Abstract
The non-classical major histocompatibility complex (MHC) class I molecule CD1d presents lipid antigens to invariant natural killer T (iNKT) cells, which are an important part of the innate immune system. CD1d/iNKT systems are highly conserved in evolution, and cross-species reactivity has been suggested to be a common feature of different animals based on research in humans and mice. However, we found that CD1d from the tree shrew (Tupaia belangeri), a close evolutionary relative of primates, failed to stimulate human iNKT cells, despite being more homologous to human CD1d than that of mouse. Sequence comparison and molecular modelling showed that two of the key amino acid residues in human CD1d proposed to be in direct contact with T-cell receptors were mutated in tree shrew CD1d. Substitution of one of the residues, but not the other, with the human residue enabled tree shrew CD1d to regain the ability to present lipid antigen to human iNKT cells. These results indicate that CD1d/iNKT recognition is species-specific, and that cross-species reactivity may be less common than currently proposed. Also, a naturally occurring CD1d mutation(s) that confers inability to stimulate iNKT cell function may have implications for future studies on CD1d/iNKT-associated diseases.
Keywords: antigen presentation, CD1d antigen, T lymphocytes, Tupaiidae
Introduction
CD1d is a non-classical major histocompatibility class I (MHC-I) molecule belonging to the CD1 family, which also includes CD1a, CD1b, CD1c and CD1e in various species.1 CD1 proteins have a similar overall structure to MHC-I molecules, in which a heavy chain encoded by CD1 genes is non-covalently linked to the light chain β2 microglobulin (β2m). However, unlike peptide-presenting MHC-I, CD1 molecules present lipid and glycolipid antigens derived either endogenously or from micro-organisms.2 A subset of CD1d-restricted T cells, invariant natural killer T (iNKT) cells, are an important component of the innate immune system. They have a limited T-cell receptor (TCR) repertoire and are reactive with α-galactosylceramide (αGalCer), a synthetic glycolipid derived from a marine sponge.3,4 iNKT cells have the unique ability to rapidly secrete both T helper type 1 (Th1) and Th2 cytokines upon antigenic stimulation, resulting in activation of other cell types such as dendritic cells (DCs), NK cells, B cells and conventional T cells. This important immunomodulatory function suggests that they are involved in host defence against bacteria and viruses, and in tumour immunity and autoimmune disease pathogenesis.5–7
The CD1d/iNKT system is conserved in evolution. CD1d genes have been found in a variety of primates, rodents and other animals,1 and iNKT cells have been identified in mice, rats and monkeys.4,8–10 It appears that cross-species reactivity is a common feature among different animals: TCRs from human iNKT cells cross-react with mouse CD1d–αGalCer complexes (and vice versa), and CD1d–αGalCer tetramers from either species stain iNKT cells of the other species.4,8,11,12 Thus it seems that this ligand recognition is highly conserved through evolution.
Tree shrews (Tupaia belangeri) are small animals originally regarded as either primates or insectivores, and later classified in a separate order, Scandentia. They are most closely related to primates, and have been successfully employed as model animals for psychosocial stress and myopia.13–15 Recently, tree shrews were found to be susceptible to human hepatitis viruses such as hepatitis B virus (HBV) and hepatitis C virus (HCV). As hepatitis viruses have very narrow host ranges (for example, HBV only infects humans and chimpanzees), virus-infected tree shrews provide an invaluable tool for studies on hepatitis virus infections and hepatitis virus-associated hepatocellular carcinoma.15 Considering the involvement of iNKT cells in host defence against viruses, especially hepatitis viruses,6,16,17 and their potential role in antitumour immunity against hepatocellular carcinoma,18–21 it is important to identify the CD1d/iNKT system in the tree shrew in order to investigate the pathogenesis of hepatitis virus infection and hepatocellular carcinoma in this model. Here we report the molecular cloning of tree shrew CD1d (tsCD1d) and examine its cross-species reactivity with human iNKT cells. We found that a single amino acid residue difference between tree shrew CD1d and human CD1d could prevent cross-species recognition of tree shrew CD1d by human iNKT cells. Cross-species recognition of CD1d by mammalian iNKT cell may therefore be less common.
Materials and methods
Animals
Tree shrews (Tupaia belangeri) were bred and maintained in Kunming Institute of Zoology, Chinese Academy of Science (CAS). All the procedures were performed in accordance with the regulations of animal care of CAS.
Cell separation, tissue isolation and RNA preparation
To extract RNA from blood samples, red blood cells (RBCs) were first removed by adding RBC lysis buffer [155 mm NH4Cl, 10 mm KCO3 and 0·1 mm ethylenediaminetetraacetic acid (EDTA)], and then total RNA was extracted from the remaining cells using Trizol Reagent (MRC, Cincinnati, OH). Brain, heart, lung, liver, spleen, stomach and small intestine samples were collected and frozen in liquid nitrogen within 12–15 min after the animals had been killed. The frozen tissue samples were then homogenized into a powder using a mortar and pestle and total RNA was isolated using an RNA Extraction Kit (Tiangen, Beijing, China) according to the manufacturer’s manual.
tsCD1d cDNA cloning and reverse transcription–polymerase chain reaction (RT-PCR)
First-strand cDNA was synthesized by oligo(dT) priming from 5 μg of total RNA using TIANScript M-MLV reverse transcriptase (Tiangen). cDNA was then amplified using Pfu DNA polymerase (Tiangen) with primers tsCD1dF0 paired and tsCD1dR0 (Table 1). The PCR conditions were: 95° for 30 seconds, 58° for 30 seconds, and 72° for 2 min. This sequence was repeated for 30 cycles followed by a further extension step for 10 min at 72°. The double-stranded cDNA was then added with an extra deoxyadenosine (A) at the 3′ end using Taq polymerase and cloned into the pMD19-T vector (Takara, Shiga, Japan). Cloned tsCD1d cDNAs from three individual animals were sequenced by Sunbiotech (Beijing, China), to confirm the sequence.
Table 1.
Nucleotide sequences
| tsCD1d primers | |
|---|---|
| cDNA | F0: 5′ TCACCGGATGTACTGATAGAAGCAGC 3′ |
| R0: 5′ ATGGGGT(AG)CCT(AG)C(CT)GT(GT)(CGT)CTG 3′ | |
| Wild type | F1: 5′ CCCAAGCTTGTCCCGCAAAGGCATTTCC 3′ |
| R1: 5′ GTCGAGGATCCCGGATGTACTGATAGAAGC 3′ | |
| Mutation Q86K | F2: 5′ GACGTACAGGAATTCGCCAAAATG 3′ |
| Mutation H89R | F3: 5′ CAGGAATTCGCCCAAATGATACGCTTAGTC 3′ |
| huCD1d primers | |
| Wild type | F1: 5′ CCCAAGCTTAGGCTTTTCCCCCTCCGC 3′ |
| R1: 5′ CGGGATCCAGGACGCCCTGATAGGA 3′ | |
| Mutation K86Q | F2: 5′ GACGTGAAGGAATTCGCCCAAATG 3′ |
| Mutation R89H | F3: 5′ AAGGAATTCGCCAAAATGCTACACTTATCC 3′ |
| CD5-FLAG sequence | |
| 5′GCTAGCTTCTAGAGTCCCTCGACCTCGAGATCCATTGTGCTCTAAAGGAGATACCCGGCCAGACACCCTCACCTGCGGTGCCCAGCTGCCCAGGCTGAGGCAAGAGAAGGCCAGAAACCATGCCCATGGGGTCTCTGCAACCGCTGGCCACCTTGTACCTGCTGGGGATGCTGGTCGCTTCCTGCCTCGGACGÿGCTAGCTTCTAGAGTCCCTCGACCTCGAGATCCATTGTGCTCTA 3′ | |
Expression vector construction
The expression vector pFLAG-GFP was generated by cloning the CD5 leader sequence linked to a FLAG tag (DYKDDDD) sequence into the pEGFP-N1 vector (Clontech, Mountain View, CA) using NheI/HindIII (Table 1). tsCD1d and huCD1d DNA sequences encoding the mature proteins (lacking the leader sequences) were amplified using Pfu DNA polyermase (Strategene, Dorchester, UK) with primers huCD1dF1/R1 and tsCD1dF1/R1 (Table 1), and then cloned into the pFLAG-GFP vector using HindIII/BamHI in frame with FLAG and green fluorescent protein (GFP) sequences. Point mutation constructs were generated by PCR-based site-directed mutagenesis and the PCR products were cloned into the pFLAG-GFP vector for expression (Table 1). The final constructs were sequenced to make sure no mutations had occurred during the PCR and cloning process.
Gene transfection, iNKT stimulation, flow cytometry analysis and cytokine detection
HEK293T cells were transfected with GeneJuice Transfection Reagent (Merck Biosciences, San Diego, CA), and cells were stained with anti-FLAG monoclonal antibody (mAb) M2 (Sigma-Aldrich, St Louis, MO), anti-huCD1d mAb CD1d42 (BD Pharmingen, Oxford, UK), 51.1 (eBioscience, San Diego, CA), NOR3.2 (AbD Serotec, Oxford, UK), or anti-msCD1d mAb 3C11 (BD Pharmingen), followed by allophycocyanin (APC)-conjugated anti-mouse secondary Ab (eBioscience) or phycoerythrin (PE)-conjugated anti-rat immunoglobulin secondary Ab (DAKO, Glostrup, Denmark). Cells were then fixed with 1% paraformaldehyde and acquired with a FACSCalibur™ (Becton Dickinson, Franklin Lakes, NJ). Fluorescence-activated cell sorter (FACS) data were analysed with Flowjo software (Tree Star, Inc., Ashland, OR).
iNKT stimulation and cytokine detection were performed as described previously.22 Briefly, iNKT cells were co-cultured with αGalCer-pulsed CD1d transfectants and treated with Brefeldin A (Sigma-Aldrich) for 5 hr before being permeabilized and stained with anti-hu-interferon (IFN)-γ, anti-hu-interleukin (IL)-4 and anti-CD3-APC mAbs (DAKO). For some experiments, iNKT cells were co-cultured with CD1d transfectants for 24 hr and the culture supernatant was then collected and analysed for cytokine secretion with an IFN-γ enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Minneapolis, MN).
Immunoprecipitation
CD1d transfected HEK293T cells were lysed with lysis buffer (50 mm HEPES, pH 7·4, 150 mm NaCl and 1% digitonin) supplemented with Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). After the removal of cell debris by spinning in a microcentrifuge, 50 μl of protein A Sepharose beads (Sigma-Aldrich) bound with 5 μg of anti-FLAG mAb was added and the samples kept rolling at 4° for 2 hr. The beads were then washed thoroughly with cold phosphate-buffered saline (PBS) before being re-suspended in loading buffer for sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were transferred to Hybond-C membrane (GE Healthcare, Amersham, UK) with a Trans-blot™ SD Semi-dry Transfer Cell (Bio-Rad, Hercules, CA), and blotted with anti-GFP mAb (Roche), and then detected with the ECL Western Blotting Detecting System (GE Healthcare).
Tree shrew CD1d modelling
The model of tree shrew CD1d was generated using the program swiss-model (Online resources)23 using the alignment interface and the human CD1d/TCR complex structure (2po6) as the template for the heavy chain. Figures were generated using CCP4mg24 and grasp.25
Results
tsCD1d is a close evolutionary relative of primate CD1ds
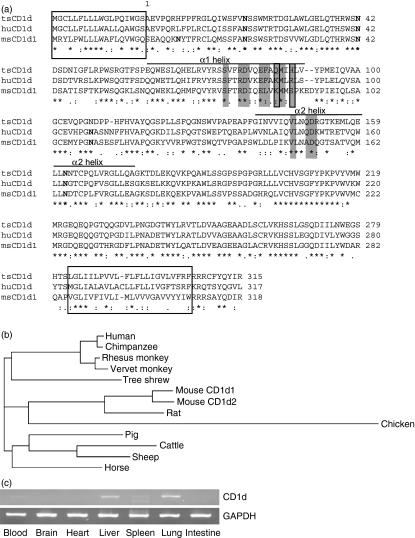
To clone the tsCD1d gene, we searched the major gene databases and found a transcript in the Ensembl Genome Browser (transcript ID: ENSTBET00000001753) containing the 3′ sequence of the tsCD1d cDNA. A 3′ primer based on this transcript was designed and paired with a degenerate 5′ primer derived from the CD1d sequences of human, rhesus monkey, mouse and rat, to perform degenerate PCR. Total RNA extracted from tree shrew spleen cells was used as a template and a band of ∼1·0 kb could be successfully amplified. PCR products from three individual animals were cloned and sequenced, and the consensus sequence showed a 1002-bp open reading frame (Genbank accession number FJ213842) with 81·7% identity to rhesus monkey CD1d, 81·3% to human CD1d, and 71·5% to mouse CD1d1. The cloned tsCD1d encoded a 333-amino acid polypeptide with 70% identity to rhesus monkey CD1d, 69% to human CD1d, and 56% to mouse CD1d1. By comparison with homologous proteins, we predicted that tsCD1d would encode a single transmembrane protein with a 18-amino acid signal peptide, followed by a 282-amino acid extracellular domain, a 23-amino acid transmembrane domain and a short, 10-amino acid cytoplasmic tail (Fig. 1a). The predicted tsCD1d protein had a predicted molecular weight of 37713 Da and pI = 6·60 as calculated using editseq software (Online resources). NetNGlyc (Online resources) predicted that it would have three potential N-linked glycosylation sites, in comparison with four and five for human and mouse CD1ds, respectively, and all three tree shrew sites were conserved in human and mouse CD1d proteins. NetOGlyc (Online resources) did not find any potential O-linked glycosylation site, nor did it find any with human or mouse CD1d.
Figure 1.
Tree shrew tsCD1d sequence and tissue distribution. (a) Amino acid sequence alignment of tree shrew CD1d with its human and mouse homologues. Signal peptide and transmembrane domain sequences are boxed and the potential N-glycosylation sites are shown in bold. Amino acid residues involved in direct contact with the T-cell receptor (TCR) are shaded and amino acids 86 and 89 are shaded and boxed. Consensus symbols: ‘*’ stands for identical substitutions, ‘:’ for conserved substitutions, and ‘.’ for semi-conserved substitutions. (b) Phylogram of CD1d amino acid sequences from different species. The alignments and phylogenic analysis were performed using CLUSTALW2 at EMBL-EBI (Online resources). (c) Tissue distribution of CD1d in tree shrew detected using semiquantitive reverse transcription–polymerase chain reaction (RT-PCR). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. The CD1d fragment is 1002 bp, and GAPDH is 669 bp.
Phylogenetic analysis of the tsCD1d amino acid sequence compared with those of other animals found that tsCD1d was more closely related to primate CD1ds (human, chimpanzee, rhesus monkey and vervet monkey) than to those of rodents (rat and mouse), artiodactyls (pig, sheep and cattle) or perissodactyls (horse) (Fig. 1b). This was consistent with the findings of previous studies which showed that tree shrew MHC-I molecules and p53 protein had higher homology with their counterparts from primates than with those from any other animals.26,27 Therefore, CD1d provides further molecular evidence for the evolutionary proximity between tree shrews and primates.
Human CD1d has been reported to be expressed on myeloid cells, such as monocytes, macrophages and DCs, as well as lymphoid cells such as B cells and activated T cells.1 In addition, CD1d is also expressed on non-haemotopoietic cells, such as epithelial cells, parenchymal cells, and vascular smooth muscle cells in the gut and liver.28 To investigate whether tsCD1d has a similar expression profile, we used RT-PCR to examine the tissue distribution of tsCD1d. We found that tsCD1d mRNA could be detected in tissues such as blood, lung, spleen and liver, with the highest levels found in the liver and lung (Fig. 1c). As iNKT cells were suggested to be involved in host defence against hepatitis virus17 and antitumour immunity against hepatocellular carcinoma,18,20,21 the expression of CD1d in tree shrew liver makes its role in liver physiology and pathophysiology more interesting.
tsCD1d does not activate human iNKT cells
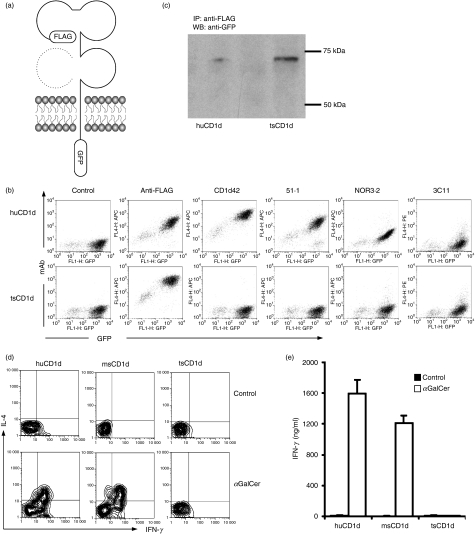
CD1d proteins present lipid antigens to iNKT cells from the cell surface of antigen presenting cells (APCs). To confirm that tsCD1 can be expressed on the cell surface, we generated an expression construct in which a FLAG tag was added to the N terminal (the extracellular region) of tsCD1d, and a GFP reporter gene to the C terminal (the cytoplasmic domain) (Fig. 2a). This construct, along with a control vector that contained huCD1d tagged in the same way, was transfected into HEK293T cells, and their expression on the cell surface was detected by immunofluorescence staining and flow cytometry analysis. huCD1d expression on the surface of transfected cells could be detected by both the huCD1d-specific mAb CD1d42 and the FLAG tag-specific mAb M2. tsCD1d, however, could only be detected by M2 mAb (Fig. 2b). M2-positive staining suggested that the transfected tsCD1d was expressed on the cell surface, and CD1d42-negative staining indicated that this mAb did not cross-react with tsCD1d. To find a specific mAb that can be used to detect tsCD1d expression on native tissues, we checked a panel of mAbs, including huCD1d-specific mAbs 51.1 and d3.2, and msCD1d-specific mAb 3C11, for their cross-reactivity with tsCD1d. However, none of them showed positive staining for tsCD1d transfectants (Fig. 2b).
Figure 2.
Expression and function of tree shrew tsCD1d. (a) Schematic illustration of FLAG-CD1d-GFP construct in which FLAG and green fluorescent protein (GFP) tags were added on either side of tsCD1d or human huCD1d for expression in mammalian cells. Dotted structure represents β2 microglobulin (β2m), which is non-covalently associated with CD1d during protein expression. (b) Fluorescence-activated cell sorter (FACS) plots showing anti-FLAG, anti-huCD1d and anti-msCD1d monoclonal antibody (mAb) staining of tsCD1d and huCD1d transfected HEK293T cells. (c) Immunoprecipitation of tsCD1d and huCD1d protein from their transfectants was performed using FLAG mAb, and western blotting was performed with GFP mAb subsequently. Molecular weight standards are labelled on the right (75 and 50 kDa). (d, e) Stimulation of human invariant natural killer T (iNKT) cells by CD1d transfectants. HEK293T cells transfected with human, mouse or tree shrew CD1d were pulsed with α-galactosylceramide (αGalCer) (10 ng/ml) to stimulate human iNKT cells. The experiment was repeated in triplicate, and each time with three different iNKT cell lines/clones (NKN, MX and MT2). Results for one representative iNKT clone from one experiment are shown. (d) Intracellular staining of interferon (IFN)-γ and interleukin (IL)-4 production from CD1d-stimulated human iNKT cells. (e) Enzyme-linked immunosorbent assay (ELISA) detection of IFN-γ release from human iNKT cells stimulated by CD1d transfectants.
To confirm the identify of the FLAG mAb-recognized protein on the cell surface of tsCD1d transfectants, the cells were subjected to immunoprecipitation with anti-FLAG mAb, and then the bound protein was detected using anti-GFP mAb in western blotting. Both huCD1d and tsCD1d showed single bands at ∼70 kDa (Fig. 2c), which was roughly the combined size of the FLAG tag (1 kDa), the GFP protein (27 kDa) and the glycosylated huCD1d (43 kDa). The fact the tsCD1d has a similar molecular weight to huCD1d, despite the fact that it has one less potential N-linked glycosylation site, suggests that either the extra potential site in the human molecule is not heavily glycosylated, or that proteins migrate differently through the gel as a result of to their differences in pI.
The CD1d/iNKT system is regarded as a conserved branch of the innate system, and cross-species reactivity between humans and mice is well documented.4,8,12 Therefore, it was considered likely that tsCD1d would be able to present antigens to iNKT cells of human or other origin. To test this hypothesis, we used expressional constructs of CD1d molecules from human, mouse and tree shrew to transfect cells, which were subsequently used as APCs to present the model antigen αGalCer to human iNKT cells. As expected, both human and mouse CD1d stimulated human iNKT cell activation, which led to the production of cytokines detected by both intracellular cytokine staining and ELISA (Fig. 2d, e). However, tsCD1d did not show any detectable stimulation of human iNKT cells. This was not a result of the variability of iNKT cells, because all three iNKT clone/cell lines tested gave the same negative results. We therefore concluded that tsCD1d could not cross-stimulate human iNKT cells.
A key amino acid determines tsCD1d cross-species antigen presentation
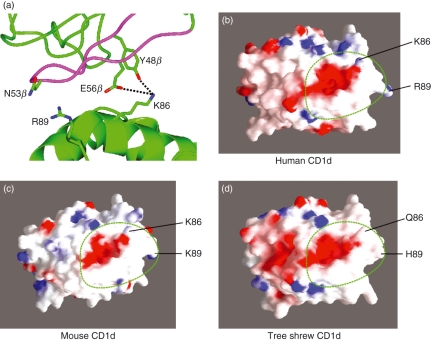
To investigate the mechanism behind the inability of tsCD1d to cross-stimulate human iNKT cells, we compared among different species the amino acid residues that were shown to be in direct TCR contact in the human iNKT–CD1d co-crystal structure.29 We found that tsCD1d and huCD1d had most of the key amino acid residues conserved, including those in the α1 helix: Ser76, Arg79, Asp80, Glu83, Phe84 and Met87, as well as those in the α2 helix: Val147, Gln150 and Asp151 (Fig. 1a). A notable difference was that at position 86, where both human and mouse CD1ds had lysine residues and tsCD1d had a Gln residue, and at position 89, where the three CD1d molecules had completely different amino acids. In the co-crystal structure of iNKT TCR with huCD1d, Lys86 was found to be in direct contact with CDR2β, interacting with the Tyr48β hydroxyl via a hydrogen bond and Glu56β via a salt bridge, while Arg89 forms van der Waals with Asn53 from CDR2β (Fig. 3a).29 Modelling the tree shrew CD1d and electrostatic molecular surface analysis of human, mouse and tree shrew CD1ds revealed that the K86Q change affected the surface structure and charge distribution within the TCR binding footprint (Fig. 3b–d). Furthermore, a report indicated that the substitution of Lys86 with Ala caused a fivefold decrease in the binding affinity of huCD1d to an iNKT TCR.30 We therefore hypothesized that the inability of tsCD1d to stimulate human iNKT was probably attributable to the naturally occurring variations at amino acid position 86, and possibly also position 89.
Figure 3.
(a) Structure of the human CD1d–T-cell receptor (TCR) complex (PDB code 2PO6), illustrating the TCR Vb domain (green) and CDR2 loop (magenta) contacts made with the two variant amino acids between human huCD1d and tree shrew tsCD1d. (b–d) Electrostatic surface representations of (b) human (PDB code 2PO6), (c) mouse (PDB code 1ZT4), and (d) tree shrew (model) CD1d structures illustrating the positions of residues 86 and 89 and the overall docking position of the invariant TCR footprint (dotted green line).
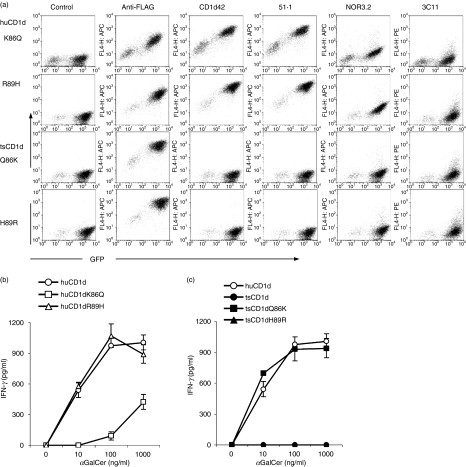
To test this hypothesis, we first generated expression constructs for huCD1d in which Lys86 and Arg89 were replaced with Gln and His, respectively, as they were in tsCD1d, and used these construct-transfected cells as APCs to stimulate human iNKT cells. These constructs were found to be expressed on the cell surface as detected by anti-FLAG mAb (Fig. 4a). They could also be stained with a panel of CD1d mAbs showing similar profiles to that of wild-type CD1d, suggesting that these mutations did not change the overall structure of the molecules (Figs. 4a and 2b). However, when these transfectants were used to stimulate human iNKT cells, it was found that the mutation at Lys86 to Gln (huCD1dK86Q) significantly reduced the ability of huCD1d to present αGalCer to human iNKT cells, especially at low αGalCer concentrations, where stimulation was almost undetectable (Fig. 4b). In contrast, the substitution of Arg89 with His (huCD1dR89H) did not cause a significant reduction in iNKT cytokine secretion (Fig. 4b). These results suggest that Lys86 is very important for recognition between CD1d and human iNKT cells and its replacement with an acidic amino acid effectively destroys this interaction. However, the replacement of Arg89 with His does not seem to be the major cause of the inability of tsCD1d to cross-stimulate human iNKT cells, perhaps because both amino acids are positively charged and therefore the substitution does not change the overall surface charge of the molecule.
Figure 4.
Amino acid 86 defines human invariant natural killer T (iNKT) reactivity. (a) Fluorescence-activated cell sorter (FACS) plots showing anti-FLAG, anti-human CD1d (huCD1d) and anti-mouse CD1d (msCD1d) monoclonal antibody (mAb) staining of tree shrew tsCD1d and huCD1d mutant transfected HEK293T cells. (b) Wild-type and mutant huCD1d (b) or tsCD1d (c) -transfected HEK293T cells were pulsed with α-galactosylceramide (αGalCer) at the indicated concentrations, before being used to stimulate human iNKT cells. Interferon (IFN)-γ concentrations from the culture supernatant were measured by enzyme-linked immunosorbent assay (ELISA). Three different iNKT cell lines/clones (NKN, MX and MT2) were used in four repeated experiments and results from one representative experiment are shown.
Next we mutated the corresponding amino acids in tsCD1d back to those of huCD1d and tested their effects on the ability of tsCD1d to cross-stimulate human iNKT cells. Again, the mutated tsCD1d was strongly expressed on the cell surface, as detected by FLAG tag-specific mAb, and it still could not be recognized by mAbs to huCD1d or msCD1d (Fig. 4a). When transfectants with similar levels of CD1d expression were used to stimulate human iNKT cells, it was found that, while wild-type tsCD1d could not stimulate human iNKT cells at all, a Gln86 to Lys mutation (tsCD1dQ86K) made tsCD1d as competent as huCD1d in presenting αGalCer to human iNKT cells. However, a His89 to Arg mutation (tsCD1dH90R) did not seem to have any impact, even when the concentration of αGalCer was as high as 1000 ng/ml (Fig. 4c). These results demonstrate that Lys86 plays an indispensable role in human CD1d–iNKT recognition, and its substitution by glutamine is the main reason for the inability of tsCD1d to cross-stimulate human iNKT cells.
Discussion
CD1 genes have been found in various primates, rodents and other species.1,31,32 While most of these animals have both group 1 CD1 (CD1a, CD1b and CD1c) and group 2 CD1 (CD1d), rodents lost the group 1 genes during evolution, apparently as a result of a chromosomal break event.33,34 This study is the first report of the CD1 system in the tree shrew, a close relative of primates that has the potential to replace them as experimental animals in certain research areas.
The sequence of tsCD1d shows higher homology to primate CD1ds than those of rodents, confirming the proximal genetic association of the tree shrew with humans. However, the inability of tsCD1d to present αGalCer to human iNKT cells suggests that the CD1d/iNKT system in the tree shrew is significantly different from those in humans and mice.
The first question is whether tsCD1d can still bind and present αGalCer in a similar conformation to that of its counterparts in other animals. The ability of the tsCD1d Q86K to stimulate human iNKT cells indicates that at least this mutant CD1d presents αGalCer in an analogous conformation to that of huCD1d. Despite numerous polymorphic differences between huCD1d and tsCD1d within the lipid-binding groove, human iNKT cells are broadly insensitive to these polymorphisms in the context of αGalCer. However, conservation of key amino acids in the TCR binding footprint is critical for functional cross-recognition of the various species’ CD1d molecules by the conserved iNKT TCR, suggesting that, in species where the conserved TCR arises, there has been evolutionary conservation of CD1d residues participating directly in TCR recognition.30 As, structurally, Lys86 lies in the TCR interface and is not involved in binding αGalCer directly,29,35 it is unlikely that this mutation affects the ability of CD1d to bind αGalCer. Therefore, it is likely that wild-type tsCD1d binds αGalCer, and its failure to stimulate human iNKT cells is attributable to the changes at the TCR interface caused by amino acid differences. At the same time, it also suggests that tree shrew iNKT cell TCRs may have significant structural differences from those of humans and mice. Previous studies have shown that the CDR3α, CDR1α and CDR2β loops are the conserved hot-spots between human and mouse iNKT TCRs, and these are the main factors enabling cross-species reactivity between these two species.11,36 The inability of tsCD1d to cross-react with human iNKT cells, and the importance of Lys86, which is in direct contact with CDR2β, suggest that if the tree shrew has iNKT cells, their TCR CDR2β region must have adapted during evolution to accommodate this difference. The identification of tsCD1d-restricted iNKT cells and elucidation of the structure of their TCRs will help to answer these questions. In a similar case, rat iNKT cells are found not to cross-react with mouse CD1d, and their CDR2β plays a decisive role in this phenomenon.36 Therefore, although CD1/iNKT systems have been broadly conserved during evolution, there are some discrepancies among species, and CDR2β may be one of the hot-spots for these variations that contribute to species-specific CD1d-mediated antigen presentation.
It is interesting to note that the single amino acid mutation of Lys86 to Gln in huCD1d can dramatically reduce its ability to stimulate iNKT cells, as shown in Fig. 4. In fact, with some human iNKT clones we tested, this mutation caused complete abolishment of iNKT activation (data not shown). HuCD1d Lys86 is one of the major amino acids involved in TCR/CD1d contact and it interacts with two different amino acids, TCR CDR2b Tyr48β and Glu56β, via a hydrogen bond and a salt bridge, respectively. Therefore, when it is replaced with Gln, whose inability to form the same hydrogen bond and salt bridge means that it loses contact with Tyr48β, and the salt bridge with TCR Glu56β, the CDR2β of the iNKT TCR consequently loses one of the major binding sites on the CD1d surface. Conventional MHC class I molecules would be more tolerant of this type of mutation because the peptide antigens are the focus of contacts with both TCR and human leucocyte antigen (HLA)-I, and consequently minor changes at the HLA-I interface could be ‘compensated’ by the binding forces between the peptide and the TCR.37,38 In the case of CD1d, however, the head group of αGalCer makes only a very minor contribution to the overall binding energy of the complex, shown also in the co-structure of CD1d-iNKT TCR,29 and therefore the interaction between the TCR and CD1d proteins is more important in maintaining the complex, and mutations affecting the interaction between these proteins will have a more disruptive effect than that for HLA-I and TCR.
Although in a previous study analysis of surface plasmon resonance suggested that Lys86 had a moderate effect on the CD1d/TCR interaction,30 our results indicate that, functionally, this mutation is much more detrimental. This is probably why Lys86 is so conserved in evolution: of all the CD1d genes cloned from different species to date, lysine is preserved except in chicken, cattle and tree shrew CD1ds (Fig. 5).
Figure 5.
Alignment of CD1d α1 helix amino acid sequences from different species. Shaded residues are amino acids predicted to be in direct contact with the T-cell receptor (TCR) based on human CD1d/TCR co-structure.
It is worth mentioning that there are significant variations among different iNKT clones in terms of their reactivity towards human and tree shrew CD1d mutants. According to our data (not shown), some clones also showed reactions to tsCD1dH89R mutants, although at much lower levels compared with tsCD1dQ86K, suggesting that the amino acid at position 89 may also contribute to the iNKT–CD1d interaction, at least within a certain subpopulation of iNKT cells. This is in agreement with findings from previous studies.11,30
The inability of tsCD1d to recognize human iNKT TCRs also raises the possibility that the corresponding T-cell population in tree shrew may fail to be selected during positive selection in the thymus, consequently are devoid in the peripheral lymphoid tissue. Another possibility is that, as a result of the mutation of the key amino acid in tsCD1d, the T-cell population positively selected by tsCD1d in the thymus possesses a different TCR structure from that of other animals, and consequently these cells may not behave like iNKT cells. These are all interesting questions that require further study.
In summary, we report the cDNA sequence of tsCD1d, which shows high homology to those of primates and rodents, but has no cross-species reactivity with human iNKT cells. The substitution of key amino acids in the CDR2β-interacting region generally restores the ability of tsCD1d to cross-react with human iNKT cells. These findings demonstrate that the interaction between CD1d and iNKT TCR is highly specific, and the cross-species reactivity observed between human and mouse is probably an exception rather than a common phenomenon. More importantly, such a naturally occurring CD1d mutation(s) that confers an inability to stimulate iNKT cell function may have implications for future studies on CD1d/iNKT-associated diseases.
Acknowledgments
This work was supported by the UK Medical Research Council, Chinese National 973 Program (2007CB512807) and Chinese Academy of Sciences grants for 100’s Talents and Overseas Scholar Team. And we thank Miss Cecilia Chui for critical reading of the manuscript.
Disclosures
The authors have no conflict of interest.
References
- 1.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–90. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–41. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 3.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–7. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 5.Li D, Xu XN. NKT cells in HIV-1 infection. Cell Res. 2008;18:817–22. doi: 10.1038/cr.2008.85. [DOI] [PubMed] [Google Scholar]
- 6.Kakimi K, Guidotti LG, Koezuka Y, Chisari FV. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J Exp Med. 2000;192:921–30. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–64. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Motsinger A, Azimzadeh A, Stanic AK, Johnson RP, Van Kaer L, Joyce S, Unutmaz D. Identification and simian immunodeficiency virus infection of CD1d-restricted macaque natural killer T cells. J Virol. 2003;77:8153–8. doi: 10.1128/JVI.77.14.8153-8158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashiwase K, Kikuchi A, Ando Y, et al. The CD1d natural killer T-cell antigen presentation pathway is highly conserved between humans and rhesus macaques. Immunogenetics. 2003;54:776–81. doi: 10.1007/s00251-002-0527-8. [DOI] [PubMed] [Google Scholar]
- 10.Gansuvd B, Hubbard WJ, Hutchings A, Thomas FT, Goodwin J, Wilson SB, Exley MA, Thomas JM. Phenotypic and functional characterization of long-term cultured rhesus macaque spleen-derived NKT cells. J Immunol. 2003;171:2904–11. doi: 10.4049/jimmunol.171.6.2904. [DOI] [PubMed] [Google Scholar]
- 11.Kjer-Nielsen L, Borg NA, Pellicci DG, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–73. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidobre S, Kronenberg M. CD1 tetramers: a powerful tool for the analysis of glycolipid-reactive T cells. J Immunol Methods. 2002;268:107–21. doi: 10.1016/s0022-1759(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 13.Vollmayr B, Mahlstedt MM, Henn FA. Neurogenesis and depression: what animal models tell us about the link. Eur Arch Psychiatry Clin Neurosci. 2007;257:300–3. doi: 10.1007/s00406-007-0734-2. [DOI] [PubMed] [Google Scholar]
- 14.Metlapally S, McBrien NA. The effect of positive lens defocus on ocular growth and emmetropization in the tree shrew. J Vis. 2008;8:1–12. doi: 10.1167/8.3.1. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Yang EB, Su JJ, Li Y, Chow P. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J Med Primatol. 2003;32:123–30. doi: 10.1034/j.1600-0684.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 16.Baron JL, Gardiner L, Nishimura S, Shinkai K, Locksley R, Ganem D. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity. 2002;16:583–94. doi: 10.1016/s1074-7613(02)00305-9. [DOI] [PubMed] [Google Scholar]
- 17.Mizrahi M, Lalazar G, Ben Ya’acov A, et al. Beta-glycoglycosphingolipid-induced augmentation of the anti-HBV immune response is associated with altered CD8 and NKT lymphocyte distribution: a novel adjuvant for HBV vaccination. Vaccine. 2008;26:2589–95. doi: 10.1016/j.vaccine.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Shibolet O, Alper R, Zlotogarov L, Thalenfeld B, Engelhardt D, Rabbani E, Ilan Y. NKT and CD8 lymphocytes mediate suppression of hepatocellular carcinoma growth via tumor antigen-pulsed dendritic cells. Int J Cancer. 2003;106:236–43. doi: 10.1002/ijc.11201. [DOI] [PubMed] [Google Scholar]
- 19.Shibolet O, Alper R, Zlotogarov L, Thalenfeld B, Engelhardt D, Rabbani E, Ilan Y. Suppression of hepatocellular carcinoma growth via oral immune regulation towards tumor-associated antigens is associated with increased NKT and CD8+ lymphocytes. Oncology. 2004;66:323–30. doi: 10.1159/000078334. [DOI] [PubMed] [Google Scholar]
- 20.Margalit M, Shibolet O, Klein A, et al. Suppression of hepatocellular carcinoma by transplantation of ex-vivo immune-modulated NKT lymphocytes. Int J Cancer. 2005;115:443–9. doi: 10.1002/ijc.20889. [DOI] [PubMed] [Google Scholar]
- 21.Ito H, Ando K, Ishikawa T, et al. Role of Valpha14+ NKT cells in the development of Hepatitis B virus-specific CTL: activation of Valpha14+ NKT cells promotes the breakage of CTL tolerance. Int Immunol. 2008;20:869–79. doi: 10.1093/intimm/dxn046. [DOI] [PubMed] [Google Scholar]
- 22.Li D, Chen N, McMichael AJ, Screaton GR, Xu XN. Generation and characterisation of CD1d tetramer produced by a lentiviral expression system. J Immunol Methods. 2008;2:57–63. doi: 10.1016/j.jim.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 24.Potterton L, McNicholas S, Krissinel E, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;1:2288–94. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–96. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 26.Flugge P, Fuchs E, Gunther E, Walter L. MHC class I genes of the tree shrew Tupaia belangeri. Immunogenetics. 2002;11:984–8. doi: 10.1007/s00251-001-0401-0. [DOI] [PubMed] [Google Scholar]
- 27.Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–54. [PubMed] [Google Scholar]
- 28.Canchis PW, Bhan AK, Landau SB, Yang L, Balk SP, Blumberg RS. Tissue distribution of the non-polymorphic major histocompatibility complex class I-like molecule, CD1d. Immunology. 1993;80:561–5. [PMC free article] [PubMed] [Google Scholar]
- 29.Borg NA, Wun KS, Kjer-Nielsen L, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–9. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 30.Wun KS, Borg NA, Kjer-Nielsen L, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–49. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angenieux C, Salamero J, Fricker D, Cazenave JP, Goud B, Hanau D, de La Salle H. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem. 2000;275:37757–64. doi: 10.1074/jbc.M007082200. [DOI] [PubMed] [Google Scholar]
- 32.Consortium CSaA Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 33.Moseley WS, Watson ML, Kingsmore SF, Seldin MF. CD1 defines conserved linkage group border between human chromosomes 1 and mouse chromosomes 1 and 3. Immunogenetics. 1989;30:378–82. doi: 10.1007/BF02425278. [DOI] [PubMed] [Google Scholar]
- 34.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–8. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 35.Koch M, Stronge VS, Shepherd D, et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–26. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 36.Pyz E, Naidenko O, Miyake S, Yamamura T, Berberich I, Cardell S, Kronenberg M, Herrmann T. The complementarity determining region 2 of BV8S2 (V beta 8.2) contributes to antigen recognition by rat invariant NKT cell TCR. J Immunol. 2006;176:7447–55. doi: 10.4049/jimmunol.176.12.7447. [DOI] [PubMed] [Google Scholar]
- 37.Ishizuka J, Stewart-Jones GB, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28:171–82. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]