Abstract
HIV-1 requires a −1 translational frameshift to properly synthesize the viral enzymes required for replication. The frameshift mechanism is dependent upon two RNA elements, a seven-nucleotide slippery sequence (UUUUUUA) and a downstream RNA structure. Frameshifting occurs with a frequency of ~5%, and increasing or decreasing this frequency may result in a decrease in viral replication. Here, we report the results of a high-throughput screen designed to find small molecules that bind to the HIV-1 frameshift site RNA. Out of 34,500 compounds screened, 202 were identified as positive hits. We show that one of these compounds, doxorubicin, binds the HIV-1 RNA with low micromolar affinity (Kd = 2.8 μM). This binding was confirmed and localized to the RNA using NMR. Further analysis revealed that this compound increased the RNA stability by approximately 5 °C and decreased translational frameshifting by 28% (±14%), as measured in vitro.
Twenty-five years after its initial identification, human immunodeficiency virus type 1 (HIV-1) continues to be a major health concern across much of the world. Although many anti-HIV-1 drugs have been developed, the virus is often able to evade these therapies owing to its high mutation rate. In addition to new classes of drugs being designed against the traditional targets (protease and reverse transcriptase) (1, 2), recent research has focused on targeting the −1 programmed translational frameshift that occurs between the gag and pol reading frames (3-8). This frameshift allows for translation of the pol genes (encoding the protease, reverse transcriptase, and integrase enzymes) in the form of a Gag-Pol fusion polyprotein via the evasion of a stop codon located at the end of the gag gene. Without this frameshift, only the Gag polyprotein (matrix, capsid, nucleocapsid, and p6 structural proteins) will be produced. The structural and enzymatic proteins are found in approximately a 20:1 molar ratio as a result of a frameshift efficiency of approximately 5% (9-15). This stoichiometry is required for appropriate packaging of virus particles, and an increase or decrease in the frameshift efficiency has been found to significantly decrease the production of infectious virions (4, 15, 16).
The frameshift event is programmed by two cis-acting RNA elements between the gag and pol genes: a seven-nucleotide slippery sequence (UUUUUUA) and a highly conserved stem-loop structure immediately downstream (9, 17). This stem-loop structure has been shown to induce ribosomal pausing, which is required for frameshifting, and its stability has been correlated with the efficiency of ribosomal frameshifting (18, 19). At the base of the stem-loop, the frameshift site RNA contains a conserved GGA bulge. This sequence has been hypothesized to interact with the translational machinery during frameshifting (20), by virtue of its predicted positioning near the mRNA entrance channel, which is located 13–15 nucleotides of the P site codon (21). Due to the distance the mRNA must traverse through the ribosome and the fact that the mRNA entrance channel is too narrow to fit a duplex, the lower helix must be denatured during frameshifting, and it is likely that the GGA sequence is positioned within the mRNA entrance channel during this time. Therefore, it is possible that small molecules that bind tightly to this sequence may interfere with the ribosome's ability to engage the stable upper stem-loop during frameshifting. Thus, the frameshift site RNA is an attractive target for drug development (4, 5, 7, 22).
Here, we report the results of a high-throughput screen for small molecules that bind the conserved GGA sequence at the base of the HIV-1 frameshift site stem-loop (Figure 1). Previously, our lab investigated the thermodynamics and determined the NMR structure of the stable HIV-1 frameshift site stem-loop RNA (23), as well as a longer RNA including the conserved GGA sequence (24) (Figure 1, panel a). The latter structure revealed a dynamic central guanosine in the GGA bulge (24), which is frequently mutated to an adenosine but never a pyrimidine (http://www.hiv.lanl.gov, Supplementary Figure 1). On the basis of these observations, the predicted location of the GGA sequence at the mRNA entrance channel during frameshifting, and prior studies showing that substitution of the three purines with pyrimidines reduces frameshift efficiency (13, 22), we reasoned that this would be an attractive site to target with small, drug-like molecules as an initial step toward development of compounds that affect the frameshifting efficiency of HIV-1. To do this, we substituted the dynamic central guanosine with a fluorescent nucleotide analogue, 2-aminopurine (Figure 1, panel b), to serve as a reporter of compounds that bind at this site. 2-Aminopurine will fluoresce when un-stacked, but its fluorescence is quenched upon stacking with other bases (25-30). This property of 2-aminopurine provides a readout of the local RNA bulge structure and was adapted for use in screening for small molecules that bind to this site on the RNA. Molecules that altered the 2-aminopurine fluorescence were then tested for binding specificity and effects on frameshifting in vitro.
Figure 1.
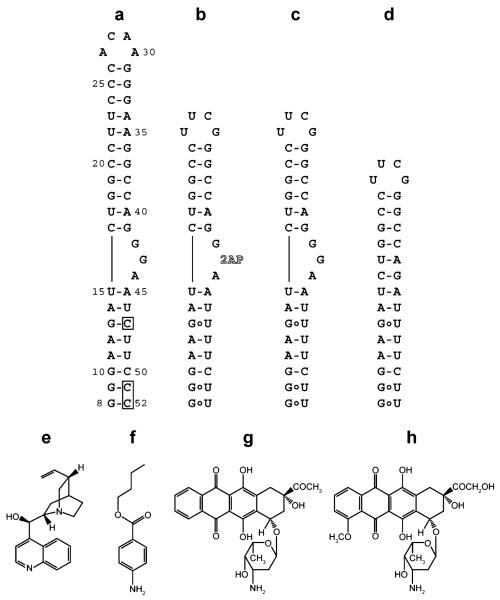
RNA constructs and selected compounds used in this study. a) HIV-1 frameshift site RNA stem-loop. Boxes indicate where the native uridines at positions 47, 51, and 52 have been changed to cytidines for NMR studies (HIV-18-52). The construct is numbered starting from the first residue of the UUUUUUA slippery sequence (not shown), which is immediately adjacent to the 5′ end of the stem-loop. b) Frameshift site RNA with a truncated upper stem, a native lower helix, and a 2-aminopurine substituted for the central guanosine in the three-purine bulge (HIV-1screen). c) Sequence as in panel b, except the central guanosine remains unaltered (HIV-1wtbulge). d) Sequence as in panel c, except the three-purine bulge has been deleted (HIV-1Δbulge). e) Cinchonidine. f) Butamben. g) Idarubicin. h) Doxorubicin.
RESULTS
Screening for Compounds Affecting the HIV-1 Bulge
The RNA sequence chosen for screening corresponds to the consensus sequence of 619 HIV-1 group M subtype B isolates (http://www.hiv.lanl.gov, Supplementary Figure 1). In order to minimize cost and prevent compounds from binding the distal ACAA tetraloop, the exceptionally stable stem-loop (23) was truncated and capped with a stable UUCG tetraloop (Figure 1, panel b). The RNA was substituted with a 2-aminopurine in place of the dynamic central guanosine in the conserved GGA sequence for use as a fluorescent readout of local structure (HIV-1screen, Figure 1, panel b). Additional RNA constructs used in this study included a control without the modified nucleotide (HIV-1wtbulge, Figure 1, panel c) and a GGA bulge deletion for testing the specificity of binding (HIV-1Δbulge, Figure 1, panel d). Based upon the fact that 2-aminopurine is a sensitive reporter of local structure (25-30), a high-throughput screen was designed using a fluorescence increase or decrease as an indicator of compounds that affect the local RNA structure upon binding.
Approximately 34,500 small molecule compounds were screened. Of these, 531 compounds showed an increase in fluorescence of between 170% and 230% of the intrinsic RNA fluorescence measured in the compound solvent control (2% DMSO) or a decrease in fluorescence to less than 35% of the control and were picked as initial hits. Repeated testing for compounds that have a high degree of autofluorescence, where compounds were tested for fluorescence under the same conditions with no RNA present, resulted in 202 compounds remaining. Twenty of these compounds (Supplementary Figure 2) were selected on the basis of their favorable spectroscopic properties, including lack of detectable autofluorescence at 360 nm and low intrinsic UV260 absorbance, to continue with follow-up studies. Compounds were titrated into 1 μM HIV-1screen RNA from 10−9 to 10−3 M final compound concentration in order to monitor dose response (Supplementary Figure 3). On the basis of the observed dose responses, four compounds were selected for further examination (Figure 1, panels e–h).
Characterization of Compound–RNA Interaction
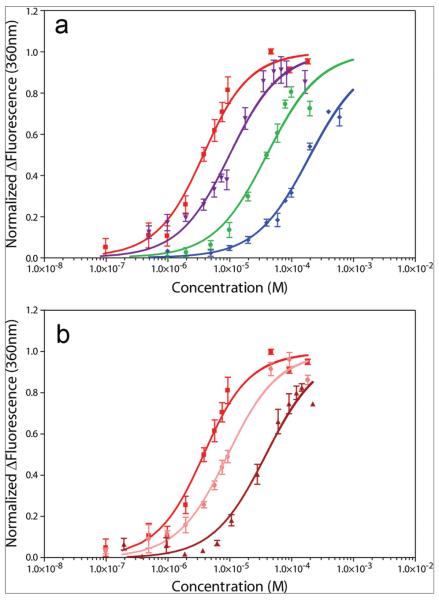
Compound affinities for RNA were measured by titration of compounds into a solution containing 2 μM HIV-1screen. Doxorubicin and idarubicin were found to decrease fluorescence as the titration progressed, whereas butamben and cinchonidine increased fluorescence. Data are plotted as magnitude change in fluorescence for clarity (Figure 2, panel a). Results show that the compounds doxorubicin, idarubicin, butamben, and cinchonidine have apparent Kd values of 2.8 ± 0.4, 9.1 ± 1.3, 39.4 ± 7.0, and 193.2 ± 21.3 μM, respectively (Table 1). Hill coefficients for these compounds ranged from 1.0 to 1.7 (Table 1).
Figure 2.
Fluorescence-monitored titrations of compounds with HIV-1screen RNA. Titrations are plotted as the normalized change in fluorescence versus compound concentration. Data represent an average of 3 titrations, with the SEM indicated. a) Doxorubicin, red squares; idarubicin, violet triangles; butamben, green circles; and cinchonidine, blue diamonds. b) Representative data from doxorubicin competition titrations. Data from titrations into HIV-1screen alone, HIV-1screen in competition with 10-fold molar excess HIV-1Δbulge, and HIV-1screen in competition with 10-fold molar excess HIV-1wtbulge are shown as red squares, pink circles, and dark red triangles, respectively.
TABLE 1.
Apparent Kd values (μM) as measured in competition experiments
| Compound |
Kd without competitorsa |
Kd with HIV-1Δbulgeb |
Kd with HIV-1wtbulgec |
Degree of nonspecificity |
Degree of direct competition |
Relative specificity for bulge |
Hill coefficientd |
|---|---|---|---|---|---|---|---|
| Cinchonidine | 193.2 ± 21.3 | n.d. | n.d. | n.d. | n.d. | n.d. | 1.5 ± 0.15 |
| Butamben | 39.4 ± 7.0 | n.d. | n.d. | n.d. | n.d. | n.d. | 1.7 ± 0.2 |
| Idarubicin | 9.1 ± 1.3 | 17.6 ± 2.4 | 81.7 ± 3.4 | 17.6 ± 3.5% | 81.6 ± 12.1% | 4.7 ± 1.2 | 1.0 ± 0.15 |
| Doxorubicin | 2.8 ± 0.4 | 8.4 ± 0.9 | 37.8 ± 6.1 | 27.3 ± 4.9% | 77.2 ± 16.7% | 2.8 ± 0.8 | 1.5 ± 0.15 |
Compounds were titrated into 2 μM HIV-1screen in 3 replicate experiments. Mean apparent Kd and SEM are indicated.
n.d. = not determined. 20 μM HIV-1Δbulge was included in the titrations. This is a 10-fold molar excess of RNA relative to the HIV-1screen RNA containing the 2-aminopurine reporter.
20 μM HIV-1wtbulge was included in the titrations. This is a 10-fold molar excess of RNA relative to the HIV-1screen RNA containing the 2-aminopurine reporter.
Hill coefficient errors are shown as standard errors resulting from the least-squares fit.
The specificity of binding was investigated for the two molecules with the highest binding affinities, doxorubicin and idarubicin, using competition experiments. Solubility limits prevented the accurate assessment of binding specificity for butamben and cinchonidine. Specificity was measured by performing the titrations in the presence of either a 10-fold molar excess of nonspecific (helical) RNA (HIV-1Δbulge) or a 10-fold molar excess of competitor RNA (HIV-1wtbulge) (Figure 2, panel b, Table 1). Upon challenge with HIV-1Δbulge, the apparent Kd values increased slightly, to 8.4 ± 0.9 and 17.6 ± 2.4 μM for doxorubicin and idarubicin, respectively, indicating some degree of nonspecific RNA interaction(s). However, the observed interactions are not completely nonspecific, as a proportional (10-fold) increase in apparent Kd was not observed upon challenge with the nonspecific RNA. The degree of nonspecificity was quantified by calculating the fractional increase in apparent Kd relative to the amount of HIV-1Δbulge added (Table 1). Conversely, upon challenge with HIV-1wtbulge, the apparent Kd values did increase approximately 10-fold (Table 1). Note that since the competitor RNA has a guanine in place of 2-aminopurine, this result indicates that the compounds are largely unable to discriminate between 2-aminopurine and guanine. The degree of direct competition by the competitor RNA is quantified (Table 1). Comparison of the degree of direct competition for the bulge and the degree of nonspecificity indicates that the compounds bind between 2.8- and 4.7-fold more specifically to the GGA bulge-containing RNA than to the RNA with the GGA deletion (Table 1).
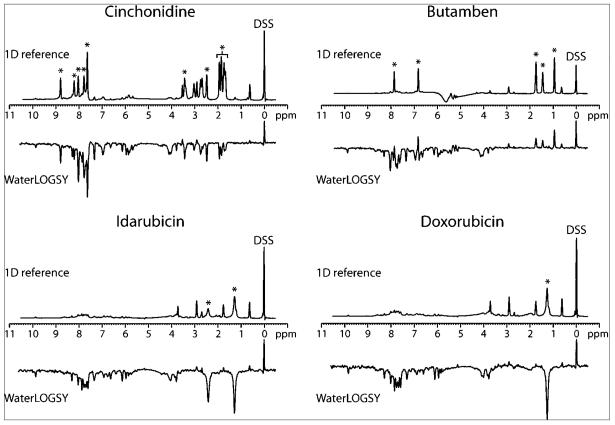
A water-ligand optimized gradient spectroscopy (WaterLOGSY) NMR experiment (31, 32) was used to directly monitor compound binding to the unmodified RNA (HIV-1wtbulge). The WaterLOGSY experiment monitors changes in the sign of the nuclear Overhauser effect (NOE) transfer of magnetization for a rapidly tumbling free ligand versus a slowly tumbling ligand interacting with a macromolecule. Previously, the WaterLOGSY experiment has been useful for investigating small molecule binding to RNA (33). The data show that all of the selected compounds except butamben bind the HIV-1wtbulge RNA, as evidenced by the change in sign of the compound peaks relative to the DSS internal control (Figure 3). This may indicate that, unlike the other 3 compounds, butamben does not bind the unmodified RNA and may require 2-aminopurine in place of guanine for it to bind. Therefore, butamben was not investigated further.
Figure 3.
NMR analysis of compound binding to RNA by WaterLOGSY. 1D reference experiments (upper spectra) and WaterLOGSY experiments (lower spectra) are grouped according to compound. The 0 ppm internal reference standard is indicated (DSS), which also serves as a control small molecule that does not bind RNA. The DSS signal is phased to be positive in both the 1D reference and the WaterLOGSY experiments. Selected peaks that are specific to each compound are indicated by asterisks.
Doxorubicin Affects –1 Translational Frameshifting in Vitro
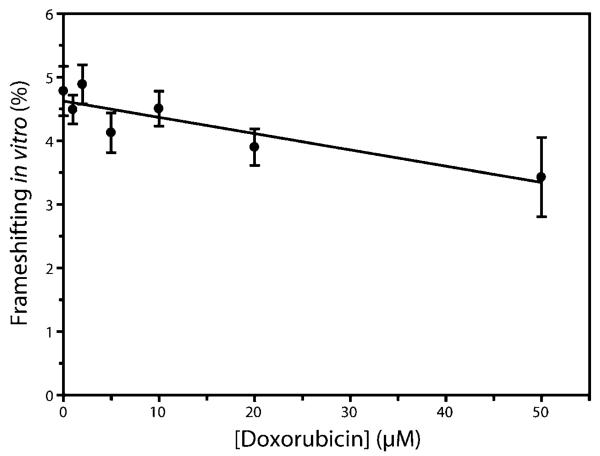
The three remaining compounds (doxorubicin, idarubicin, and cinchonidine) were tested for effects on −1 translational frameshifting using an in vitro assay system (34). Results represent an average of 10 independent experiments, with the SEM indicated. The addition of doxorubicin showed a general trend of decreasing frameshifting with increasing compound concentration, with a 28% (±14%) decrease in frameshifting observed at 50 μM compound (Figure 4). This decrease is significantly different from the level of frame-shifting in the untreated control (p < 0.05). Idarubicin showed a similar trend of decreasing frameshifting, although the data were not statistically significant, and cinchonidine did not have an effect on frameshifting (data not shown).
Figure 4.
In vitro –1 translational frameshifting efficiency as a function of doxorubicin concentration. Frameshifting efficiencies at each compound concentration were determined by an average of at least 10 independent experiments. Data were fit to a linear regression line with a best-fit slope of −0.0256, an ordinate-intercept of 4.63 and an overall r2 value of 0.787. Error bars indicate the SEM for each data point.
Addition of doxorubicin was also observed to decrease overall translation by approximately 60% at 50 μM, as measured by a change in Renilla luciferase luminescence (data not shown). Doxorubicin is therefore likely to have a general inhibitory effect on translation. This inhibition is likely due to interactions with rRNA as a consequence of the modest 2.8-fold specificity for the HIV-1 RNA bulge over the control RNA (Table 1). In this respect, the action of doxorubicin may be similar to anisomycin, which also affects frameshifting efficiency (35). Further study must be done to better clarify the role of doxorubicin in translational inhibition.
Mapping of Doxorubicin Interactions with the HIV-18-52 RNA
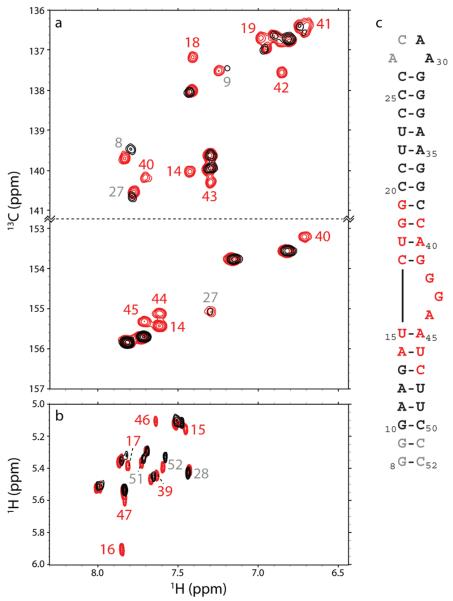
Titration of doxorubicin into 13C/15N-labeled HIV-18-52 RNA (Figure 1, panel a) from a molar ratio of 0:1 to 10:1 compound:RNA was monitored by NMR using 2D 1H–13C-TROSY-HSQC (Figure 5, panel a) and 2D TOCSY (Figure 5, panel b) experiments. As the compound is titrated into the RNA, a subset of the peaks become selectively broadened beyond detection. This line broadening can be attributed to binding kinetics occurring on a similar time scale as the NMR experiment (a process termed intermediate exchange). This is consistent with the observed apparent Kd of 2.8 μM. These residues involved in binding doxorubicin were mapped to the HIV-1 structure (24) (Figure 5, panel c, red). A second subset of peaks showed changes in chemical shift, indicative of weaker binding involving kinetics in fast exchange relative to the time scale of the NMR experiment. Residues experiencing this fast exchange are located at the termini of the molecule (Figure 5, panel c, gray).
Figure 5.
Localization of doxorubicin to the HIV-18-52 RNA by NMR. Peaks are labeled by residue number, as in Figure 1, panel a. Peaks labeled in red are selectively line broadened by doxorubicin, and peaks in gray show chemical shift changes. a) Region of the TROSY-modified 2D 1H–13C HSQC spectra acquired showing the C8–H8 and C2–H2 correlations of purines. b) Region of the 2D TOCSY spectra showing the H5–H6 correlations of pyrimidines. c) HIV-1 RNA structure as in Figure 1, panel a, with residues selectively line broadened by doxorubicin indicated in red, residues experiencing chemical shift changes indicated in gray, and residues not affected shown in black.
Thermodynamic Stability of HIV-1 RNA Is Increased by Doxorubicin Binding
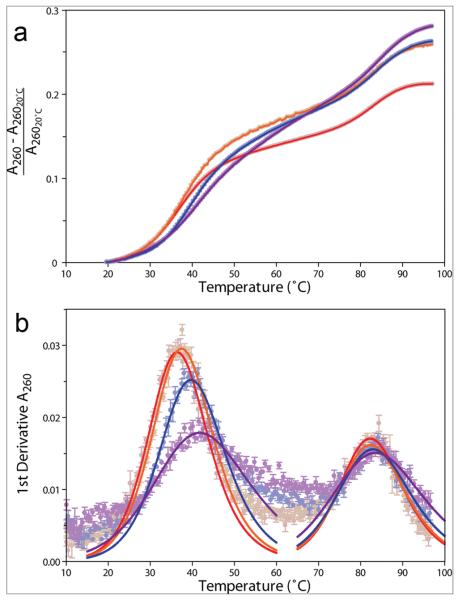
The effects of doxorubicin on RNA stability were assayed by performing UV-monitored RNA thermal denaturation experiments at varying compound concentrations (Figure 6, Table 2). The contribution of doxorubicin to the overall UV260 absorbance was less than 30% of the RNA contribution, and no temperature-dependent change in absorbance was observed for doxorubicin alone (data not shown). This allowed the overall UV260 absorbance to be corrected for the doxorubicin contribution. Results show that adding doxorubicin produces a significantly (p < 0.001) larger hyperchromic effect than is observed when just DMSO is added (Table 2). This suggests that the addition of doxorubicin increases the number and/or quality of base stacking in the RNA. The observed decrease in HIV-1screen fluorescence upon compound binding (described earlier), indicative of increased 2-aminopurine base stacking, is consistent with this suggestion. In accordance with this observation, doxorubicin increases the stability of the lower helix of the RNA by up to 5 °C at 50 μM (Figure 6, panel b, Table 2); however, it does not appear to have any effect on the melting transition of the upper stem-loop.
Figure 6.
UV-monitored thermal denaturation profiles for HIV-1wtbulge in the presence of varying concentration of doxorubicin. Molar ratios of doxorubicin:RNA of 0:1, 1:1, 5:1, and 10:1 are indicated as red, orange, blue, and violet circles and lines, respectively. Data points represent the average of 5 independent replicates, and error bars correspond to the SEM. a) Data are plotted as fractional hyperchromicity relative to the absorbance at 20 °C at 260 nm. Error bars have been omitted for clarity. b) First-derivative plot of data in panel a, with fitted curves showing the deconvoluted transitions as solid lines.
TABLE 2.
Thermodynamic effects of doxorubicin binding on HIV-1posctrlRNAa
| Ratio of compound to RNA | Tm1 (°C) | Tm2 (°C) | Hyperchromicity increase (%)b |
|---|---|---|---|
| 0:1 | 36.1 ± 0.3 | 82.0 ± 0.3 | 21.3 ± 0.6 |
| 1:1 | 37.1 ± 0.3 | 82.1 ± 0.3 | 26.0 ± 0.7 |
| 5:1 | 39.3 ± 0.3 | 82.5 ± 0.3 | 26.4 ± 0.4 |
| 10:1 | 41.0 ± 0.3 | 83.0 ± 0.3 | 28.2 ± 1.0 |
Values listed represent the best-fit solutions to the van't Hoff equation. Fitting errors for Tm were less than the nominal error of the temperature probe (0.3 °C, Varian); therefore all Tm errors were set to 0.3 °C.
Percentage increase in UV absorbance observed at 260 nm from 20 to 95 °C.
Other Uses of Doxorubicin
In addition to the effects of doxorubicin on RNA reported here, doxorubicin is an U.S. Food and Drug Administration-approved chemotherapy drug used for the treatment of several cancers (36, 37). It is believed to act by intercalating into DNA (38, 39) and has been observed to do so via crystallography (40). The affinity of doxorubicin for DNA is approximately 40 nM (41), which is significantly better than the affinity for the HIV-1 RNA observed in this study. Additionally, doxorubicin has been shown to bind the Iron-Responsive Element RNA with a similar affinity as observed for the HIV-1 RNA (42). This may indicate an approximate affinity of 1 μM for the interaction of doxorubicin with ideal sites on RNA molecules, such as internal bulges, loops, and U-G wobble pairs (42).
Doxorubicin has also been shown to inhibit HIV replication in monocyte-macrophage cells, although this effect was limited to inhibition of initial infection rather than decreased viral replication in chronically infected cells (43). Therefore, it is likely that doxorubicin has some effect on the HIV-1 reverse-transcribed DNA integration into the cellular genome, as suggested by the authors (43), rather than having a downstream effect on the viral replication cycle at the doses tested. It should be noted, however, that since the levels of the protease, reverse transcriptase, and integrase proteins were not measured relative to the levels of p24 (capsid protein), a direct effect of doxorubicin on frameshifting in monocyte-macrophage cells has not been demonstrated.
The high affinity of doxorubicin for DNA, in combination with limited RNA specificity, poses a significant challenge for the further optimization of this compound as a possible RNA-targeting therapeutic; however, several analogues of doxorubicin have only a moderate affinity for DNA (10–50 μM) (41). It is possible that chemical modification of doxorubicin or its analogues may confer additional specificity and affinity for the HIV-1 RNA, and efforts to examine the structure–function relationships of doxorubicin analogues are ongoing.
CONCLUSION
In this study, we have screened approximately 34,500 compounds for binding to the conserved three-purine bulge of the HIV-1 frameshift-inducing RNA. A total of 202 compounds were identified, representing a diverse set of chemical structures that share the common properties of aromaticity and capacity to form hydrogen bonds, as would be expected for molecules that bind RNA. These compounds were grouped into 20 structurally similar classes (Supplementary Figure 4), in addition to 148 compounds that were not sufficiently similar to be clustered into representative classes. These classes may represent compounds that can bind to RNA and provide an array of structural scaffolds upon which more specific RNA-binding molecules could potentially be designed.
Doxorubicin, a representative of one of these classes, has been shown to bind to the HIV-1 frameshift-inducing RNA with low micromolar affinity. This affinity is comparable to the best binder selected from an 11,325-member combinatorial library (5). Fluorescence and thermodynamic data suggest that the compound may stabilize the lower helix of the RNA by intercalating into the bulge and altering its structure. The observed decrease in fluorescence upon doxorubicin titration indicates a stabilization of a stacked conformation for the central base in the bulge. This increased stacking is also observed in UV-monitored RNA denaturation experiments by an increase in overall hyperchromicity when doxorubicin is present, as well as by the increase in lower helix stability. NMR data indicated that doxorubicin binding is primarily localized to the three-purine GGA bulge and also interacts weakly with the helical termini. On the basis of these data, we generated a structural model for the interaction of doxorubicin with the HIV-1 frameshift site RNA (Supplementary Figure 5). This model is consistent with the observed chemical shift changes and suggests several testable hypotheses regarding the binding mode of doxorubicin to the HIV-1 RNA, which are in the process of being examined. For example, modification of the amino group on doxorubicin into a more extended guanidinium group may allow for more hydrogen bonds to other nucleoside bases in the bulge, or to the phosphate backbone itself, which may increase the affinity and specificity of the small molecule–RNA complex. Additionally, modification of the hydroxyethanone into a more cationic, longer chain (such as a linkage to arginine) could allow more contacts with the lower helix of the RNA.
Doxorubicin-induced alterations in the stability and structure of the RNA may contribute to the observed decrease in frameshifting in vitro. However, we cannot rule out the possibility that the compound may exert this effect through other interactions (e.g., with rRNA). We do, however, find that doxorubicin is modestly (approximately 3-fold) more specific for the GGA sequence than for the rest of the RNA. Efforts to improve upon the specificity and affinity of binding of doxorubicin to the HIV-1 RNA via investigation and chemical modification of the drug and its analogues are ongoing.
METHODS
High-Throughput Screening
Screening was performed at the Keck-UW Comprehensive Cancer Center Small Molecule Screening Facility, utilizing a Tecan Safire II plate reader to measure fluorescence and a BioMek FK robot for liquid handling. Approximately 34,500 small molecule compounds from the Chem-bridge DIVERSet, Maybridge HitFinder, Prestwick Chemical Library, Microsource SPECTRUM Collection, and Sigma LOPAC1280 commercially available libraries were screened in 384- and 1536-well formats. Screening samples were 500 nM (final concentration) HIV-1screen RNA (Dharmacon, Inc.) in 10 mM HEPES (pH 7.0). The HIV-1screen RNA sequence is shown in Figure 1. Compounds were added to a final concentration of 20 μM from 1 mM compound stocks in dimethyl sulfoxide (DMSO). Plates were then placed at 30 °C for 5 min prior to measurement.
Fluorescence measurements were performed by excitation of the 2-aminopurine at 309 nm and measuring emission at 363 nm. Plates contained a minimum of 4 replicates of a compound solvent control (DMSO) at 2% final concentration. This control was used as a reference to identify changes in fluorescence due to ligand binding. Initial lead compounds were tested for autofluorescence at 363 nm and UV absorbance at 260 nm in 10 mM HEPES (pH 7.0) without RNA added, and the initial assay was repeated to reduce false-positives. Compounds identified as hits were obtained (Ryan Scientific, Inc., Sigma-Aldrich) and dissolved to stock concentrations of 100 mM in either 2H2O or d6-DMSO.
RNA Synthesis and Purification
Milligram quantities of RNA suitable for NMR and UV spectroscopy methods were transcribed in vitro using purified His6-tagged T7 RNA polymerase, synthetic DNA oligonucleotides (Integrated DNA Technologies, Inc.), and ribonucleotide triphosphates (Sigma-Aldrich) as described previously (44). Briefly, RNA was purified by denaturing 20% (w/v) (HIV-1Δbulge), 15% (HIV-1wtbulge), or 10% (HIV-18-52) polyacrylamide gel electrophoresis, recovered by diffusion into 0.3 M sodium acetate, precipitated with ethanol, purified by anion exchange chromatography, again precipitated with ethanol, and desalted. RNA modified with 2-aminopurine (HIV-1screen) waspurchased from Dharmacon, deprotected according to the protocol provided, and desalted. The purified RNA was lyophilized, resuspended in water, and brought to pH 7.0 by the addition of 1 M NaOH or by solvent exchange using Amicon spin-filters (Millipore) into a buffer containing either 10 mM HEPES (pH 7.0 or 7.4) or 10 mM d11-Tris (pH 6.9).
Fluorescence-Monitored Titrations
Fluorescence measurements were performed using a QuantaMaster Model C-60/2000 spectrofluorimeter (Bioinstrumentation Facility, UW-Madison). A 160 μL sample cell was used, and fluorescence was measured for 10 s at 30 °C for each sample. Fluorescence was measured at 360 nm after excitation of the 2-aminopurine of HIV-1screen at 309 nm. Compounds were titrated in triplicate from 10−9 to 10−3 M into 1–2 μM HIV-1screen in 10 mM HEPES (pH 7.0). Competition titrations in identical conditions were also performed in the presence of either 20 μM HIV-1Δbulge or 20 μM HIV-1wtbulge.
Data were initially fit to the quadratic solution to the law of mass action (eq 1)
| (1) |
using Prism 4.3 (GraphPad), where fB is the fraction of RNA bound by the small molecule, Bmax is a scaling parameter representing the response at maximal binding, [R]tot is the total concentration of RNA molecules, [X] is the concentration of compound, and Kd is the dissociation constant for the small molecule-RNA complex. Data were then normalized to the initial Bmax fits and fit to eq 1 again. Normalized data were also fit to the modified Hill equation (45) (eq 2)
| (2) |
where nH is the Hill coefficient, and fB, Bmax, Kd, and [X] are as described above.
Data derived from competition experiments were analyzed as degree of nonspecificity, degree of direct competition, and relative specificity for the GGA bulge. The degree of nonspecificity (degns) is defined as
| (3) |
where 11 represents the fold increase of RNA upon addition of 10 equiv of competitor RNA (HIV-1Δbulge) to 1 equiv of HIV-1screen, and Δ(fold)Kd represents the ratio of apparent Kd with HIV-1Δbulge to Kd without competitor. The degree of direct competition (degdc) is defined as in eq 3, except the competitor RNA is HIV-1wtbulge and Δ(fold)Kd represents the ratio of apparent Kd with HIV-1wtbulge to Kd without competitor. The relative specificity for the bulge is defined as the ratio of the degree of direct competition to the degree of nonspecificity. Results are reported in Table 1.
Statistical Analyses
See Supporting Information online.
Frameshifting Assay
RNA samples were translated in vitro using the Flexi Rabbit Reticulocyte Lysate System (Promega). Translation reactions were conducted in 96-well plates (Costar) and contained 1.5 μg of RNA per reaction mixture (12.5 μL total volume), 1–2 mM Mg2+, 70 mM KCl, and 20 μM of each amino acid except methionine and leucine (10 μM each). Compounds (0.5 μL well−1) were added to both the experimental and positive control reactions at varying concentrations, and reactions were incubated at 37 °C for 90 min. The DMSO concentration was kept under 1% for all reactions. For each compound concentration, a minimum of 10 replicates each of the experimental and positive control reactions were assayed using the Dual-Luciferase Reporter Assay System (Promega). Luminescence was measured and frameshifting efficiencies were calculated as previously described (44). For additional method information see Supporting Information.
UV Spectroscopy
Thermal denaturation of HIV-1wtbulge RNA was performed in the presence and absence of small molecule compounds using a Cary Model 100 Bio UV–visible spectrophotometer equipped with a Peltier heating accessory and temperature probe. All samples contained 10 mM HEPES (pH 7.6–6.4 over the range of the assay), 20 mM KCl, 0.4% DMSO, and 2 μM HIV-1wtbulge RNA in a volume of 1 mL. Prior to data collection, samples were heated to 95 °C and cooled to 10 °C to ensure homogeneous folding. Compound was then added, samples were mixed and then heated from 10 to 95 °C at a rate of 1 °C min−1, and absorbance data were collected at 260 nm in 0.5 °C increments. A minimum of five replicates were acquired for each compound concentration. The data were corrected for compound contributions to UV260 absorbance, normalized at 20 °C, derivatized, and fit using a nonlinear least-squares fit to solve for Tm as previously described (46). Data analysis was performed using Prism 4.3 (GraphPad) and thermodynamic parameters for each compound concentration are reported (Table 2).
NMR Spectroscopy
All NMR spectra were obtained on a Bruker Avance DMX 750 MHz spectrometer at the National Magnetic Resonance Facility at Madison (NMRFAM). The spectrometer was equipped with a single z-axis gradient proton, carbon, nitrogen cryogenically cooled probe. Spectra were acquired in 90% H2O/10% 2H2O, 10 mM d11-Tris (temperature-corrected pH 7.3) at 283 K or in 100% 2H2O, 10 mM d11-Tris (temperature-corrected pH 6.7) at 303 K.
Secondary screening of small molecule binding to RNA was performed in 90% H2O/10% 2H2O, 10 mM d11-Tris (temperature-corrected pH 7.3) at 283 K using a water-ligand optimized gradient spectroscopy experiment (WaterLOGSY) (31, 32) in conjunction with acquisition of 1D reference spectra of the RNA-compound mixtures. Water suppression for these experiments was achieved using a WATERGATE water suppression scheme (47). WaterLOGSY spectra were collected with 1 s mixing time, 48.7 ms acquisition time, 1 s recycle delay, 14 ppm sweep width and 1024 scans. All spectra were referenced to a 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS, Sigma) internal control.
Chemical shift mapping of small molecule interactions with RNA was performed by monitoring compound titrations with 2D TOCSY and TROSY-modified 2D 1H–13C HSQC spectra at 303 K; 100 μM 13C/15N-labeled HIV-18-52 in 100% 2H2O, 10 mM d11-Tris (pH 6.7) was used for these experiments, and compound was titrated from a 0:1 to a 10:1 molar ratio of compound:RNA. Spectral data were normalized to levels that were equal to 10 times the estimated experimental noise for each experiment.
All data were processed using XwinNMR (Bruker) software, and assignments were made using Sparky (http://www.cgl.ucsf. edu/home/sparky/).
Supplementary Material
Acknowledgment
We thank Dr. Lawrence Clos II, Jordan Halsig, Stephen Martin-Tumasz, Ashley Richie, Dr. Dipali Sashital, and Dr. Kirk Vander Meulen for helpful discussions. We also thank Noël Peters and Lane Milde at the Keck-UW Comprehensive Cancer Center (Keck-UWCCC) Small Molecule Screening Facility for assistance with high-throughput screening, Prof. A. C. Palmenberg and her laboratory (University of Wisconsin-Madison) for equipment use, and Dr. W. Milo Westler and the staff at the National Magnetic Resonance Facility at Madison (NMRFAM) for assistance with NMR data collection. All NMR studies were carried out at NMRFAM (http://www.nmrfam.wisc.edu/) with support from U.S. National Institutes of Health (NIH) [P41RR02301 (Biomedical Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences)]. Equipment funding for the facility was provided by the University of Wisconsin, the NIH [P41GM66326, P41RR02301, RR02781, RR08438], the National Science Foundation [DMB-8415048, OIA-9977486, BIR-9214394], and the U.S. Department of Agriculture. The KeckUWCCC Small Molecule Screening Facility was established with grants from the W. M. Keck Foundation and University of Wisconsin Comprehensive Cancer Center. This work was supported by NIH grant [GM072447 to S.E.B.], NIH Predoctoral Training Grant [T32 GM08349 to R.J.M.], an E.W. Hopkins Predoctoral Fellowship [to R.J.M], and by the Wisconsin Alumni Research Foundation through the Lead Discovery Initiative.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lefebvre E, Schiffer CA. Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir. AIDS Rev. 2008;10(3):131–142. [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller DS, Youssef-Bessler M. Etravirine: a second-generation nonnucleoside reverse transcriptase inhibitor (NNRTI) active against NNRTI-resistant strains of HIV. Clin. Ther. 2009;31(4):692–704. doi: 10.1016/j.clinthera.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Dinman JD, Ruiz-Echevarria MJ, Peltz SW. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 1998;16(4):190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung M, Patel P, Davis S, Green SR. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998;72(6):4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNaughton BR, Gareiss PC, Miller BL. Identification of a selective small-molecule ligand for HIV-1 frameshift-inducing stem-loop RNA from an 11,325 member resin bound dynamic combinatorial library. J. Am. Chem. Soc. 2007;129(37):11306–11307. doi: 10.1021/ja072114h. [DOI] [PubMed] [Google Scholar]
- 6.Staple DW, Venditti V, Niccolai N, Elson-Schwab L, Tor Y, Butcher SE. Guanidinoneomycin B recognition of an HIV-1 RNA helix. ChemBioChem. 2008;9(1):93–102. doi: 10.1002/cbic.200700251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiya S, Koh CS, Matsufuji S, Harada K. Analysis of the interaction between selected RNA-binding peptides and a target RNA containing a bulge and a GNRA-type tetraloop. Nucleic Acids Symp. Ser. 2008;(52):209–210. doi: 10.1093/nass/nrn106. [DOI] [PubMed] [Google Scholar]
- 8.Dulude D, Theberge-Julien G, Brakier-Gingras L, Heveker N. Selection of peptides interfering with a ribosomal frameshift in the human immunodeficiency virus type 1. RNA. 2008;14(5):981–991. doi: 10.1261/rna.887008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 10.Parkin NT, Chamorro M, Varmus HE. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66(8):5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Morrow CD. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 1991;65(9):5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YG, Maas S, Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29(5):1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leger M, Sidani S, Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10(8):1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed –1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30(23):5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345(1):127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 16.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75(4):1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frame-shifting. Cold Spring Harbor Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 18.Telenti A, Martinez R, Munoz M, Bleiber G, Greub G, Sang-lard D, Peters S. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J. Virol. 2002;76(15):7868–7873. doi: 10.1128/JVI.76.15.7868-7873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset JP. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3(10):1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudin C, Mazauric MH, Traikia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349(5):1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 21.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106(2):233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 22.Baril M, Dulude D, Gendron K, Lemay G, Brakier-Gingras L. Efficiency of a programmed –1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA. 2003;9(10):1246–1253. doi: 10.1261/rna.5113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staple DW, Butcher SE. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31(15):4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349(5):1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 25.Kirk SR, Luedtke NW, Tor Y. 2-Aminopurine as a real-time probe of enzymatic cleavage and inhibition of hammer-head ribozymes. Bioorg. Med. Chem. 2001;9(9):2295–2301. doi: 10.1016/s0968-0896(01)00123-7. [DOI] [PubMed] [Google Scholar]
- 26.Holmen A, Norden B, Albinsson B. Electronic transition moments of 2-aminopurine. J. Am. Chem. Soc. 1997;119(13):3114–3121. [Google Scholar]
- 27.Xu D, Evans KO, Nordlund TM. Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry. 1994;33(32):9592–9599. doi: 10.1021/bi00198a027. [DOI] [PubMed] [Google Scholar]
- 28.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine ribo-side, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244(5):1228–1237. [PubMed] [Google Scholar]
- 29.Menger M, Tuschl T, Eckstein F, Porschke D. Mg(2+)-dependent conformational changes in the hammerhead ribozyme. Biochemistry. 1996;35(47):14710–14716. doi: 10.1021/bi960440w. [DOI] [PubMed] [Google Scholar]
- 30.Lacourciere KA, Stivers JT, Marino JP. Mechanism of neomycin and Rev peptide binding to the Rev responsive element of HIV-1 as determined by fluorescence and NMR spectroscopy. Biochemistry. 2000;39(19):5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 31.Dalvit C, Pevarello P, Tato M, Veronesi M, Vulpetti A, Sundstrom M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR. 2000;18(1):65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 32.Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR. 2001;21(4):349–359. doi: 10.1023/a:1013302231549. [DOI] [PubMed] [Google Scholar]
- 33.Johnson EC, Feher VA, Peng JW, Moore JM, Williamson JR. Application of NMR SHAPES screening to an RNA target. J. Am. Chem. Soc. 2003;125(51):15724–15725. doi: 10.1021/ja037499s. [DOI] [PubMed] [Google Scholar]
- 34.Grentzmann G, Ingram JA, Kelly PJ, Gestaland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(04):479–486. [PMC free article] [PubMed] [Google Scholar]
- 35.Dinman JD, Ruiz-Echevarria MJ, Czaplinski K, Peltz SW. Peptidyl-transferase inhibitors have antiviral properties by altering programmed –1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl. Acad. Sci. U.S.A. 1997;94(13):6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arcamone F. Doxorubicin: Anticancer Antibiotics. Vol. 17. Academic Press; New York, NY: 1981. [Google Scholar]
- 37.Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of pegylated liposomal doxorubicin: review of animal and human studies. Clin. Pharmacokinet. 2003;42(5):419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 38.Momparler RL, Karon M, Siegel SE, Avila F. Effect of adriamycin on DNA, R. N. A. and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36(8):2891–2895. [PubMed] [Google Scholar]
- 39.Fornari FA, Randolph JK, Yalowich JC, Ritke MK, Gewirtz DA. Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol. Pharmacol. 1994;45(4):649–656. [PubMed] [Google Scholar]
- 40.Frederick CA, Williams LD, Ughetto G, van der Marel GA, van Boom JH, Rich A, Wang AH. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29(10):2538–2549. [PubMed] [Google Scholar]
- 41.Chaires JB, Satyanarayana S, Suh D, Fokt I, Przewloka T, Priebe W. Parsing the free energy of anthracycline antibiotic binding to DNA. Biochemistry. 1996;35(7):2047–2053. doi: 10.1021/bi952812r. [DOI] [PubMed] [Google Scholar]
- 42.Canzoneri JC, Oyelere AK. Interaction of anthracyclines with iron responsive element mRNAs. Nucleic Acids Res. 2008;36(21):6825–6834. doi: 10.1093/nar/gkn774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergamini A, Perno CF, Balzarini J, Capozzi M, Marinelli L, Milanese G, Pesce CD, Calio R, Rocchi G. Selective inhibition of HIV replication by adriamycin in macrophages but not in lymphocytes. AIDS Res. Hum. Retroviruses. 1992;8(7):1239–1247. doi: 10.1089/aid.1992.8.1239. [DOI] [PubMed] [Google Scholar]
- 44.Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373(3):652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindslev N. Drug-Acceptor Interactions: Modeling Theoretical Tools To Test and Evaluate Experimental Equilibrium Effects. Co-Action Publishing; Sweden: 2008. p. 428. [Google Scholar]
- 46.John DM, Weeks KM. van't Hoff enthalpies without baselines. Protein Sci. 2000;9(07):1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.