Abstract
The clinical need for both three-dimensional (3D) soft tissue replacements and in vitro adipose tissue models continues to grow. In this study, we evaluated structural and functional characteristics of an in vitro 3D coculture model of vascularized adipose tissue. Tomato red–infected human adipose tissue–derived mesenchymal stem cells (hASCs) and green fluorescence protein–infected human umbilical vein endothelial cells were cocultured on 3D aqueous-derived silk scaffolds for 2 weeks. Confocal microscopy images demonstrated viability of cocultures and organization of both cell types over time. Endothelial cells aligned with time, and further histological analyses revealed continuous endothelial lumen formation in both differentiated and undifferentiated cocultures. Differentiated adipose cocultures secreted significantly higher levels of leptin than undifferentiated cocultures at 1 and 2 weeks. Additionally, lipid accumulation was demonstrated with Oil Red O staining, where positive staining was higher in the differentiated cocultures. A promising in vitro approach for the vascularization of tissue-engineered adipose tissue, and the ability to vascularize a construct containing hASCs was demonstrated. The strategy outlined provides a basis for the formation of other in vitro vascularized tissues as well as a path forward for the sustainable formation of soft tissue due to the use of slowly degrading silk scaffolds.
Introduction
An in vitro model of adipose tissue that mimics aspects of the native tissue would offer major benefits to the biomedical research community. The increased incidence of adipose-related diseases such as obesity and insulin resistance associated with type 2 diabetes prompts rising interest in the development of models to allow the more systematic study of disease mechanisms. These models can serve to elucidate mechanisms of disease origin and progression, as well as for therapeutic screening and diagnostic tools. According to the National Center for Health Statistics, 30% of adults in the United States are considered obese.1 Obesity has severe consequences, such as increased risk of coronary heart disease, type 2 diabetes, and hypertension.2
Adipose tissue is highly vascularized.3 White adipose tissue, the predominant type of fat in humans, is composed of preadipocytes, differentiated adipocytes, interstitial cells, and a microvascular system that is organized within an extracellular matrix that consists of collagen types I, III, IV, V, and VI as well as other extracellular matrix proteins.4,5 Recent studies have shown that a close correlation exists between adipogenesis and angiogenesis in which specific adipokines, or cytokines secreted by adipocytes, play a role in vascular homeostasis.6 These adipokines include leptin, adiponectin, tumor necrosis factor-α, plasminogen activator inhibitor type 1, and resistin.6
One approach for an adipose tissue model is to tissue engineer adipose tissue in vitro to mimic native tissue. However, a major obstacle to this approach is the lack of a vascular supply.7 Therefore, a current need in tissue engineering rests with the formation of a vascular supply in tissue-engineered constructs.8 Incorporating vasculature in vitro would contribute to viability during tissue growth and induce structural organization in vitro as well as in vivo, as well as provide more relevance to in vitro models of tissues.9
Current in vitro adipose tissue models involve monocultures of differentiated bone marrow–derived mesenchymal stem cells, differentiated adipose tissue–derived stem cells, as well as embryonic stem cells cultured on synthetic or natural polymeric matrices or scaffolds.4,10–13 Coculture models for adipose tissue engineering have been explored to a limited extent in prior reports.14–17 Recently, Frerich et al. developed an in vitro coculture model using human adipose stromal cells and human umbilical vein endothelial cells (HUVECs), where perfused tubes demonstrated capillary-like networks that were observed sprouting from the central lumen wall.14 Borges et al. cocultured preadipocytes from human adipose tissue with endothelial cells in a fibrin glue matrix on the chorioallantoic membrane, and after 7 days culture on the CAM, positive CD31 stain was evident in lumen-like structures in the fibrin matrix.16 While these studies, among others, show promise toward vascularized engineered tissue, we are interested in establishing a functional model such that adipose-derived stromal cells are differentiated toward adipose tissue specifically, cocultured with endothelial cells on a material that has shown promise for the development of adipose tissue, and exhibit adipose tissue functionality. Further, our long-term goal is a sustainable coculture system, such that the three-dimensional (3D) vascularized tissue can be utilized as sustainable disease models in vitro, or for long-term soft tissue repairs in vivo. Thus, biomaterial scaffolds that degrade slowly over months to years are required.18
Therefore, the goals of this study were to develop a vascularized human adipose tissue system through a coculture approach. Undifferentiated and differentiated human adipose–derived mesenchymal stem cells (hASCs) cocultured with human umbilical cord endothelial cells on porous silk protein scaffolds were utilized to address the major needs of the system. The 3D in vitro model for human adipose tissue described here represents a first step toward the generation of long-term sustainable tissue systems.
Materials and Methods
Materials
Cocoons from Bombyx mori silkworm were supplied by Tajimia Shoji Co. (Yokohama, Japan). hASCs were isolated according to previously published methods from subcutaneous adipose tissue donated with written consent by healthy volunteers undergoing elective liposurgery.19 The work was reviewed and approved by the Pennington Biomedical Research Center Institutional Review Board. HUVECs and endothelial growth medium (EGM) were purchased from Cambrex (East Rutherford, NJ). Dulbecco's modified Eagle's medium–Nutrient mix F-12 (1:1) containing L-glutamine (DMEM/F-12), fetal bovine serum (FBS), trypsin-EDTA, phosphate buffered saline, human recombinant insulin, and antibiotic–antimycotic (10,000 U/mL penicillin, 10,000 μg/mL streptomycin, and fungizone) were purchased from Gibco–Invitrogen (Carlsbad, CA); Biotin, 2,3-thiazolidinedione (TZD), dexamethasone, 3-isobutyl-1-methylxanthine (IBMX), and pantothenate were purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture
Isolated hASCs, as described by McIntosh et al.,20 were cultured in DMEM/F-12 supplemented with 10% FBS and 1% antibiotic–antimycotic. Media were replenished every 3 days, and cells were passaged at 80% confluency using trypsin–EDTA and frozen using cell culture growth medium (GM) containing 10% DMSO. HUVECs were cultured according to company protocols. Briefly, HUVECs were expanded in EGM supplemented with EGM-bullet kit. Media were replenished every 2 days and followed the same passaging/freezing protocols as for the hASCs.
Lentivirus transduction
hASCs and HUVECs were labeled with tomato red and green fluorescence protein (GFP), respectively, using a lentivirus delivery system. Viral particles were produced via a human embryonic kidney cell line (293FT), modified to express the large T antigen. LentiLox 3.7 plasmid, along with three packaging plasmids (pRRE, VSV, and pREV), were delivered to the 293FT cells using Lipofectamine 2000. Virus was collected 72 h later, and quantified via p24 antigen ELISA. Virus was delivered to both hASCs and HUVECs, mediated by protamine sulfate (6 μg/mL). After transduction, cells underwent fluorescent-activated cell sorting (FACS) to produce homogenously labeled cell populations.
Preparation of aqueous silk solution
Aqueous silk solution was prepared as previously described.21 Briefly, silk fibroin from silkworm (B. mori) cocoons was extracted with 0.2 M sodium carbonate solution, rinsed in distilled water, dissolved in a 9.3 M lithium bromide solution, and dialyzed for 48 h against distilled water using a Slide-a-Lyser dialysis cassette. Dissolved silk was then collected and centrifuged twice for 20 min at 5–10°C (9000 rpm). The resulting solution (7–8% w/v) was further diluted with distilled water to yield a 6% aqueous silk solution.
Preparation of aqueous silk scaffolds
Aqueous silk scaffolds were prepared by adding 4 g of sieved granular sodium chloride (particle size: 500–600 μm) to 2 mL 6% aqueous silk solution in a cylindrical container as previously described.21 Containers were covered and kept at room temperature for 72 h. Opened containers were then rinsed against distilled water for 2 days. Once NaCl particles were leached out, scaffolds were removed from the containers and an 8-mm biopsy punch was utilized to obtain 8 × 4 mm (d × h) scaffolds.
Research design
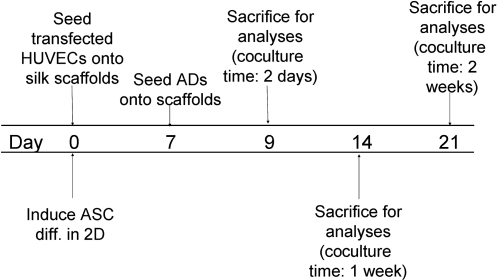
Cells were transfected using a lentivirus delivery system such that HUVECs were labeled with GFP, and hASCs were labeled with tomato red. On day 0, scaffolds were seeded with HUVECs according to the cell seeding protocol described below. Additionally, hASCs were induced in 2D culture flasks on day 0. Differentiated adipocytes were then seeded onto HUVEC-seeded scaffolds on day 7. Cocultures were cultivated for up to 14 days, in which scaffolds were assayed on days 2, 7, and 14 (Fig. 1). Control cocultures were maintained in parallel such that HUVECs were cocultivated with undifferentiated hASCs.
FIG. 1.
Research design for cocultivation of HUVECs and hASCs/differentiated adipocytes on silk scaffolds.
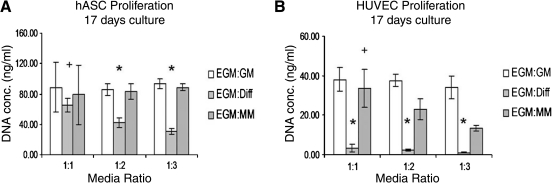
DNA assay
To determine optimal coculture media, DNA content was measured after 17 days of culture in various medium compositions. HUVECs and hASCs were individually cultured in various ratios of EGM, hASC GM, hASC differentiation medium (Diff), and adipocyte maintenance medium (MM). Three medium ratios of EGM:GM, EGM:Diff, and EGM:MM (1:1, 1:2, and 1:3) were investigated. After 17 days' culture, cells were rinsed three times with phosphate buffered saline, incubated with 0.25% trypsin–EDTA for 5 min at 37°C/5% CO2/95% RH, and centrifuged at 155 g for 10 min at 4°C. Cells were lysed in 200 μL 0.2% (v/v) Triton X-100 and 5 mM MgCl2 for 10 min to release DNA. After centrifugation for 10 min at 15,700 g and 4°C, supernatants were collected for assay. DNA content was determined fluorometrically at 480 nm/525 nm (ex/em) using a Fluoroskan Ascent FL spectrofluorometer (Thermo Life Sciences, Basingstoke, UK). The amount of DNA was determined by interpolation from a standard curve prepared using lambda DNA in 10 mM Tris-HCl (pH 7.4), 5 mM NaCl, and 0.1 mM EDTA over a range of concentrations.
Cell seeding
Aqueous silk scaffolds were dried, autoclaved, and presoaked in media overnight at 37°C (5% CO2/95% relative humidity). Media were then aspirated from scaffolds the following day, and 60 μL cell suspensions were pipetted onto the scaffolds. Cells were allowed to attach for 1.5–2 h at 37°C (5% CO2/95% RH). Seven mL of media was then added to each scaffold (one scaffold/well of six-well plate) and replenished every 2 days.
Adipocyte differentiation
hASCs seeded in 2D culture flasks were grown for 1 week in DMEM/F-12 media supplemented with 10% FBS and 1% penicillin–streptomycin (GM). Differentiation was induced for 7 days, in DMEM/F-12 media containing 3% FBS, 1% penicillin–streptomycin, 33 μM biotin, 17 μM pantothenate, 1 μM insulin, 1 μM dexamethasone, 500 μM IBMX, and 5 μM TZD (Diff). Adipocytes were maintained in differentiation medium minus IBMX and TZD for the remainder of culture (MM).
Coculture on silk scaffolds
HUVECs were seeded on silk scaffolds (800,000 cells/scaffold) on day 0. Seven-day differentiated adipocytes (375,000 cells/scaffold) were added to HUVEC-seeded scaffolds 1 week later. Scaffolds were cultured in 1:1 EGM:MM for 2 weeks in which media were replenished every 2 days.
Confocal microscopy
Cell organization on scaffolds was characterized via confocal microscopy on days 2, 7, and 14. GFP-labeled HUVECs were detected via 483/520 (ex/em), and tomato red–labeled hASCs were detected at 483/600 (ex/em).
Immunohistochemistry
Scaffolds were fixed in 10% formalin at 4°C for up to 3 weeks before automated paraffin processing and embedding. In brief, fixed samples are dehydrated through a graded ethanol series (80%, 95%, and 100%) and cleared with three xylene washes. Sections were then automatically infiltrated with paraffin for 1.5 h in vacuum. After processing, scaffolds were embedded in paraffin, allowed to cool overnight, and frozen. Frozen paraffin blocks were then sectioned (5 μm sections) and stained for anti-CD31 (Invitrogen, Carlsbad, CA). On the first day, sections underwent 3 xylene washes, dehydration through a reversed graded ethanol series (100%, 95%, and 70%), and microwave pretreatment in 0.01 M citric acid buffer (pH 6). This was followed by quenching of endogenous peroxidase activity in 3% hydrogen peroxide in methanol for 15 min, blocking of unspecified sites with normal goat serum (1:20 in 1.5% milk), and finally incubating in a 1:50 dilution of primary anti-human CD31 antibody in BC-11 buffer overnight. The next day, sections were incubated with biotin-labeled goat anti-mouse secondary antibody for 1 h. After incubation, the reaction was developed by incubation in 1:150 extravadin–peroxidase complex in 1% BSA for 30 min followed by incubation in a DAB/urea solution for 10 min. Sections were then counterstained with hematoxylin and mounted according to standard protocols.
Lumen analysis
Lumens, as specified by positive CD31 sections, were quantified. Lumens were quantified on four cross-sectional areas of each scaffold, and the average number of lumens per cross-section was determined. Continuous lumen morphology through sections was also qualitatively assessed via Zeiss Axiovert S100 light microscope (Carl Zeiss, Jena, Germany).
Oil Red O
Accumulated lipid was stained using Oil Red O. Scaffolds were fixed in 2 M sucrose overnight. The next day, scaffolds were embedded in OCT medium and frozen over dry ice. Frozen scaffolds were stored at −80°C, sectioned (10 μm sections), and subsequently stained with Oil Red O. Percentage of lipid accumulating cells of total cells per area observed was determined by counting number of Oil Red O–positive cells and total cells in four areas per scaffold cross section (three sections/sample, n = 3).
Leptin quantification
Secreted leptin was quantified using a quantitative sandwich enzyme immunoassay technique (ELISA) for human leptin. Medium samples were collected from cocultures on days 2, 7, and 14, centrifuged at 15,700 g for 10 min, and stored at −80°C until the Quantikine human leptin immunoassay was performed (R&D Systems, Minneapolis, MN).
Statistical analysis
All reported values were averaged (n = 3) and expressed as mean ± standard deviation (SD). Statistical differences were determined by Student's two-tailed t-test, and differences were considered statistically significant at p < 0.05.
Results
Preliminary screening to establish coculture conditions
An important aspect of developing the coculture model was to determine optimal culture conditions. Coculture media along with appropriate cell seeding protocols were addressed initially. hASCs and HUVECs were cultured separately in various medium compositions and ratios for 17 days (Fig. 2). Final DNA quantification results demonstrated significant proliferation for both hASCs and HUVECs in EGM:GM and EGM:MM media when compared with growth in EGM:Diff media. With increasing ratio of either EGM:GM or EGM:MM, hASC proliferation was not affected, whereas HUVEC proliferation showed a significant decrease. As both hASCs and HUVECs had significantly decreased proliferation in EGM:Diff media, subsequent experiments were conducted such that hASC differentiation was induced in monocultures, rather than in the presence of HUVECs. Therefore, control cocultures of uninduced hASCs and HUVECs were cultured in 1:1 EGM:GM media, and induced cocultures were conducted such that hASC differentiation was separately induced in tissue culture flasks (in differentiation medium for 7 days), and then added to seeded HUVECs, where together they were cultured in 1:1 EGM:MM media.
FIG. 2.
DNA content results for optimization of coculture media. hASCs and HUVECs were grown in three types of media: EGM:GM, EGM:Diff, and EGM:MM in 1:1, 1:2, and 1:3 medium ratios. EGM, endothelial growth medium; GM, growth medium; Diff, differentiation medium; MM, maintenance medium. hASCs exhibited significantly greater proliferation in 1:1 EGM:Diff than 1:2 or 1:3 (+p < 0.05) and in EGM:GM and EGM:MM at 1:2 and 1:3 ratios (*p < 0.05). Additionally, HUVECs exhibited significantly less proliferation in EGM:Diff media at all three ratios (*p < 0.05), and 1:1 EGM:MM showed significantly more proliferation than 1:3 EGM:MM (+p < 0.05).
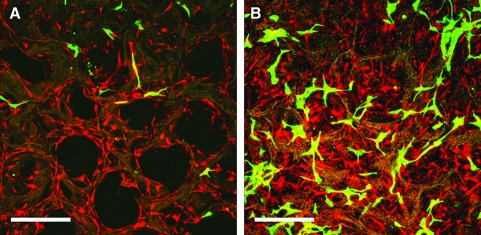
In addition to coculture media, preliminary work was performed to determine optimal seeding order. hASCs and HUVECs were cocultured and seeded together on the scaffolds on day 0, and confocal microscopy was utilized to view cell distribution on days 4 and 8 after seeding. Preliminary data showed delayed growth of HUVECs in the presence of hASCs, whereas hASCs continued to proliferate at both days 4 and 8 (Fig. 3). Subsequent experiments were conducted such that HUVECs were seeded and cultured for 7 days before the addition of either hASCs (uninduced coculture) or differentiated adipocytes (induced coculture) so that the endothelial cells could first form a niche within the scaffold environment before the addition of the more proliferative hASCs and adipocytes.
FIG. 3.
Confocal microscopy images for hASC and HUVEC cocultures on day 4 (A) and day 8 (B). hASCs and HUVECs were coseeded on day 0. HUVECs, GFP labeled; hASCs, tomato red labeled. Scale bar = 375 μm. Color images available online at www.liebertonline.com/ten.
Coculture organization
Based on the preliminary experiments, HUVECs were seeded onto the scaffolds on day 0 and cultured in monoculture for 7 days. Undifferentiated (uninduced control) or adipogenic (induced) hASCs were then seeded directly onto HUVEC-seeded scaffolds. Confocal microscopy was utilized for both uninduced and induced cocultures; however, due to the similarity of results, only representative images for induced cocultures are reported here. Successful seeding onto scaffolds was confirmed with confocal microscopy on day 2 coculture (Fig. 4A, B). Both cell types were detected on the surface of both uninduced and induced cocultures. By day 7, GFP-HUVECs began to show increased organization such that 3D stacked images showed aligned endothelial cells in a bed of adipocytes (Fig. 4C, D). Finally, by day 14, more extensive alignment of HUVECs amidst adipocytes was observed (Fig. 4E–H).
FIG. 4.
Confocal images of induced coculture on days 2 (A, B), 7 (C, D), and 14 (E–H). HUVECs, GFP labeled; hASCs, tomato red labeled. Scale bars as marked. Color images available online at www.liebertonline.com/ten.
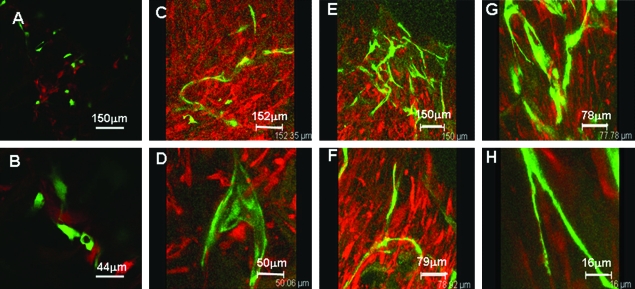
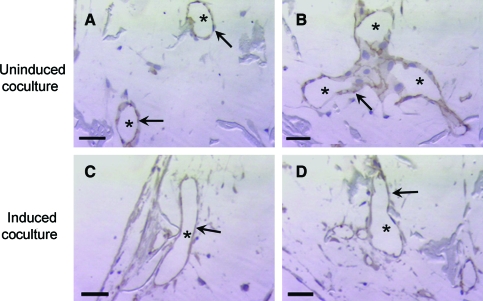
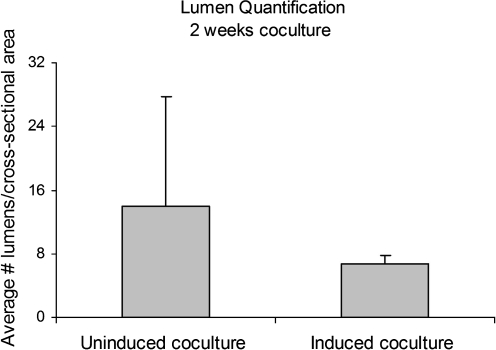
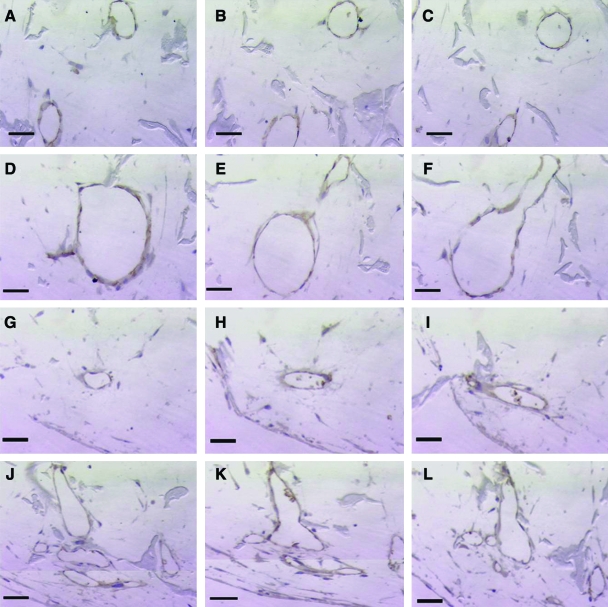
For further structural analysis, histological characterization of cocultures was conducted with immunohistochemistry for CD31 (or platelet–endothelial cell adhesion molecule, PECAM), an endothelial cell–specific membrane marker. Lumen formation was observed in both uninduced and induced cocultures, as indicated by the aligned CD31-positive endothelial cells that form lumen structures within the pores of the scaffolds (Fig. 5). The average number of lumens per cross-sectional area was determined at three depths in both uninduced and induced scaffolds (Fig. 6). Continuity of lumen structures through the scaffold constructs was also investigated. Both uninduced and induced cocultures demonstrated lumen extension through several consecutive sections (Fig. 7A–C, G–I). In addition to lumen extension, branching and converging lumen structures were observed through the consecutive sections (Fig. 7D–F, J–L).
FIG. 5.
CD31 ICC with hematoxylin counterstain for uninduced (A, B) and induced (C, D) cocultures after 14 days. Lumen organization evident in both uninduced and induced cocultures as marked by both aligned endothelial cells (arrows) and open lumens (*). Scale bar = 50 μm. Original magnification, × 320. Color images available online at www.liebertonline.com/ten.
FIG. 6.
Lumen quantification of uninduced and induced cocultures after 2 weeks of culture. Uninduced cocultures did not exhibit statistically greater number of lumens per scaffold cross-sectional area than induced cocultures.
FIG. 7.
Continuity of lumen structures through consecutive sections of scaffolds. Both uninduced (A–C) and induced (G–I) cocultures show lumen continuity through consecutive sections. Additionally, converging of lumen, as shown in this representative uninduced coculture sample (D–F), and branching of lumen, as shown by this representative induced coculture sample (J–L), have both been observed. Scale bars: (A–C, G–I) 100 μm (original magnification, ×100); (D–F, J–L) 50 μm (original magnification, × 320). Color images available online at www.liebertonline.com/ten.
Functional attributes of adipose tissue from cocultures
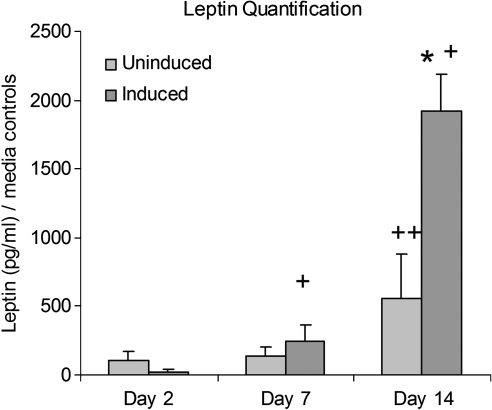
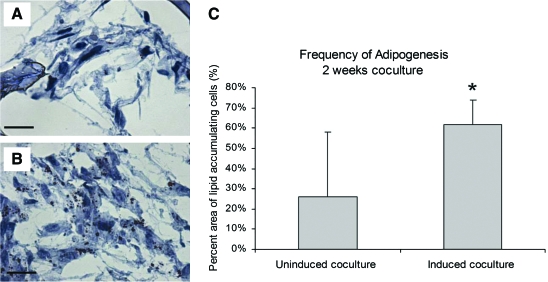
Cocultures were evaluated for adipose tissue function by measuring secreted leptin levels at days 2, 7, and 14 (Fig. 8). While there were no significant differences between uninduced and induced cocultures in leptin levels at day 2, induced cocultures showed significantly greater leptin levels by days 7 and 14. Another important attribute of adipose tissue is lipid accumulation. The ability of differentiated adipocytes to accumulate lipid in the presence of endothelial cells is crucial as this is representative of the in vivo environment. Scaffolds were stained with Oil Red O to target lipid. Although Oil Red O–positive cells were evident in both uninduced and induced cocultures, a significantly greater amount was evident in induced cocultures (Fig. 9).
FIG. 8.
Secreted leptin levels in both uninduced and induced cocultures. Induced cocultures secreted significantly more leptin than uninduced at day 14 (*p < 0.05) and increased secretion at days 7 and 14 (+p < 0.05), indicative of the presence of more mature adipocytes. Uninduced cocultures increased leptin section at day 14 as well (++p < 0.05).
FIG. 9.
Oil Red O–stained frozen sections after 2 weeks of coculture. Uninduced cocultures (A) exhibited less frequent Oil Red O stain, whereas all induced cocultures (B) were positive for lipid. (C) Frequency of adipogenesis was determined, and induced cocultures contained higher percentages of lipid-accumulating cells than uninduced cocultures (*p < 0.0001). Scale bars = 50 μm. Original magnification, × 40. Color images available online at www.liebertonline.com/ten.
Discussion
The present study demonstrates the coculture of both undifferentiated and adipocyte-differentiated hASCs with endothelial cells in 3D silk scaffolds. Other recent adipose tissue models have included the cultivation of murine embryonic stem cells, as well as both hASCs and bone marrow–derived hMSCs, on a variety of biopolymers and synthetic materials. Reported coculture models, as mentioned previously, have shown degrees of success with respect to cocultivation of adipose tissue stromal cells with endothelial cells and endothelial lumen formation.14–16 However, the formation of endothelial lumens within a tissue-engineered adipose tissue construct has not been observed entirely in vitro. In addition to cocultivation in vitro, our study focused on functionally characterizing our engineered adipose tissue along with confirmation of lumen formation. Finally, a distinguishing aspect of our model is that we conducted our cocultures on 3D silk fibroin scaffolds. Previous studies in our laboratory have shown the potential of these protein scaffolds to support the growth of adipose-like tissue both in vivo and in vitro.22 Briefly, both hASCs and bone marrow–derived hMSCs were induced toward adipogenesis on 3D silk scaffolds as well as collagen and PLA scaffolds for comparison.22 After 21 days in vitro, all scaffolds with adipogenic (induced) cells exhibited significant lipid accumulation and upregulation of adipogenesis-specific mRNA transcript levels compared with uninduced controls.22 In addition, silk scaffolds that were implanted for 4 weeks in a rat muscle pouch defect model remained viable, while both collagen and PLA scaffolds were undetectable due to their rapid degradation rate.22 Other common biomaterials for adipose tissue engineering include injectable gels, gelatin, and fibrin matrices, and 3D PGA/PLA-based matrices.13–17,22–27 However, while these materials supported the cultivation of adipose-like tissue, they lack the mechanical integrity and therefore slow degradability that silk possesses, leading to limitations with long-term maintenance of such systems in vitro or in vivo. Our laboratory also recently demonstrated the potential of 3D porous silk scaffolds as a biocompatible biomaterial that can maintain mechanical integrity while degrading slowly in vivo.28 The promising nature of silk fibroin as a biomaterial for 3D matrices in tissue engineering applications prompted us to further investigate the potential of this material for developing a 3D in vitro coculture adipose tissue model that more closely mimics in vivo adipose tissue.
To further study and develop a relevant in vitro model, our goal was to incorporate relevant vascular cells into the system as a first step toward vascularized systems. Thus, we successfully incorporated human adipocyte cells (differentiated hASCs) along with a human endothelial cell source (HUVECs). One of the goals was to develop coculture conditions to promote the formation of a vascular network within a bed of adipose tissue. Preliminary experiments were conducted to determine a coculture medium that promoted cell viability and proliferation of both cell types in coculture, as well as gain insight regarding the appropriate cocultivation seeding protocol. As a result of testing various medium compositions and ratios, we found that a 1:1 ratio of EGM to both hASC GM and differentiated adipocyte MM promoted viability and proliferation after 17 days in 2D culture. Based on this, we then cocultivated both hASCs and HUVECs in a 1:1 medium to further investigate how the cells behaved in the 3D scaffold environment. Delayed growth of HUVECs in coculture was observed, leading to the strategy to initially seed HUVECs in the matrices 1 week before adding the hASCs/adipocytes.
Following these protocols, we cocultivated endothelial cells with either undifferentiated hASCs (uninduced) or differentiated adipocytes (induced) in the 3D silk fibroin scaffolds for 2 weeks. Confocal microscopy and histology were used to characterize the structural organization of cocultures; leptin and accumulated lipid, via ELISA and Oil Red O staining, respectively, were used to evaluate functional attributes. Both confocal microscopy and histological analyses revealed endothelial alignment and organization in both uninduced and induced cocultures. Three-dimensional confocal image stacks showed that by days 7 and 14, endothelial cells had aligned, and immunohistochemical staining for an endothelial-specific marker, CD31, revealed that the endothelial cells had oriented to form open lumens within the pores of the silk matrix. This finding supports the potential of this in vitro system not only for the vascularization of adipose tissue, but also for other tissue-engineered constructs. Monocultures of each cell type were also cultivated on silk fibroin scaffolds. When endothelial cells were seeded alone on silk scaffolds, CD31-positive lumens were not detected after 2 weeks (data not shown), suggesting that the coculture of endothelial cells with hASCs or adipocytes played a role in endothelial lumen formation. Further studies, however, need to be conducted to further investigate the cell–cell communication and mechanisms involved in the observed lumen formation.
In addition to the structural components of cocultures, the constructs were also evaluated for specific adipose tissue function. Several studies have shown an inhibitory effect of HUVECs on osteogenic differentiation of mesenchymal stem cells.29,30 Contrary to these findings, we demonstrate the ability to coculture endothelial cells with differentiated adipocytes, while retaining adipose functionality through both leptin secretion and lipid accumulation. A sign of maturing adipose tissue is the secretion of leptin, an important adipokine known to play a vital role in adipose–hypothalamus cross-talk, as well as in energy homeostasis of adipose tissue.31As mentioned earlier, the secretion of adipokines has been shown to contribute to vascular formation and homeostasis in adipose tissue.6 Secreted leptin was monitored in both uninduced and induced cocultures at days 2, 7, and 14, and induced cocultures showed significantly higher leptin levels than uninduced cultures, as well as increased leptin levels over time. These results suggest that the adipogenic (induced) cocultures were continuing to mature over time in conjunction with endothelial cell alignment and lumen formation. Induced cocultures secreted more leptin than both hASC and adipocyte monocultures at days 2 and 7. However, by day 14, induced cocultures secreted leptin at similar levels like adipocyte monocultures (data not shown). Physiological leptin levels are dependent on body mass index, such that plasma concentrations can range from 1.9 ng/mL in lean individuals to 9.3 ng/mL in obese individuals.32 Our induced cocultures and adipocyte monocultures fell within this physiological range, such that by day 14, a 2 ng/mL leptin concentration was observed. This demonstrates that our culture system is physiologically and functionally relevant with respect to secreted leptin.
Another indicator of mature adipose tissue formation is lipid accumulation. Cocultures were evaluated for lipid formation through Oil Red O staining at day 14 culture. Although positive Oil Red O staining was evident in both uninduced and induced cocultures, a significantly greater percentage of Oil Red O was found in induced cultures. It is not uncommon to observe some lipid accumulation in uninduced cultures due to the presence of serum factors in GM, and therefore it is important to note the significantly larger percentage of positive Oil Red O stain in the induced cultures. Similar to leptin secretion, detected Oil Red O levels in uninduced and induced cocultures were similar to levels observed in both hASC and adipocyte monocultures (data not shown), respectively.
While our results show evidence for a relationship between endothelial lumen formation, leptin secretion, lipid accumulation, and monoculture versus coculture conditions, further studies are needed to characterize specific signaling mechanisms responsible for our findings in the coculture system. Specifically, it is of interest and importance to determine how the presence of endothelial cells further enhances adipose tissue functionality, and likewise whether the presence of either hASCs or adipocytes significantly improves formation and stability of endothelial lumen formation.
Conclusions
Both structural and functional characteristics of a coculture system related to adipogenesis were evaluated. A promising in vitro approach for the vascularization of tissue-engineered adipose tissue, as well as the ability to vascularize a construct containing hASCs, was demonstrated. This strategy offers new options for adipose tissue engineering, as 3D sustainable models can be considered for the study of disease formation and disease progression and for therapeutic screening. Further, the system described can also be considered for soft tissue regeneration needs. Finally, the strategy outlined provides a basis for the formation of other in vitro vascularized tissues. Further studies are required to optimize coculture distribution, to understand coculture signaling, and to characterize the long-term culture potential of this system both in vitro and in vivo.
Acknowledgments
The authors would like to thank Laurence Daheron for her assistance with lentivirus labeling, Silva Krause for her assistance in histology staining, and William Rice for his assistance in confocal imaging. We would like to acknowledge support from the Tissue Engineering Resource Center (TERC)–NIH P41 EB002520 from the National Institute of Biomedical Imaging and Biomedical Engineering.
Footnotes
This work was presented as an abstract at the Biomedical Engineering Society Annual Meeting on October 2, 2008, in St. Louis, Missouri.
Disclosure Statement
No competing financial interests exist.
References
- 1.Atlanta, GA: Department of Health and Human Services, Centers for Disease Control and Prevention; 2008. Overweight and Obesity (in Introduction) [Google Scholar]
- 2.Neels J.G. Thinnes T. Loskutoff D.J. Angiogenesis in an in vivo model of adipose tissue development. FASEB J. 2004;18:983. doi: 10.1096/fj.03-1101fje. [DOI] [PubMed] [Google Scholar]
- 3.Hausman G.J. Richardson R.L. Adipose tissue angiogenesis. J Anim Sci. 2004;82:925. doi: 10.2527/2004.823925x. [DOI] [PubMed] [Google Scholar]
- 4.Saiki A. Watanabe F. Murano T. Miyashita Y. Shirai K. Hepatocyte growth factor secreted by cultured adipocytes promotes tube formation of vascular endothelial cells in vitro. Int J Obes. 2006;30:1676. doi: 10.1038/sj.ijo.0803316. [DOI] [PubMed] [Google Scholar]
- 5.Flynn L. Semple J. Woodhouse K. Decellularized placental matrices for adipose tissue engineering. J Biomed Mater Res A. 2006;79:359. doi: 10.1002/jbm.a.30762. [DOI] [PubMed] [Google Scholar]
- 6.Mertsching H. Walles T. Hofmann M. Schanz J. Knapp W. Engineering of a vascularized scaffold for artificial tissue and organ generation. Biomaterials. 2005;26:6610. doi: 10.1016/j.biomaterials.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 7.Sieminski A.L. Hebbel R.P. Gooch K.J. Improved microvascular network in vitro by human blood outgrowth endothelial cells relative to vessel-derived endothelial cells. Tissue Eng. 2005;11:1332. doi: 10.1089/ten.2005.11.1332. [DOI] [PubMed] [Google Scholar]
- 8.Levenberg S. Rouwkema J. Macdonald M. Garfein E.S. Kohane D.S. Darland D.C. Marini R. van Blitterswijk C.A. Mulligan R.C. D'Amore P.A. Langer R. Engineering vascularized skeletal muscle tissue. Nat Biotechnol. 2005;23:879. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 9.Sodian R. Lemke T. Fritsche C. Hoerstrup S.P. Fu P. Potapov E.V. Hausmann H. Hetzer R. Tissue-engineering bioreactors: a new combined cell-seeding and perfusion system for vascular tissue engineering. Tissue Eng. 2002;8:863. doi: 10.1089/10763270260424222. [DOI] [PubMed] [Google Scholar]
- 10.Neumann T. Nicholson B.S. Sanders J.E. Tissue engineering of perfused microvessels. Microvasc Res. 2003;66:59. doi: 10.1016/s0026-2862(03)00040-2. [DOI] [PubMed] [Google Scholar]
- 11.Walton R.L. Beahm E.K. Wu L. De novo adipose formation in a vascularized engineered construct. Microsurgery. 2004;24:378. doi: 10.1002/micr.20056. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama T. Ohashi K. Kuge H. Kanehiro H. Iwata H. Yamato M. Nakajima Y. In vivo engineering of metabolically active hepatic tissues in a neovascularized subcutaneous cavity. Am J Transplant. 2006;6:50. doi: 10.1111/j.1600-6143.2005.01155.x. [DOI] [PubMed] [Google Scholar]
- 13.Aoki S. Toda S. Sakemi T. Sugihara H. Coculture of endothelial cells and mature adipocytes actively promotes immature preadipocyte development in vitro. Cell Struct Funct. 2003;28:55. doi: 10.1247/csf.28.55. [DOI] [PubMed] [Google Scholar]
- 14.Frerich B. Zuckmantel K. Winter K. Muller-Durwald S. Hemprich A. Maturation of capillary-like structures in a tube-like construct in perfusion and rotation culture. Int J Oral Maxillofac Surg. 2008;37:459. doi: 10.1016/j.ijom.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich F. Lelkes P.I. Fine-tuning of a three-dimensional microcarrier-based angiogenesis assay for the analysis of endothelial-mesenchymal cell co-cultures in fibrin and collagen gels. Angiogenesis. 2006;9:111. doi: 10.1007/s10456-006-9037-x. [DOI] [PubMed] [Google Scholar]
- 16.Borges J. Mueller M.C. Padron N.T. Tegtmeier F. Lang E.M. Stark G.B. Engineered adipose tissue supplied by functional microvessels. Tissue Eng. 2003;9:1263. doi: 10.1089/10763270360728170. [DOI] [PubMed] [Google Scholar]
- 17.Montesano R. Pepper M.S. Orci L. Paracrine induction of angiogenesis in vitro by Swiss 3T3 fibroblasts. J Cell Sci. 1993;105:1013. doi: 10.1242/jcs.105.4.1013. [DOI] [PubMed] [Google Scholar]
- 18.Wang X. Zhang X. Castellot J. Herman I. Iafrati M. Kaplan D.L. Controlled release from multilayer silk biomaterial coatings to modulate vascular cell responses. Biomaterials. 2008;29:894. doi: 10.1016/j.biomaterials.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois S.G. Floyd E.Z. Zvonic S. Kilroy G. Wu X. Carling S. Halvorsen Y.D. Ravussin E. Gimble J.M. Isolation of human adipose-derived stem cells from biopsies and liposuction specimens. Methods Mol Biol. 2008;449:69. doi: 10.1007/978-1-60327-169-1_5. [DOI] [PubMed] [Google Scholar]
- 20.McIntosh K. Zvonic S. Garrett S. Mitchell J. Floyd E. Hammill L. Kloster A. Di Halvorsen Y. Ting J. Storms R. Goh B. Kilroy G. Wu X. Gimble J. The immunogenicity of human adipose-derived cells: temporal changes in vitro. Stem Cells. 2006;24:1246. doi: 10.1634/stemcells.2005-0235. [DOI] [PubMed] [Google Scholar]
- 21.Kim U.-J. Park J. Kim H.J. Wada M. Kaplan D.L. Three-dimensional aqueous derived biomaterial scaffolds from silk fibroin. Biomaterials. 2005;26:2775. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 22.Mauney J.R. Nguyen T. Gillen K. Kirker-Head C. Gimble J.M. Kaplan D.L. Engineering adipose-like tissue in vitro and in vivo utilizing human bone marrow and adipose-derived mesenchymal stem cells with silk fibroin 3D scaffolds. Biomaterials. 2007;28:5280. doi: 10.1016/j.biomaterials.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marra K.G. Defail A.J. Clavijo-Alvarez J.A. Badylak S.F. Taieb A. Schipper B. Bennett J. Rubin J.P. FGF-2 enhances vascularization for adipose tissue engineering. Plast Reconstr Surg. 2008;121:1153. doi: 10.1097/01.prs.0000305517.93747.72. [DOI] [PubMed] [Google Scholar]
- 24.Rubin J.P. Bennett J.M. Doctor J.S. Tebbets B.M. Marra K.G. Collagenous microbeads as a scaffold for tissue engineering with adipose-derived stem cells. Plast Reconstr Surg. 2007;120:414. doi: 10.1097/01.prs.0000267699.99369.a8. [DOI] [PubMed] [Google Scholar]
- 25.Vashi A.V. Abberton K.M. Thomas G.P. Morrison W.A. O'Connor A.J. Cooper-White J.J. Thompson E.W. Adipose tissue engineering based on the controlled release of fibroblast growth factor-2 in a collagen matrix. Tissue Eng. 2006;12:3035. doi: 10.1089/ten.2006.12.3035. [DOI] [PubMed] [Google Scholar]
- 26.Weiser B. Prantl L. Schubert T.E. Zellner J. Fischbach-Teschl C. Spruss T. Seitz A.K. Tessmar J. Goepferich A. Blunk T. In vivo development and long-term survival of engineered adipose tissue depend on in vitro precultivation strategy. Tissue Eng Part A. 2008;14:275. doi: 10.1089/tea.2007.0130. [DOI] [PubMed] [Google Scholar]
- 27.Frye C.A. Patrick C.W. Three-dimensional adipose tissue model using low shear bioreactors. In Vitro Cell Dev Biol Anim. 2006;42:109. doi: 10.1290/0509055.1. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y. Rudym D. Walsh A. Abrahamsen L. Kim H. Kim H. Kirker-Head C. Kaplan D. In vivo degradation of three-dimensional silk fibroin scaffolds. Biomaterials. 2008;29:3415. doi: 10.1016/j.biomaterials.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y. Abe M. Hiasa M. Oda A. Amou H. Nakano A. Takeuchi K. Kitazoe K. Kido S. Inoue D. Moriyama K. Hashimoto T. Ozaki S. Matsumoto T. Myeloma cell-osteoclast interaction enhances angiogenesis together with bone resorption: a role for vascular endothelial cell growth factor and osteopontin. Clin Cancer Res. 2007;13:816. doi: 10.1158/1078-0432.CCR-06-2258. [DOI] [PubMed] [Google Scholar]
- 30.Meury T. Verrier S. Alini M. Human endothelial cells inhibit BMSC differentiation into mature osteoblasts in vitro by interfering with osterix expression. J Cell Biochem. 2006;98:992. doi: 10.1002/jcb.20818. [DOI] [PubMed] [Google Scholar]
- 31.Rupnick M.A. Panigrahy D. Zhang C.-Y. Dallabrida S.M. Lowell B.B. Langer R. Folkman J.M. Adipose tissue mass can be regulated through the vasculature. Proc Natl Acad Sci USA. 2002;99:10730. doi: 10.1073/pnas.162349799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal K. Landt M. Klein S. Relationship between insulin sensitivity and plasma leptin concentration in lean and obese men. Diabetes. 1996;45:988. doi: 10.2337/diab.45.7.988. [DOI] [PubMed] [Google Scholar]