) images of polylactic acid (PLA) fibers. (B) Average fiber diameter was determined from image analysis of SEM micrographs. (C) Brightfield (4×) and fluorescent image of PLA scaffold cultured with HepG2 hepatocytes pre-stained using CellTracker Orange CMTMR. Fluorescent image shows islands of HepG2 cells cultured into an 8 × 8 grid pattern. (D) SEM micrograph of an electrospun mesh of poly-lactic acid printed with extracellular matrix (ECM) and seeded with cells. Inset shows depression made on mesh after ECM printing using a manual arrayer. Scale bar of inset is 500 μm. Color images available online at www.liebertonline.com/ten.
PLA scaffolds were mounted to glass slides using 1% (w/v) agarose as an adhesive. The PLA scaffolds were printed with an 8 × 8 microarray consisting of alternating rows of Fn and C1 with a center-to-center spacing of 500 μm. HepG2-CMTMRs and BAECs were seeded sequentially on the printed scaffolds to establish patterned co-cultures. Cells cultured onto these scaffolds could not be visualized using brightfield microscopy (Fig. 6C, brightfield), which mainly revealed the architecture of the mounted scaffold. Using fluorescence microscopy, we were able to observe intense fluorescent spots corresponding to cultures of patterned HepG2-CMTMR. The cells formed islands that were spatially distributed in the 8 × 8 microarray pattern where Fn and C1 were deposited. The pattern of fluorescence indicated that HepG2 attached to Fn and C1, and the scaffolds probably contained both cell types at each of these ECM positions. SEM micrographs of PLA scaffolds printed with ECM proteins and then seeded with cells show clearly the depression made on the scaffold upon protein printing and the plaques of ECM or cells that were established in the area of protein deposition (Fig. 6D).
Discussion
Many commercial microarray surfaces are available for immobilizing proteins, but for these slides to be valuable for cell patterning, the surfaces must confine cell attachment to areas where adhesive substrates have been deposited. Surfaces resisting non-specific adsorption of serum proteins that are commonly present in cell culture media show the greatest promise for establishing patterned 2D cell cultures. We selected the commercially available HydroGel™ slides for our work because they did not require a blocking step to prevent non-specific protein adsorption and because the immobilized protein spots and the resulting cellular islands were more reproducible on the HydroGel surface than on other commercial protein microarrays.
Characterizing the immobilization and spreading of C1 on our ECM microarrays showed that the presence of other ECM proteins in the mixtures did not significantly alter antigenic recognition and spatial distribution of C1 (Fig. 2A). We observed that each of the C1–containing ECM spots promoted cell attachment and the growth of localized microcultures of HepG2 hepatocytes, LX-2 HSCs, PFs, and BAECs. In the case of HepG2 hepatocytes, we observed that the cells remained viable within these microcultures. Although we only characterized the immobilization and spreading of C1, protein deposits of pure Fn, Ln, EHS, C3, or C4 also produced localized microcultures for each of the four cell types. Thus, we observed that immobilization in spots on the slide did not prevent the ECM materials from acting as substrates for cell attachment. We also evaluated cell attachment using a centrifugation assay. In this assay, HepG2 cells were allowed to establish overnight on the ECM microarrays, after which time the medium was removed, and the slides were affixed to the bottom of a chamber. The sealed chamber was inverted and centrifuged using a plate rotor. Cell attachment and the integrity of the arrays were evaluated qualitatively using light microscopy. Under these conditions, we did not observe a significant difference between HepG2 cell attachment before and after centrifugation, and the integrity of the array was not altered (data not shown). Differences in attachment might be found in longer-term studies, but because the integrity of the microarray substrate was severely compromised after 7 d in culture medium, improvements must first be made in this area. We found that the HydroGel™ pad became detached from the glass support at incubation times of >48 h, and complete detachment occured at 7 d. Therefore, the HydroGel™ surface is not adequate for following long-term cultures.
Contact printing using the robotic arrayer produced reproducible ECM islands on the HydroGel™ substrate as long as fewer than five microarrays were printed. We occasionally observed missing ECM mixtures on our printed arrays and speculated that this arose from inadequate protein acquisition by the pins before deposition. Printing larger numbers of microarrays resulted in lower amounts of protein deposited onto slides positioned later in the print sequence, which required the pins to be re-dipped into the protein mixtures. Therefore, we would normally print only five microarrays using a single dip of the split pins.
In this work, we screened a limited number of ECM proteins for their ability to elicit differential attachment by cell types relevant in liver culture systems. Immunohistochemical staining of cryosectioned liver tissue shows heterogeneous staining for collagen I, collagen IV, laminin, and Fn. Therefore, the ECM proteins that we chose to evaluate on our arrays mostly represent major matrix proteins present in the liver. Thus, it is not surprising that the liver cell types (HepG2 cells, LX-2 HSCs, PFs) attached to most of the ECM combinations deposited on our arrays. The observation that HepG2 cells did not attach to regions deposited only with Fn was unexpected because Fn-coated microarray slides and tissue culture plates have been used to support HepG2 cultures.20
To explore this observation further, we deposited the nearest neighbors to Fn (red box, Fig. 1A) in the pattern of the ECM array and seeded co-cultures of HepG2 cells with BAECs or LX-2 HSCs. The pattern of the array preserved the Fn neighbors but deposited this pattern in multiple positions to test whether the location of the Fn spot and its neighboring ECM material may have altered the attachment phenotype of HepG2 cells. Preliminary results showed that HepG2 cells do not attach to Fn as long as the ECM composition and proximity of the nearest neighbors is preserved. We also observed that the nearest neighbor pattern, similar to the ECM microarray pattern, supported co-cultures of HepG2 cells with BAECs or LX-2 HSCs. The attachment phenotype of HepG2 cells to Fn on our arrays appears to be preserved under multiple contexts that we tested. This interesting result should be further tested with primary hepatocytes. We suspect that a combination of the local microenvironment and the transformed HepG2 phenotype may contribute to our observed results because we are confident that Fn was deposited at these positions at a surface density sufficient for mediating attachment and spreading of hepatocytes (Fig. 2).9 Others have shown that the cell-surface receptor for Fn is saturated by soluble Fn and its peptide fragments, leading to inhibition of cell attachment to Fn as well as to collagen.23,24 However, we do not expect that this mechanism is leading to the phenotype that we observed for HepG2 cell attachment to Fn.
Sequentially seeding BAECs and HepG2-CMTMRs onto 8 × 8 arrays printed with alternating rows of Fn and C1 did not produce patterned co-cultures. Instead, HepG2-CMTMR appeared to attach and spread equally on Fn and C1 positions, and we were unable to identify microcultures that consisted solely of a single cell type. We also printed arrays with alternating Fn and C1 rows in a 4 × 4 grid. These arrays, where the center-to-center spacing between the Fn and rat collagen I islands was twice the distance of the 8 × 8 arrays, appeared to be more conducive to forming isolated microcultures of BAECs and HepG2-CMTMR. These observations may cumulatively suggest that the location of the Fn spot on the ECM array and the neighboring ECM material may alter the attachment phenotype of HepG2 cells to Fn. We are interested in continuing to explore the rules that govern HepG2 cell interaction with Fn and developing broader platforms that allow us to co-culture multiple cell types.
Patterning cell cultures and co-cultures onto biodegradable scaffolds provides opportunities to explore the influence that 3D environments exert on cell function. We fabricated biodegradable scaffolds by electrospinning PLA fibers into 3D matrices suitable for depositing ECM proteins and for cell culture. Contact printing on the PLA matrices using the robotic arrayer required that scaffolds be mounted onto glass slides. We cut the scaffolds to the same geometry as the HydroGel™ pad (Fig. 1C) and affixed them to standard microscope slides. The PLA scaffolds were printed with an 8 × 8 microarray composed of alternating rows of Fn and C1 and seeded sequentially with BAECs and HepG2-CMTMR. Because we were unable to visualize the cell cultures on the scaffolds using brightfield microscopy, it was not possible to compare the spatial distribution of fluorescence with the brightfield image of the cells. Fluorescence images of the cultured scaffolds indicate that HepG2-CMTMR established on the Fn and C1 spots, which is consistent with the results that we observed for identical arrays printed onto the HydroGel™ slides. The spatial distribution of cell fluorescence on the scaffolds shows that cells attached and established microcultures to regions of ECM deposition rather than forming random cultures across the entire 3D matrix. Cell spreading on the scaffolds appeared to be greater than on the 2D HydroGel™ surface, but the underlying adhesive substrates still appeared to confine the boundary of the microcultures. We expect that this platform will be useful for understanding the role that localized and insoluble cues have on cell function in three dimensions versus two dimensions. Protein arrays printed onto 3D substrates, such as our electrospun scaffolds, could also be a new direction for high-throughput cell-microenvironment studies.
Conclusions
In contrast to photolithographic techniques, robotic microarray printing enables patterning of biomolecules in a high-density format with minimal surface modifications such as exposure to photoresists and organic solvents. Establishing micropatterned co-cultures in this way also allowed us to simultaneously evaluate many spatial configurations of cellular islands and hence types and degrees of cell–cell interactions while using significantly fewer reagents than with typical culture techniques. Ease of patterning multiple biomolecules on the same surface is also an advantage of this approach.
In this work, we screened multiple cell types for attachment to ECM protein microarrays and identified a new approach for establishing co-cultures with HepG2 hepatocytes. We also developed methods for monitoring hepatocyte viability on 2D microarray substrates. Finally, we provided data showing that biodegradable electrospun meshes deposited with ECM proteins support patterned cell cultures.
Acknowledgments
We thank K. Nelson (YCGP) for printing the ECM microarrays, C. Saroka (J.L. Boyer Lab) for providing the HepG2 hepatocytes, and N. Sheung (J.A. Dranoff Lab) for providing the LX-2 HSC line and primary PFs. Funding was generously provided by a pilot grant from the Yale Liver Center (DK34989) and a grant from the Human Frontier Science Program (RGP5/2006, co-investigator: F. Breitling). KAW acknowledges the fellowship support from a YCGP postdoctoral training fellowship (T32-HG003198-02). MJW was funded partially by a Yale Science and Engineering Association grant and by the Yale Perspectives on Science Program.
References
- 1.Musat A.I. Sattler C.A. Sattler G.L. Pitot H.C. Reestablishment of cell polarity of rat hepatocytes in primary culture. Hepatology. 1993;18:198. [PubMed] [Google Scholar]
- 2.Moghe P.V. Coger R.N. Toner M. Yarmush M.L. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- 3.Berthiaume F. Moghe P.V. Toner M. Yarmush M.L. Effect of extracellular matrix topology on cell structure, function, and physiological responsiveness: hepatocytes cultured in a sandwich configuration. FASEB J. 1996;10:1471. doi: 10.1096/fasebj.10.13.8940293. [DOI] [PubMed] [Google Scholar]
- 4.Bhandari R.N.B. Riccalton L.A. Lewis A.L. Fry J.R. Hammond A.H. Tendler S.J.B. Shakesheff K.M. Liver tissue engineering: a role for co-culture systems in modifying hepatocyte function and viability. Tissue Eng. 2001;7:345. doi: 10.1089/10763270152044206. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia S.N. Balis U.J. Yarmush M.L. Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: role of homotypic cell interactions. Biotechnol Progr. 1998;14:378. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia S.N. Balis U.J. Yarmush M.L. Toner M. Probing heterotypic cell interactions: hepatocyte function in microfabricated co-cultures. J Biomater Sci Polym Ed. 1998;9:1137. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia S.N. Balis U.J. Yarmush M.L. Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia S.N. Yarmush M.L. Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J Biomed Mater Res. 1997;34:189. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Revzin A. Rajagopalan P. Tilles A.W. Berthiaume F.O. Yarmush M.L. Toner M. Designing a hepatocellular microenvironment with protein microarraying and poly(ethylene glycol) photolithography. Langmuir. 2004;20:2999. doi: 10.1021/la035827w. [DOI] [PubMed] [Google Scholar]
- 10.Guguen-Guillouzo C. Clement B. Baffet G. Beaumont C. Morelchany E. Glaise D. Guillouzo A. Maintenance and reversibility of active albumin secretion by adult-rat hepatocytes co-cultured with another liver epithelial-cell type. Exp Cell Res. 1983;143:47. doi: 10.1016/0014-4827(83)90107-6. [DOI] [PubMed] [Google Scholar]
- 11.Hui E.E. Bhatia S.N. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104:5722. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui E.E. Bhatia S.N. Microscale control of cell contact and spacing via three-component surface patterning. Langmuir. 2007;23:4103. doi: 10.1021/la0630559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J.L. Liu W. Nelson C.M. Raghavan S. Chen C.S. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 14.Lahann J. Balcells M. Rodon T. Lee J. Choi I.S. Jensen K.F. Langer R. Reactive polymer coatings: a platform for patterning proteins and mammalian cells onto a broad range of materials. Langmuir. 2002;18:3632. [Google Scholar]
- 15.Barron J.A. Wu P. Ladouceur H.D. Ringeisen B.R. Biological laser printing: a novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed Microdevices. 2004;6:139. doi: 10.1023/b:bmmd.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 16.Roth E.A. Xu T. Das M. Gregory C. Hickman J.J. Boland T. Inkjet printing for high-throughput cell patterning. Biomaterials. 2004;25:3707. doi: 10.1016/j.biomaterials.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 17.Sanjana N.E. Fuller S.B. A fast flexible ink-jet printing method for patterning dissociated neurons in culture. J Neurosci Methods. 2004;136:151. doi: 10.1016/j.jneumeth.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Douglas E.S. Chandra R.A. Bertozzi C.R. Mathies R.A. Francis M.B. Self-assembled cellular microarrays patterned using DNA barcodes. Lab Chip. 2007;7:1442. doi: 10.1039/b708666k. [DOI] [PubMed] [Google Scholar]
- 19.Chandra R.A. Douglas E.S. Mathies R.A. Bertozzi C.R. Francis M.B. Programmable cell adhesion encoded by DNA hybridization. Angew Chem Int Edit. 2006;45:896. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 20.Saito S. Sakagami K. Miyazaki M. Matsuoka J. Shiozaki S. Orita K. Basic study for hybrid artificial liver by combination of hepatocytes and biomatrix. Artif Organs. 1987;11:312. [Google Scholar]
- 21.Jhandier M.N. Kruglov E.A. Lavoie E.G. Sevigny J. Dranoff J.A. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem. 2005;280:22986. doi: 10.1074/jbc.M412371200. [DOI] [PubMed] [Google Scholar]
- 22.Flaim C.J. Chien S. Bhatia S.N. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 23.Yamada K.M. Kennedy D.W. Dualistic nature of adhesive protein function - fibronectin and its biologically-active peptide fragments can autoinhibit fibronectin function. J Cell Biol. 1984;99:29. doi: 10.1083/jcb.99.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engvall E. Ruoslahti E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int J Cancer. 1977;20:1. doi: 10.1002/ijc.2910200102. [DOI] [PubMed] [Google Scholar]