Abstract
Premutation CGG repeat expansions (55–200 CGG repeats; preCGG) within the fragile X mental retardation 1 (FMR1) gene give rise to the neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS), primary ovarian insufficiency and neurodevelopmental problems. Morphometric analysis of Map2B immunofluorescence reveals that neurons cultured from heterozygous female mice with preCGG repeats in defined medium display shorter dendritic lengths and fewer branches between 7 and 21 days in vitro compared with wild-type (WT) littermates. Although the numbers of synapsin and phalloidin puncta do not differ from WT, preCGG neurons possess larger puncta. PreCGG neurons display lower viability, and express elevated stress protein as they mature. PreCGG neurons have inherently different patterns of growth, dendritic complexity and synaptic architecture discernable early in the neuronal trajectory to maturation, and may reflect a cellular basis for the developmental component of the spectrum of clinical involvement in carriers of premutation alleles. The reduced viability of preCGG neurons is consistent with the mRNA toxicity and neurodegeneration associated with FXTAS.
INTRODUCTION
Fragile X syndrome (FXS) is the most common inherited form of cognitive impairment and a leading single-gene disorder associated with autism (1,2). FXS is caused by trinucleotide CGG repeat expansions within the 5′ non-coding region of the fragile X mental retardation 1 (FMR1) gene that exceed 200 repeats (full mutation), at which point the promoter region of the gene generally becomes hypermethylated, leading to gene silencing and absence of the fragile X mental retardation protein (FMRP) (3–5). The frequency of full mutation alleles in the general population is estimated at approximately 1 out of 2500 individuals (male and female) (6,7). Individuals with smaller (premutation) expansions (55–200 CGG) are typically unaffected by FXS. However, such carriers do display a range of clinical features, including behavioral and cognitive involvement in children (8–11), primary ovarian insufficiency (POI) in women (12) and a late-adult-onset neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS) (13–15).
At least 40% of male premutation carriers and ∼8–16% of female carriers over 50 years of age, in families ascertained through a child with FXS, will develop clinical features of FXTAS, with phenotypic penetrance increasing with age (6,16–18). Clinical manifestations of FXTAS include the core features of progressive intention tremor and/or gait ataxia, frequently accompanied by Parkinsonism, neuropathy and progressive cognitive impairment (13,19,20) in addition to brain atrophy and white matter disease on magnetic resonance imaging (MRI) (21). In addition to the major premutation-associated disorders, POI and FXTAS, more recently described clinical features have been described in association with premutation alleles, particularly among females; such features include thyroid disease, hypertension, seizures, peripheral neuropathy and fibromyalgia (16,17). Importantly, both POI and FXTAS are unique to the premutation range, which implies that FMRP deficiency, per se, is not responsible for those disorders. Instead, evidence to date from both human and animal studies indicates that these premutation-specific disorders are caused by a direct toxic gain-of-function of the premutation CGG (preCGG) repeat FMR1 mRNA (4,20,22,23). Reductions in hippocampal volume, activation and associated memory deficit, as well as reduced amygdala activation (24–27) and psychopathology (28) appear much earlier in adulthood than do the symptoms of FXTAS (20), suggesting that the processes that ultimately will lead to FXTAS may be operating at a much earlier age. The reports of attention-deficit hyperactivity disorder (ADHD) and autism spectrum disorders (ASD) in young boys with the premutation also suggest a neurodevelopmental component to the premutation (10,29,30).
A knock-in (KI) mouse model with preCGG repeat expansion (∼70–135 CGG repeats) in the homologous Fmr1 gene was shown to express 2∼3.5-fold elevated Fmr1 mRNA, a reduced FMRP level and ubiquitin-positive inclusion bodies in the brain (31–34). Although this mouse does not recapitulate the human disease FXTAS (35), the mice do show age-dependent cognitive decline and neuromotor disturbances (36). In addition, several hormonal abnormalities were reported, including higher serum stress hormone levels and altered regulation of the HPA axis (31).
Here, we report that hippocampal neurons cultured from heterozygous female mice with one Fmr1 allele in the high-premutation range (155–200 CGG repeats, designated preCGG) exhibit elevated Fmr1 mRNA but only modest reductions of FMRP. Compared with neurons cultured from WT littermates, preCGG neurons have (i) early developmental deficits in attaining dendritic complexity, (ii) altered synaptic morphology, (iii) elevated mRNA and proteins expression levels of Cryab (alpha B-crystallin gene), Hsp27 and Hsp70 stress markers by 14 days in vitro (DIV), (iv) decreased viability that becomes apparent at 21 DIV and is pronounced by 28 DIV, and (v) incomplete X-chromosome inactivation (XCI) expression of the affected allele in enough cells to permit significant phenotypic penetrance of the premutation allele in females. Low-density hippocampal neuronal cultures from FXTAS mice recapitulate neurodevelopmental and neurodegenerative aspects of FXTAS in vivo and have several major theoretical and practical implications.
RESULTS
PreCGG neurons express elevated Fmr1 mRNA but only modestly reduced levels of FMRP
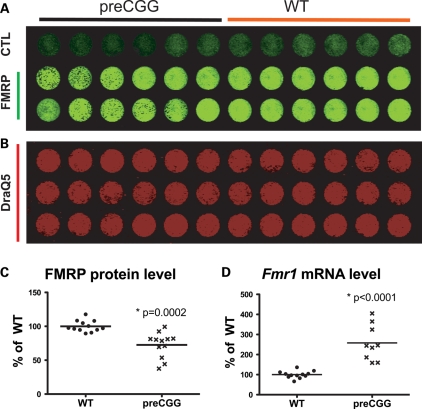
Primary hippocampal cultures grown in serum-free medium can serve as a model system to study neuronal development, synapse formation and neurotransmission (37,38). Murine hippocampal neurons were prepared on postnatal day 0∼2 from heterozygous preCGG (155–200 repeats) and their WT littermates. Hemizygous male with WT female mice serve as our breeding pairs. This allows us to identify heterozygous female litters as KI heterozygous preCGG and male litter as WT the day they are born without prerequisite genotyping prior to making low-density cultures of primary hippocampal cultures. The design also permits evaluation of the preCGG allele's phenotypic penetrance, or whether XCI is sufficiently skewed against the preCGG allele to mask the phenotype. Hippocampal cultures grown in microtiter plates were fixed at 14 DIV and stained with a polyclonal chicken antibody that binds FMRP. Utilizing an in situ western blotting method, commonly termed ‘in-cell western’ (39), we quantified the level of FMRP expression within its neuronal context. Using this method, the ratio of each FMRP signal to that of nuclear staining with DraQ5 (40) provides a quantitative way to normalize differences in cell density among culture wells. As shown in Figure 1A–C, heterozygous preCGG neurons express 72.6 ± 5.4% of the FMRP levels found in WT littermate neurons. Results from RT–PCR analyses show that heterozygous preCGG neurons (155–200 CGG repeats) show 2.6 ± 0.32-fold higher Fmr1 mRNA levels than the WT littermate controls (Fig. 1D). These observations are consistent with the previous findings of reduced FMRP and elevated Fmr1 mRNA levels in brain lysates prepared from heterozygous preCGG KI mice expressing 170 heterozygous preCGG (4,31,32), and also consistent with findings from human premutation carriers and FXTAS patients (9,23). Therefore, the presence of both significantly elevated Fmr1 mRNA and mildly reduced FMRP establishes the heterozygous preCGG KI neurons a good model to study the neuropathology and the underlying molecular pathogenic mechanism(s) involved with FXTAS and earlier manifestations of the expression of premutation alleles.
Figure 1.
Heterozygous preCGG neurons express more Fmr1 mRNAs and less FMRP proteins compared with WT littermate neurons. Fourteen DIV hippocampal neurons from heterozygous preCGG and WT littermate mice were fixed in a 96-well plate, and in-cell western analysis performed using chicken anti-FMRP antibody. (A) The 800 nm infrared wavelength signal detected by the LI-COR Odyssey scanner. The first row was stained with a secondary antibody (goat anti-chicken 800) without a primary antibody (chicken FMRP antibody). Rows 2 and 3 show FMRP protein levels expressed by the heterozygous preCGG (left six columns) and WT (right six columns) neuron-enriched low-density cultures. (B) DraQ5 nuclear staining at 700 nm, which serves to normalize FMRP signals to cell density. (C) Quantification of the signal ratio of the two channels (800:700 nm) using Odyssey 2.1 software, and the heterozygous preCGG results are presented as percentage of WT. Heterozygous preCGG KI neurons show 72.6 ± 5.4% of total FMRP level compared with WT littermate neurons 100.0 ± 2.4% (N = 3 separate cultures, four determinations per culture) (P = 0.0002). (D) Fmr1 mRNA-level comparison between heterozygous preCGG KI neurons and WT at 14 DIV after plating. Quantitative RT–PCR was performed and Fmr1 mRNA level was measured; heterozygous preCGG neurons show 2.6 ± 0.3-fold higher Fmr1 mRNA level than the WT littermate control (P < 0.0001). The P-values are calculated using two-way ANOVA based on RT–PCR results obtained from two independent cultures derived from WT and preCGG littermates (N = 2), each performed in duplicate with three starting RNA concentrations (see Materials and Methods for details).
PreCGG neurons have reduced dendritic complexity throughout in vitro development
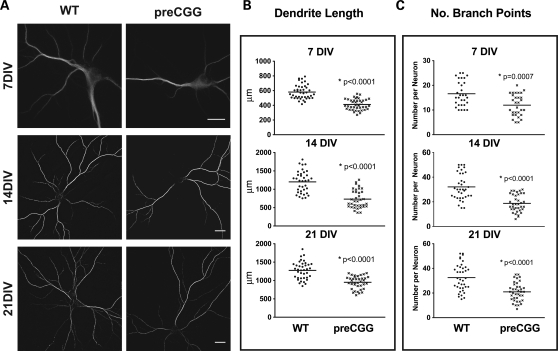
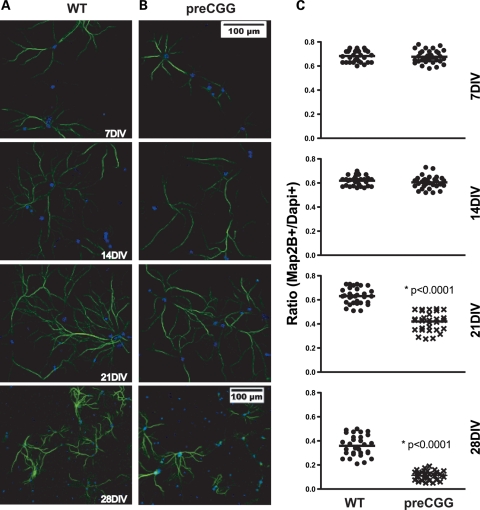
We investigated whether hippocampal neurons from premutation mice exhibited patterns of dendritic growth and complexity different from those of WT littermates by staining the cells with an antibody that specifically recognizes the microtubule-associated protein Map2B (Fig. 2A). The heterozygous preCGG neurons consistently showed reduced dendritic growth and branching complexity that manifests early in their developmental trajectory. Figures 2B and C are the results of morphometric analyses at 7, 14 and 21 DIV. At 7 DIV, the average dendritic length is 35% shorter (P < 0.0001), with 39% fewer branching points (P = 0.0007) in preCGG neurons compared with WT. By 21 DIV, preCGG neurons extend their total dendritic length 2.5-fold and increase their number of dendritic branch points 2-fold, indicating an inherent ability of these neurons to achieve a growth trajectory in vitro. Nevertheless, preCGG neurons lagged behind WT neurons, only reaching 80% of the total dendritic length and 57% of the dendritic branch points measured in WT neurons isolated from littermates by 21 DIV (Fig. 2 B and C). These results represent the first demonstration of inherent impairments in the developmental trajectory of neurons cultured from preCGG repeat expansion mice.
Figure 2.
Heterozygous preCGG KI neurons show significantly reduced dendritic complexity throughout in vitro development. Hippocampal neuronal cultures from heterozygous preCGG mice and WT littermates were fixed weekly after plating. Immunofluorescent labeling was performed using Map2B antibody. (A) Representative images from the two different genotypes at three time points. (B and C) Quantification of dendrite length and the number of branch points of individual neurons, respectively, calculated with Imaris FilamentTracer software as described in Materials and Methods. The heterozygous preCGG neurons show delayed dendritic growth from an early developmental stage (7 DIV) throughout their maturation (21 DIV). N = 3 separate cultures with 30–38 randomly selected neurons for each time points for each genotype. Scale bar: 20 µm.
PreCGG neurons have increased presynaptic synapsin and postsynaptic phalloidin puncta volumes
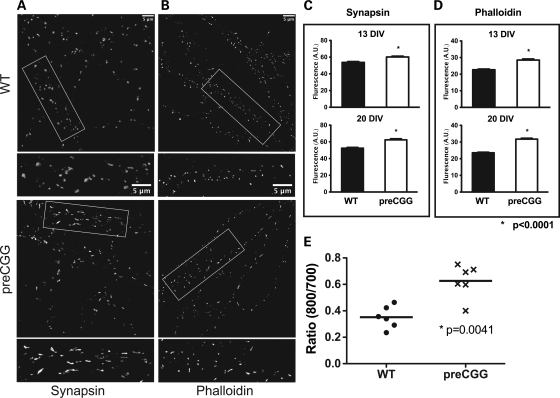
Synaptic architecture of cultured neurons at 14 and 21 DIV was examined by morphometric analysis for patterns of synapsin and phalloidin labeling. Synapsin is a presynaptic vesicle protein that labels presynaptic termini, whereas phalloidin binds to F-actin, a key component of the postsynaptic compartment. The synapsin and phalloidin puncta were quantified blinded to genotype. Heterozygous preCGG neurons were found to have larger synapsin and phalloidin puncta compared with the WT at both 14 and 21 DIV (Fig. 3), whereas the average intensity and total counts did not differ between genotype (data not shown). Interestingly, the divergence between heterozygous preCGG KI and WT in mean synapsin and phalloidin puncta size remained similar between 14 and 21 DIV, suggesting that alterations in presynaptic and postsynaptic architecture associated with heterozygous preCGG neurons manifest early in the maturation process and are sustained. Figure 3E summarizes results from in-cell western experiments showing that, at 14 DIV, heterozygous preCGG hippocampal neurons have higher levels of synapsin expression than WT neurons cultured from littermates.
Figure 3.
Neurons from heterozygous preCGG KI mice have increased presynaptic synapsin and postsynaptic phalloidin puncta volume. Mature 14 and 21 DIV neurons from heterozygous preCGG and WT littermate mice were analyzed for patterns of synapsin (A and C) and phalloidin (B and D) labeling. (A and B) Representative images from 21 DIV neurons labeled with synapsin and phalloidin (scale bar: 5 µm). Boxed regions are shown in higher magnification below each lower magnification image. The synapsin and phalloidin puncta were quantified, and summary data shown in (C) and (D) reveal that heterozygous preCGG neurons have significantly bigger synapsin and phalloidin puncta compared with the WT (P < 0.0001). The puncta average intensity and total counts did not differ between genotype at either 14 or 18 DIV (data not shown). N = 3 individual cultures with a total of 30 randomly selected neurons measured per genotype for each synapsin and phalloidin puncta quantification. (E) Results from in-cell western experiments showing that, at 14 DIV, heterozygous preCGG hippocampal neurons have higher synapsin levels than WT neurons cultured from littermates (P = 0.0041).
PreCGG neurons possess elevated heterochromatin at 14 and 21 DIV
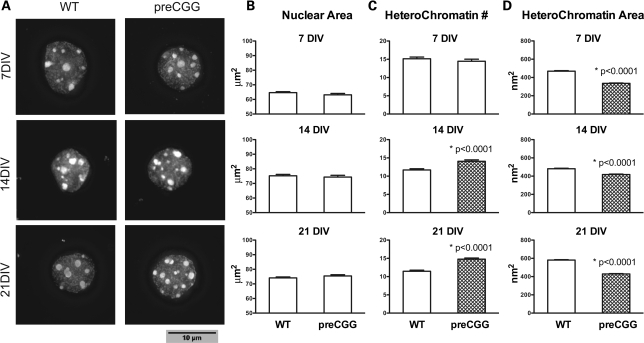
To determine whether heterozygous preCGG neurons display an immature phenotype, we analyzed nuclear size and morphology of heterochromatin. Hippocampal neurons cultured from heterozygous preCGG KI and WT littermates were stained with the nuclear stain Dapi (4′,6-diamidino-2-phenylindole). Compared with WT neurons, heterozygous preCGG neurons have similar nuclear size (area) at 7, 14 and 21 DIV (Fig. 4 A and B). However, by 14 and 20 DIV, heterozygous preCGG neurons have a significantly greater amount of heterochromatin per nucleus (Fig. 4C) than the WT neurons, associated with smaller heterochromatin size (area) compared with WT at all three DIVs (Fig. 4D).
Figure 4.
Neurons cultured from heterozygous preCGG KI mice possess elevated heterochromatin at 14 and 21 DIV. Hippocampal neurons from littermate heterozygous preCGG and WT mice were stained with DAPI (4',6-diamidino-2-phenylindole) to label the nucleus. (A) Representative Dapi nuclear staining at three time points for each genotype; (B) the average (mean ± SE) nuclear area; (C) the number; and (D) the area of heterochromatin in each individual nucleus. Heterozygous preCGG neurons have similar nuclear area, but more abundant and smaller heterochromatin compared with WT at 14 and 21 DIV. Data are from N = 3 separate cultures with 90 randomly selected nuclei analyzed from each experimental group. Scale bar: 10 µm.
PreCGG neurons show decreased survival between 21 and 28 DIV
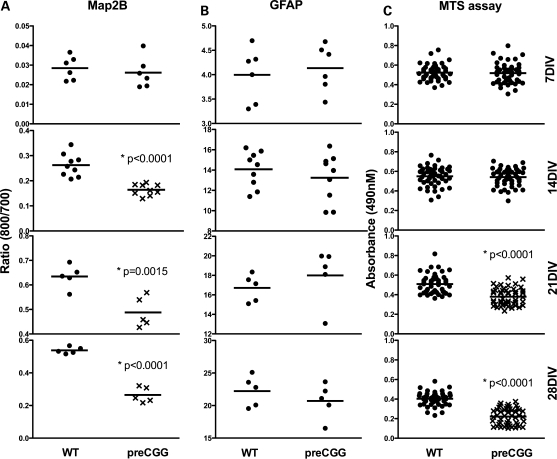
Neuronal cultures from heterozygous preCGG mice have lower viability upon maturation compared with those cultured from their WT littermates. Heterozygous preCGG and WT neurons (7–28 DIV) were examined for the evidence of dendritic retraction and loss of viability by performing morphometric analysis of cells that stained for both Map2B and Dapi, relative to the total number of Dapi-positive nuclei. Figure 5 shows that 7 and 14 DIV neuronal cultures from heterozygous preCGG and WT mice display similar ratios of Map2B-/DAPI-positive cells. However, by 21 DIV, cultures obtained from heterozygous preCGG mice exhibited significantly lower ratios of Map2B-/Dapi-positive staining compared with WT, and declined to 10% (compared with 40% in WT) at 28 DIV (Fig. 5B). Figure 6 shows weekly in-cell western analysis of Map2B fluorescence normalized to cell number with the nuclear stain DraQ5. These results show that although heterozygous preCGG and their corresponding WT littermates have similar Map2B staining densities at 7 DIV, preCGG neurons lag behind WT in their ability to gain Map2B protein at 14 and 21 DIV (Fig. 6A). By 28 DIV, preCGG cultures lose a significant amount of Map2B staining compared with WT (Fig. 6 A, lower panel), suggesting a significant loss in viability or reduced overall complexity. The low-density culture system we used in this study has a relatively low composition of glia cells (<10% around 10 DIV (41)). To rule out the possibility that glia cell overgrowth in the preCGG cultures might contribute to neuronal loss at the later stages of development (21–28 DIV), we performed dual in-cell western analysis with antibodies that recognize GFAP and Map2B. Interestingly, both preCGG and WT cultures have comparable number of GFAP-positive cells (Fig. 6B), suggesting that glia cell overgrowth in the preCGG cultures is not a major contributor to premature loss of viability but is more likely to be the result of inherent deficits in the preCGG neurons themselves.
Figure 5.
Heterozygous preCGG KI neurons show decreased viability from 21 DIV. Separate cultures of hippocampal neurons from littermate heterozygous preCGG and WT mice were fixed weekly after plating, and Map2B and Dapi immunofluorescent staining were quantified. Map2B and Dapi fluorescent staining are shown in (A) (WT) and (B) (preCGG). Images are taken using low magnification (×20). (C) Quantification of the number of Map2B-positive neurons divided by the number of Dapi-positive cells as a measure of viability. preCGG neurons have a higher fractions of Dapi-positive nuclei that lack Map2B staining on 21 and 28 DIV (P < 0.0001). N = 3 separate cultures and approximately 30 randomly selected images for each experimental group. Scale bar: 100 µm.
Figure 6.
Heterozygous preCGG KI neurons show decreased survival independent of glia cell composition. Separate cultures of hippocampal neurons from littermate heterozygous preCGG and WT mice were fixed weekly, and Map2B and GFAP in-cell western analysis performed. MTS assay was performed weekly to study the survival rate of the neurons. (A and B) Results of Map2B and GFAP in-cell western analyses, respectively, demonstrating decreased Map2B protein on 14, 21 and 28 DIV, whereas GFAP protein from the glia cell population remained comparable between genotype. Data are from N = 3 separate cultures with two to three wells for each experimental group. (C) The relative survival rate of the heterozygous preCGG and WT neurons using MTS assay. Data from N = 3 separate cultures with 16 wells from each experimental group.
Cell viability was also independently measured by the reduction of [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) as previously described for serum-free neuronal cultures (42). Figure 6C displays weekly MTS assays of heterozygous preCGG neurons and WT littermates. At 7 and 14 DIV, preCGG and WT neurons did not show differences in cell viability. But at 21 and 28 DIV, preCGG neuronal cultures showed significantly reduced viability (26 and 45% lower, respectively; Fig. 6C) compared with those cultured from their corresponding WT littermates.
PreCGG neurons express elevated stress proteins and their mRNAs
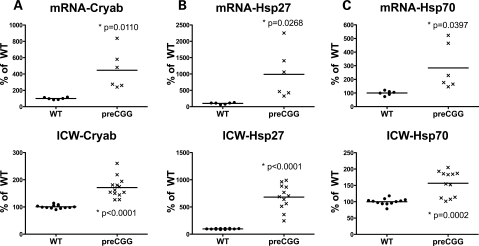
The neuropathologic hallmark of FXTAS is the presence of ubiquitin-positive intranuclear inclusions, in both neurons and astrocytes, and in broad distribution throughout the cerebrum and brainstem (43,44). Double immunofluorescence staining demonstrates co-localization of several stress proteins, including alpha B-crystallin, Hsp27 and Hsp70, with ubiquitin-positive inclusions (45). Since we did not observe ubiquitin-positive intranuclear inclusions in our low-density hippocampal cultures from preCGG or WT mice (data not shown), we determined whether these cultures differentially expressed markers for the stress proteins associated with inclusions at 14 DIV. Quantitative RT–PCR was performed to measure mRNA levels of Cryab (alpha B-crystallin gene), Hsp27 and Hsp70. Significantly higher mRNA levels were found in 14 DIV heterozygous preCGG KI neurons (compared with WT control) for Cryab (4.46 ± 0.96-fold, P = 0.0110) (Fig. 7A), Hsp27 (9.89 ± 0.31, P = 0.0268) (Fig. 7B) and Hsp70 (2.84 ± 0.68, P = 0.0397) (Fig. 7C). The elevated mRNA levels for these stress proteins in preCGG neuronal cultures mirrored elevated levels of their corresponding proteins measured by in-cell western analysis (Fig. 7 A–C, lower panels).
Figure 7.
Heterozygous preCGG KI neurons express elevated stress protein and mRNAs. Hippocampal neurons from littermate heterozygous preCGG and WT mice were collected 14 DIV after plating. Upper panels: Levels of mRNA of the stress proteins alpha B-crystallin (Cryab), Hsp27 and Hsp70 were measured by RT–PCR and normalized to expression of GUS. mRNA signals were normalized to expression of the GUS gene. Significantly higher mRNA levels for alpha B-crystallin (Cryab) (4.5 ± 0.9-fold), Hsp27 (9.9 ± 0.3), and Hsp70 (2.8 ± 0.7) were detected in 14 DIV preCGG KI neurons compared with WT control neurons. P-values are calculated on the basis of RT–PCR results obtained from two independent cultures derived from WT and preCGG littermates (N = 2), each performed in duplicate with three starting RNA concentrations (see Materials and Methods for details). Lower panels: In-cell western analysis of the corresponding proteins in the upper panels. Signals were normalized to cell number using DraQ5 staining of nuclei as described in Materials and Methods. Data from N = 2 separate cultures with six wells from each experimental group.
DISCUSSION
In the current study, we mated XpreCGGY male with XwtXwt female mice to yield littermates of two defined genotypes; heterozygous female offspring that carried an expanded CGG repeat in the high premutation range (155–200 CGG repeats) on one allele, and WT male offspring. Using this breeding strategy, preCGG and WT mice could be separated by gender at birth, allowing for the preparation of serum-free low-density primary cultures (38) segregated into preCGG and WT hippocampal neurons from littermates. Although this experimental approach was utilized to minimize external influences introduced by an across-litter design, it would be expected to yield smaller effect sizes than would a comparison between hemizygous males and WT non-littermate males, since the latter strategy would not be subject to the attenuating effects of inactivation of the preCGG allele. Unlike the case of mice with mutant Mecp2 alleles, where the huge phenotype variability and neuronal culture survival studies indicate X-inactivation in these mice highly favors expression of the wild-type (WT) allele (46), no significant skewing of X-inactivation is expected for the Fmr1 premutation mouse model. There is also evidence that females who harbor premutation alleles do not display significant skewing against the expression of the X allele with the CGG expanded repeat and toward the normal repeat (18). Indeed, heterozygous mouse neurons that harbor preCGG expansions in the upper portion of the premutation range express nearly 3-fold higher levels Fmr1 mRNA, yet levels of FMRP are reduced by only ∼25% compared with those from WT littermates. The patterns of these two key markers mirror those observed in preCGG carriers and FXTAS in humans (9,23). Along with elevated mRNA and protein level of stress proteins (including alpha B-crystallin, Hsp27 and Hsp70), the findings of the elevated Fmr1 mRNA and reduced FMRP protein levels, which mimic the human disease, make the hippocampal neuronal culture a very good model for studying mechanism and develop treatment for FXTAS. The recent finding that preCGG expressed ectopically in Purkinje neurons, outside the context of Fmr1 mRNA, can result in neuronal pathology in a mammalian system demonstrates that expanded CGG repeats containing RNA are the likely cause of the neurodegeneration in FXTAS (47).
A priori, the abnormal cellular and molecular phenotypes (including puncta volumes, altered mRNA and protein expression levels, etc.) observed for pooled neuronal cultures from heterozygous female mice are expected to be about half of the magnitude of the differences expected for hemizygous mice (assuming a linear model), since in the absence of skewing, the X-activation ratio should tend toward 0.5. Nevertheless, the importance of the current observations is the unequivocal observation of a neuropathology in the heterozygote, thus reinforcing the expectation based on clinical involvement in female carriers (16–18), that abnormal neural cell function would be manifest in heterozygous mice and cultured neurons. The variation observed in the levels of Fmr1 mRNA and FMRP among culture wells containing preCGG neurons within replicates and among independent cultures (Fig. 1) is likely to reflect an XCI pattern that cannot totally favor against the mutant allele. Instead, our findings suggest that XCI does not abrogate the phenotype of the mutation. Heterozygous female hippocampal neuronal culture showed significant deficits intrinsically in both neurodevelopmental and neurodegenerative aspects.
Morphometric analysis of dendritic complexity and nuclear architecture indicates the impaired trajectory of in vitro neuronal maturation between 7 and 21 DIV. Dendritic pathology is indicated in a number of syndromic disorders with cognitive impairment, such as Down's syndrome as well as those arising from environmental insults including prenatal alcohol exposure, drug exposure, intrauterine cytomegalovirus and rubella infection (48). Deficits commonly include reduced dendritic complexity. The properties of dendritic signaling are remarkably diverse and dynamic, and chemical and electrical compartmentalization in dendrites defines rules for synaptic plasticity and enhances the overall computational capacity of dendrites. The diversity of dendritic excitability arises from variations in dendritic morphology and channel distributions, whereas the modulation of synaptic and voltage-gated channels results in dynamic, state-dependent changes in function. Therefore, dendritic branching complexity is crucial to synaptic integration (49). Heterozygous preCGG neurons show inherent impairments during the entire in vitro developmental trajectory, suggesting one possible cellular mechanism, reduced dendritic complexity, significantly contribute to increased risk for neurodevelopmental and cognitive deficits in premutation carriers and why these deficits become progressively severe over time (27).
Altered dendritic spine number and structure are associated with cognitive impairment, FXS and autism (50–54). However, unlike FXS, where the FMR1 gene is silenced and FMRP is deficient, preCGG neurons studied here exhibited only a modest reduction in FMRP levels. Upon evaluating their dendritic spine properties, we found that the heterozygous preCGG neurons have increased presynaptic synapsin punctum volume (average number of voxels per punctum), broader and wider spine heads, but similar puncta intensity (average intensity per punctum) and total puncta count. The in-cell western measures the total level of the synapsin protein of a large number of neurons per well, and is the product of puncta intensity, puncta volume and the number of puncta. Larger synapsin bouton size in primary hippocampal neuronal culture has also been reported in the β2 m/TAP1 knock-out (KO) mice, which have reduced MHC-I surface levels (55). The SynGAP KO mouse also shows significantly larger postsynapstic protein puncta and synapsin puncta in primary hippocampal neuronal culture. These neurons also have larger dendritic protrusions (56).
Trinucleotide expansion repeats are responsible for a heterogeneous number of disorders including some myotonic dystrophy type 1 and 3, Friedreich ataxia and FXS. Although the repeats are located in different parts of different genes, they may share a common property, the ability to initiate repeat-mediated epigenetic changes that result in the formation of heterochromatin (57,58). Results from neurons cultured from mice expressing the preCGG expansion support this hypothesis in that their nuclei contain smaller and more abundant heterochromatin than neurons cultured from their WT littermates. Similar to other neurodegenerative disorders such as Parkinson disease, survival in FXTAS appears to be shortened (59). In accord with this clinical observation, our current study has provided the first evidence that heterozygous preCGG neurons have reduced survival relative to WT control neurons under culture conditions that use serum-free-defined medium (NS21; (38)), and these impairments do not appear to be associated with over-growth of the cultures with glia. The cellular basis (apoptosis, authophagy or paraptosis) contributing to the reduced neuronal viability with heterozygous preCGG KI neurons remains to be determined. However, the observation of reduced viability may reflect similar processes that are taking place in patients with FXTAS, who demonstrate reductions in brain volume as well as substantial dropout of cerebellar Purkinje cells (43,60,61). Alpha B-crystallin, a small heat shock protein (HSP), is a component of the Lewy bodies of Parkinson disease (62) and of Rosenthal fibers, a cytoplasmic inclusion found in the astrocytes of Alexander disease patients (63). Although it is unknown what role these proteins play in the disease formation of FXTAS, we postulate that these elevated mRNAs might contribute to the early death of the hippocampal neurons from the heterozygous preCGG KI mice.
In summary, our findings show for the first time that hippocampal neurons cultured from mice expressing Fmr1 CGG expansion repeats within the premutation range found in humans (preCGG mutations) have developmental and degenerative impairments compared with cultures from WT littermates. In this regard, early impairments in the developmental trajectory of dendritic growth and complexity, as well as altered synaptic architecture, have not been previously identified for preCGG mutations in Fmr1 gene. Moreover, the cultures of enriched, low-density neurons from preCGG mice identify an inherent abnormality of the neurons expressing CGG expansion repeats. The discovery that early developmental impairments precede premature loss of viability is a significant new finding since it paves the way for novel insights into potential therapeutic interventions. Such interventions in patients with the premutation may include neuroprotective agents, antioxidants, psychopharmacologic agents or targeted treatments in addition to intensive early interventions for learning and behavior (64).
Our findings show that neurons cultured from mice expressing Fmr1 CGG expansion repeats have significantly higher Fmr1 RNA but only modest reductions in FMRP, which accurately models the human premutation condition. Our findings clearly demonstrate that mice expressing Fmr1 CGG expansion repeats do not show skewed XCI to the extent of abrogating either the developmental or the degenerative phenotypes we have identified. For the first time, we show that females heterozygous for the CGG expansion repeat produce hippocampal neurons that have intrinsic impairments in both their developmental trajectory and show premature neurodegeneration relative to those cultured from WT littermates. Cultures from Mecp2 mutation mice, a model of Rett syndrome, were shown to have significantly skewed XCI that increases the phenotypic variability and significantly diminishes phenotypic severity (46). Our results with neurons from female mice carrying the premutation on one allele indicate that XCI is not complete and has significant consequences on neuronal development and survival. These results are consistent with random XCI recently identified in human female premutation carriers (18).
The present study is also the first to identify the presence of elevated stress markers (alpha B-crystallin, Hsp27 and Hsp70) mRNA and protein levels, directly in affected neurons. These findings are significant since elevated levels of these very same stress markers have been identified in human fibroblast from patients with the FMR1 premutations (Garcia-Arocena et al., submitted for publication).
MATERIALS AND METHODS
Mouse models
Both the expanded CGG repeat KI (preCGG KI) mice and the WT mice with an endogenous (CGG)8 repeat were housed under standard conditions. Hippocampal primary neuronal cultures were prepared from the brains of 0–2-day-old (P 0–2) WT mouse pups or female pups carrying one expanded (CGG)n repeat (range: 155 < n < 200; designated preCGG) on a C57BL/6J background. Pups were obtained by breeding male hemizygous mice for a CGG expansion ranging 150–180 on the Fmr1 gene to WT females. This yielded heterozygous female offspring that carried one expanded preCGG repeat and male WT offspring. Using this breeding strategy, WT and preCGG mouse pups could be identified and separated by sex at birth, allowing for the successful preparation of primary hippocampal cultures from WT and preCGG KI mice pups. Genotypes were later verified by PCR from the remaining tissues from the pups as described previously (31). Founder mice with expanded CGG trinucleotide repeats were obtained from Erasmus University and have been described previously (34,65). These mice were originally on a mixed FVB/N × C57BL/6J background. The mice used in the present study to generate pups for tissue culture were bred from these founder mice onto a C57BL/6J background over at least seven generations to be >98% C57BL/6J by microsatellite genotyping.
Genotyping of the mice
DNA was extracted from mouse-tail snips by incubating with 10 mg/ml Proteinase K (Roche Diagnostics) in 300 µl lysis buffer containing 50 mm Tris–HCl, pH 7.5, 10 mm EDTA, 150 mm NaCl, 1% SDS overnight at 55°C. An amount of 100 µl saturated NaCl was then added, and the suspension was centrifuged. One volume of 100% ethanol was added and gently mixed, the DNA was pelleted by centrifugation and the supernatant discarded. The DNA was washed and centrifuged in 500 µl 70% ethanol. The DNA was then dissolved in 100 µl milliQ-H20. CGG repeat lengths were determined by PCR using the Expanded High Fidelity Plus PCR System (Roche Diagnostics). Briefly, ∼500–700 ng of DNA was added to 50 µl of PCR mixture containing 2.0 µm of each primer, 250 µm of each dNTP (Invitrogen), 2% DMSO (Sigma), 2.5 m Betaine (Sigma), 5 U Expand HF buffer with Mg (7.5 µm). The forward primer was 5′-GCTCAGCTCCGTTTCGGTTTCACTTCCGGT-3′ and the reverse primer was 5′-AGCCCCGCACTTCCACCACCAGCTCCTCCA-3′. PCR steps were 10 min denaturation at 95°C, followed by 34 cycles of 1 min denaturation at 95°C, annealing for 1 min at 65°C, and elongation for 5 min at 75°C to end each cycle. PCR ends with a final elongation step of 10 min at 75°C. Band sizes were determined by running DNA samples on a 2.5% agarose gel and staining DNA with ethidium bromide.
Primary cultures of mouse hippocampal neurons
Low-density cultures of dissociated hippocampal neurons were prepared as described (38,66). Briefly, hippocampi from P0-2 postnatal mice (WT and heterozygous preCGG KI mice were described earlier) were incubated in HEPES buffered saline (Ca2+/Mg2+ free; HBS 0/0), containing trypsin (0.03%), at 37°C for 20 min and washed three times with HBS 0/0 before dissociating the cells with a fire-polished Pasteur pipette. After non-dispersed pieces had settled, cells in the supernatant were spun down (1100 r.p.m., 200g, 1 min), resuspended, counted and plated at a density of 3–6 × 103 cm−1 on coverslips (Warner Instruments, Hamden, CT, USA) coated with 0.1% (w/v) poly-l-lysine (Peptides International, Louisville, KY, USA) in neurobasal medium (Invitrogen, Carlsbad, CA, USA) containing NS21 supplement (38), 0.5 mm glutamine and 5% fetal bovine serum (FBS). After 4 h, the medium was replaced with serum-free neurobasal medium supplemented with NS21 and 0.5 mm glutamine. Cells were maintained at 37°C in a humidified environment of ambient air/5% CO2. One-third of the medium was changed after 4 DIV and weekly thereafter. Cultures were used weekly until 4 weeks old.
In-cell western analysis
In-cell western analyses were done according to the manufacturer's specifications. Primary hippocampal cultures were plated on poly-l-lysine-coated 96-well plates at a density of 6000 cells/well. On 14 DIV, cells were fixed with 4% formaldehyde/4% sucrose for 15 min, washed with PBS with 0.1% Triton X-100 five times and incubated in blocking solution (LI-COR Biotechnology, Lincoln, NE, USA) for 90 min. Cells were then incubated with rabbit anti-synapsin (Synaptic Systems GmbH, Germany, 1:1000) and chicken anti-FMRP (made in the P. Hagerman laboratory, 1:2000) (68) in the blocking solution for 2 h. After washing with PBS+0.1% Tween 20, cells were incubated with IRDye 800CW-labeled goat anti-rabbit antibody or IRDye 800CW-labeled goat anti-chicken, as well as IRDye 700CW-labeled DraQ5 (LI-COR Biotechnology) for 2 h. After washing with PBS+0.1% Tween 20, plates were allowed to air-dry and scanned with the LI-COR Odyssey Infrared Imaging System. Controls for all cells were stained with secondary antibody only, and this background was subtracted from the signal. Additional controls to assess the specificity of the FMRP polyclonal antibody were performed by pre-blocking with recombinant FMRP (1:250) prior to performing the in-cell western protocol. IRDye 700CW-labeled DraQ5 signals serve as plating control, and all the IRDye 800 signals were normalized using LI-COR Odyssey Infrared Imaging System application software 2.1.
Quantification of mRNA levels
Primary hippocampal cultures were plated on poly-l-lysine-coated 96-well plates at a density of 6000 cells/well (two plates for each sample: two normal male controls and two female KI mice). From each genotype, 14 DIV cultures were pooled together, and total RNA was isolated using standard methods (Trizol, Invitrogen). Quantitative RT–PCR was performed on a TaqMan real-time PCR instrument (ABI 7900, Applied Biosystems, Foster City, CA, USA) using TaqMan PCR chemistry that specifically utilizes the 5′-nuclease activity of the Taq polymerase to cleave a reporter probe to release a fluorescent dye from its quencher molecule. The change in fluorescence can be measured in real-time. Quantitative RT–PCRs were performed from two independent cultures derived from WT and preCGG littermates (N = 2). Each RT–PCR was performed in duplicate with three starting RNA concentrations (500, 250 and 125 ng), for both the target gene and the GUS amplicons (a total of 12 reactions). Control reactions (for both the target genes and the reference gene) were run in parallel with the use of a standard normal control. The specific probe and the primer set for each gene used in the analysis (Fmr1, Hsp27, Hps70 and Cryab; target genes) spanned an exon–exon boundary in order to exclude genomic DNA contamination and were obtained from Applied Biosystems (AOD, Applied Biosystems, Foster City, CA, USA). The Glucoronidase (GUS) gene was used as a reference gene. Details and conditions are as described in Tassone et al. (68).
Celltiter 96® AQueous non-radioactive cell proliferation assay (MTS)
Primary hippocampal neurons were cultured at a density of 6000 cells per well in flat-bottomed 96-well plates. After 14 days, CellTiter 96® Aqueous One Solution Reagent (Promega, Madison, WI, USA) was added to each well according to the manufacturer's instructions. In brief, the reagent is composed of solutions of a novel tetrazolium compound MTS (inner salt) and an electron-coupling reagent PMS (phenazine methosulfate). MTS and PMS detection reagents were mixed, using a ratio of 20:1 (MTS:PMS), immediately prior to the addition to the cell culture at a ratio of 1:5 (detection reagents:cell culture medium). The first and last rows of the plate were left blank as the plate background control. The WT and heterozygous preCGG neurons were cultured on the same plate side by side in the middle rows of a 96-well plate. After 2 h in culture, the cell viability was determined by measuring the absorbance at 490 nm using a computer-connected Spectramax Plus 384 (Molecular Devices, CA, USA). The Softmax Pro software generates the reading of the 490 nm absorbance and can be exported to Excel for analysis and graphing.
Immunocytochemistry and antibodies
Following fixation, cells were thoroughly washed with PBS, permeabilized with 0.05% Triton X-100 (20 min), blocked (PBS containing 2% glycerol, 0.05 m NH4Cl, 5% FBS, 2% goat serum; 2 h), incubated with primary antibodies (1.5 h at room temperature or overnight at 4°C), washed, incubated with Alexa 488- or Alexa 568-conjugated secondary antibodies (Molecular Probes, Eugene, OR, USA; goat anti-rabbit and goat anti-mouse diluted 1:500; 1 h, room temperature), washed and mounted in Prolong Gold Antifade mounting media with DAPI (Molecular Probes). Primary antibodies were rabbit anti-synapsin (Synaptic Systems GmbH, 1:1000); mouse monoclonal anti-Map2B (Transduction Laboratories, Lexington, KY, USA, 1:500); Alexa 488-conjugagted phalloidin (Molecular Probes, 1:200); rabbit polycolonal anti-Cryab (Santa Cruz Biotechnology, Inc., Santa Crutz, CA, USA); rabbit polycolonal anti-Hsp70 (Santa Cruz Biotechnology); rabbit polycolonal anti-Hsp27 (Santa Cruz Biotechnology).
Image acquisition
Neurons were fixed and stained weekly. Fluorescence microscopy was performed using The DeltaVision Core Imaging System (Applied Precision, LLC, WA, USA) for Alexa 488 (490 nm band-pass excitation, 528 nm long-pass emission) and Alexa 568 (555 nm band-pass excitation, 617 nm long-pass emission). All images were taken as 3D stacks for further deconvolution and better analysis. The acquisition and analysis of immunofluorescence was conducted with the experimenter blinded to genotype. For each culture experiment, neurons (N = 5–7) were picked at random from two coverslips for each genotype for quantitative analysis. Each experiment was replicated on three separate culture days (i.e. separate litters).
Image analysis and statistics
In all cases, the image analysis was blinded to the researcher of the genotype. Images were deconvoluted and processed using Softworx software (Applied Precision, LLC). The synapsin and phalloidin puncta analysis and the nuclear heterochromatin quantification were performed using a customized Matlab image analysis toolbox (The Mathworks, Natick, MA, USA). The size (area) of nuclei and the heterochromatin contained within were computed using NIH ImageJ. The dendritic analysis of the neurons was performed using Imaris FilamentTracer manual mode according to the software manual (Bitplane AG, Zurich, Switzerland). In all cases, statistics are done with two-way ANOVA among different culture days between genotypes using statistical software Prism® (GraphPad Software, Inc., CA, USA). P < 0.05 indicated a statistically significant difference between genotypes.
FUNDING
This work was supported by National Institutes of Health (MH073124, RL1 AG032119, RL1 NS062411 and UL1 DE19583). Additional support came from the National Institute of Environmental Health Sciences (P01ES011269), the US Environmental Protection Agency (R833292 and R829388) to the UC Davis Center for Children's Environmental Health, and Autism Speaks (Environmental Innovator Award to I.N.P.).
ACKNOWLEDGEMENTS
The authors wish to thank Lee Rognlie for the mouse breeding; Yoelia Perez, Eitan Kaplan and Kyungho Kim for helping with the data collection and analysis; Dr. W.W. Zhang for writing the customized Matlab imaging analysis program.
Conflict of Interest statement. R.J.H. receives funding for trials in Fragile X syndrome from Roche, Seaside Therapeutics and Neuropharm. She has consulted with Novartis for treatment trials. P.J.H. is a consultant (uncompensated) for Asuragen, Inc. for FMR1 genotype analysis.
REFERENCES
- 1.Jacquemont S., Hagerman R.J., Hagerman P.J., Leehey M.A. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 2007;6:45–55. doi: 10.1016/S1474-4422(06)70676-7. [DOI] [PubMed] [Google Scholar]
- 2.Hagerman R., Rivera S.M., Hagerman P. The fragile X family of disorders: a model for autism and targeted treatments. Curr. Pediatr. Rev. 2008;4:40–52. [Google Scholar]
- 3.Oostra B.A., Willemsen R. FMR1: a gene with three faces. Biochim. Biophys. Acta. 2009;1790:467–477. doi: 10.1016/j.bbagen.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouwer J.R., Huizer K., Severijnen L.A., Hukema R.K., Berman R.F., Oostra B.A., Willemsen R. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J. Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Jr, Warren S.T., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 6.Hagerman P.J. The fragile X prevalence paradox. J. Med. Genet. 2008;45:498–499. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford D.C., Meadows K.L., Newman J.L., Taft L.F., Scott E., Leslie M., Shubek L., Holmgreen P., Yeargin-Allsopp M., Boyle C., et al. Prevalence of the fragile X syndrome in African-Americans. Am. J. Med. Genet. 2002;110:226–233. doi: 10.1002/ajmg.10427. [DOI] [PubMed] [Google Scholar]
- 8.Goodlin-Jones B.L., Tassone F., Gane L.W., Hagerman R.J. Autistic spectrum disorder and the fragile X premutation. J. Dev. Behav. Pediatr. 2004;25:392–398. doi: 10.1097/00004703-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Hessl D., Tassone F., Loesch D.Z., Berry-Kravis E., Leehey M.A., Gane L.W., Barbato I., Rice C., Gould E., Hall D.A., et al. Abnormal elevation of FMR1 mRNA is associated with psychological symptoms in individuals with the fragile X premutation. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;139B:115–121. doi: 10.1002/ajmg.b.30241. [DOI] [PubMed] [Google Scholar]
- 10.Farzin F., Perry H., Hessl D., Loesch D., Cohen J., Bacalman S., Gane L., Tassone F., Hagerman P., Hagerman R. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J. Dev. Behav. Pediatr. 2006;27:S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 11.Hagerman R.J. Lessons from fragile X regarding neurobiology, autism, and neurodegeneration. J. Dev. Behav. Pediatr. 2006;27:63–74. doi: 10.1097/00004703-200602000-00012. [DOI] [PubMed] [Google Scholar]
- 12.Wittenberger M.D., Hagerman R.J., Sherman S.L., McConkie-Rosell A., Welt C.K., Rebar R.W., Corrigan E.C., Simpson J.L., Nelson L.M. The FMR1 premutation and reproduction. Fertil. Steril. 2007;87:456–465. doi: 10.1016/j.fertnstert.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Amiri K., Hagerman R.J., Hagerman P.J. Fragile X-associated tremor/ataxia syndrome: an aging face of the fragile X gene. Arch. Neurol. 2008;65:19–25. doi: 10.1001/archneurol.2007.30. [DOI] [PubMed] [Google Scholar]
- 14.Brouwer J.R., Willemsen R., Oostra B.A. The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:782–798. doi: 10.1002/ajmg.b.30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tassone F., Adams J., Berry-Kravis E.M., Cohen S.S., Brusco A., Leehey M.A., Li L., Hagerman R.J., Hagerman P.J. CGG repeat length correlates with age of onset of motor signs of the fragile X-associated tremor/ataxia syndrome (FXTAS) Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:566–569. doi: 10.1002/ajmg.b.30482. [DOI] [PubMed] [Google Scholar]
- 16.Coffey S.M., Cook K., Tartaglia N., Tassone F., Nguyen D.V., Pan R., Bronsky H.E., Yuhas J., Borodyanskaya M., Grigsby J., et al. Expanded clinical phenotype of women with the FMR1 premutation. Am. J. Med. Genet. A. 2008;146A:1009–1016. doi: 10.1002/ajmg.a.32060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Revenga L., Madrigal I., Pagonabarraga J., Xuncla M., Badenas C., Kulisevsky J., Gomez B., Mila M. Penetrance of FMR1 premutation associated pathologies in fragile X syndrome families. Eur. J. Hum. Genet. 2009 doi: 10.1038/ejhg.2009.51. epub ahead of print April, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Revenga L., Madrigal I., Badenas C., Xuncla M., Jimenez L., Mila M. Premature ovarian failure and fragile X female premutation carriers: no evidence for a skewed X-chromosome inactivation pattern. Menopause. 2009;16:944–949. doi: 10.1097/gme.0b013e3181a06a37. [DOI] [PubMed] [Google Scholar]
- 19.Hagerman P.J., Hagerman R.J. Fragile X-associated tremor/ataxia syndrome (FXTAS) Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:25–30. doi: 10.1002/mrdd.20005. [DOI] [PubMed] [Google Scholar]
- 20.Hagerman P.J., Hagerman R.J. The fragile-X premutation: a maturing perspective. Am. J. Hum. Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams J.S., Adams P.E., Nguyen D., Brunberg J.A., Tassone F., Zhang W., Koldewyn K., Rivera S.M., Grigsby J., Zhang L., et al. Volumetric brain changes in females with fragile X-associated tremor/ataxia syndrome (FXTAS) Neurology. 2007;69:851–859. doi: 10.1212/01.wnl.0000269781.10417.7b. [DOI] [PubMed] [Google Scholar]
- 22.Jacquemont S., Farzin F., Hall D., Leehey M., Tassone F., Gane L., Zhang L., Grigsby J., Jardini T., Lewin F., et al. Aging in individuals with the FMR1 mutation. Am. J. Ment. Retard. 2004;109:154–164. doi: 10.1352/0895-8017(2004)109<154:AIIWTF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tassone F., Iwahashi C., Hagerman P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 24.Hessl D., Rivera S., Koldewyn K., Cordeiro L., Adams J., Tassone F., Hagerman P.J., Hagerman R.J. Amygdala dysfunction in men with the fragile X premutation. Brain. 2007;130:404–416. doi: 10.1093/brain/awl338. [DOI] [PubMed] [Google Scholar]
- 25.Moore C.J., Daly E.M., Schmitz N., Tassone F., Tysoe C., Hagerman R.J., Hagerman P.J., Morris R.G., Murphy K.C., Murphy D.G. A neuropsychological investigation of male premutation carriers of fragile X syndrome. Neuropsychologia. 2004;42:1934–1947. doi: 10.1016/j.neuropsychologia.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Koldewyn K., Hessl D., Adams J., Tassone F., Hagerman P., Hagerman R., Rivera S. Reduced hippocampal activation during recall is associated with elevated FMR1 mRNA and psychiatric symptoms in men with the fragile X premutation. Brain Imaging Behav. 2008;2:105–116. doi: 10.1007/s11682-008-9020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornish K.M., Li L., Kogan C.S., Jacquemont S., Turk J., Dalton A., Hagerman R.J., Hagerman P.J. Age-dependent cognitive changes in carriers of the fragile X syndrome. Cortex. 2008;44:628–636. doi: 10.1016/j.cortex.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts J., Bailey D., Mankowski J., Ford A., Sideris J., Weisenfeld L., Heath T.M., Golden R. Mood and anxiety disorders in females with the FMR1 premutation. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:130–139. doi: 10.1002/ajmg.b.30786. [DOI] [PubMed] [Google Scholar]
- 29.Aziz M., Stathopulu E., Callias M., Taylor C., Turk J., Oostra B., Willemsen R., Patton M. Clinical features of boys with fragile X premutations and intermediate alleles. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;121B:119–127. doi: 10.1002/ajmg.b.20030. [DOI] [PubMed] [Google Scholar]
- 30.Bailey D.B., Jr, Raspa M., Olmsted M., Holiday D.B. Co-occurring conditions associated with FMR1 gene variations: findings from a national parent survey. Am. J. Med. Genet. A. 2008;146A:2060–2069. doi: 10.1002/ajmg.a.32439. [DOI] [PubMed] [Google Scholar]
- 31.Brouwer J.R., Severijnen E., de Jong F.H., Hessl D., Hagerman R.J., Oostra B.A., Willemsen R. Altered hypothalamus–pituitary–adrenal gland axis regulation in the expanded CGG-repeat mouse model for fragile X-associated tremor/ataxia syndrome. Psychoneuroendocrinology. 2008;33:863–873. doi: 10.1016/j.psyneuen.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwer J.R., Mientjes E.J., Bakker C.E., Nieuwenhuizen I.M., Severijnen L.A., Van der Linde H.C., Nelson D.L., Oostra B.A., Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated fragile X full mutation. Exp. Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willemsen R., Oostra B.A., Bassell G.J., Dictenberg J. The fragile X syndrome: from molecular genetics to neurobiology. Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:60–67. doi: 10.1002/mrdd.20010. [DOI] [PubMed] [Google Scholar]
- 34.Willemsen R., Hoogeveen-Westerveld M., Reis S., Holstege J., Severijnen L.A., Nieuwenhuizen I.M., Schrier M., van Unen L., Tassone F., Hoogeveen A.T., et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 35.Berman R.F., Willemsen R. Mouse models of fragile X-associated tremor ataxia. J. Investig. Med. 2009 doi: 10.231/JIM.0b013e3181af59d6. epub ahead of print July, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Dam D., Errijgers V., Kooy R.F., Willemsen R., Mientjes E., Oostra B.A., De Deyn P.P. Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (FXTAS) Behav. Brain Res. 2005;162:233–239. doi: 10.1016/j.bbr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Brewer G.J. Serum-free B27/neurobasal medium supports differentiated growth of neurons from the striatum, substantia nigra, septum, cerebral cortex, cerebellum, and dentate gyrus. J. Neurosci. Res. 1995;42:674–683. doi: 10.1002/jnr.490420510. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Stevens B., Chang J., Milbrandt J., Barres B.A., Hell J.W. NS21: re-defined and modified supplement B27 for neuronal cultures. J. Neurosci. Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H., Kovar J., Sissons S., Cox K., Matter W., Chadwell F., Luan P., Vlahos C.J., Schutz-Geschwender A., Olive D.M. A cell-based immunocytochemical assay for monitoring kinase signaling pathways and drug efficacy. Anal. Biochem. 2005;338:136–142. doi: 10.1016/j.ab.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Patel T.P., Cain W.J., Duncan M.K. Production of monoclonal antibodies against Prox1. Hybridoma (Larchmt) 2006;25:27–33. doi: 10.1089/hyb.2006.25.27. [DOI] [PubMed] [Google Scholar]
- 41.Kaduce T., Chen Y., Hell J., Spector A. Docosahexaenoic acid synthesis from n-3 fatty acid precursors in rat hippocampal neurons. J. Neurochem. 2008;105:1525–1535. doi: 10.1111/j.1471-4159.2008.05274.x. [DOI] [PubMed] [Google Scholar]
- 42.Wong P.W., Garcia E.F., Pessah I.N. Ortho-substituted PCB95 alters intracellular calcium signaling and causes cellular acidification in PC12 cells by an immunophilin-dependent mechanism. J. Neurochem. 2001;76:450–463. doi: 10.1046/j.1471-4159.2001.00022.x. [DOI] [PubMed] [Google Scholar]
- 43.Greco C.M., Hagerman R.J., Tassone F., Chudley A.E., Del Bigio M.R., Jacquemont S., Leehey M., Hagerman P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 44.Greco C.M., Berman R.F., Martin R.M., Tassone F., Schwartz P.H., Chang A., Trapp B.D., Iwahashi C., Brunberg J., Grigsby J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 45.Iwahashi C.K., Yasui D.H., An H.J., Greco C.M., Tassone F., Nannen K., Babineau B., Lebrilla C.B., Hagerman R.J., Hagerman P.J. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 46.Young J.I., Zoghbi H.Y. X-chromosome inactivation patterns are unbalanced and affect the phenotypic outcome in a mouse model of Rett syndrome. Am. J. Hum. Genet. 2004;74:511–520. doi: 10.1086/382228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashem V., Galloway J.N., Mori M., Willemsen R., Oostra B.A., Paylor R., Nelson D.L. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dierssen M., Ramakers G.J. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5(Suppl. 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 49.Hausser M., Spruston N., Stuart G.J. Diversity and dynamics of dendritic signaling. Science. 2000;290:739–744. doi: 10.1126/science.290.5492.739. [DOI] [PubMed] [Google Scholar]
- 50.Koekkoek S.K., Yamaguchi K., Milojkovic B.A., Dortland B.R., Ruigrok T.J., Maex R., De Graaf W., Smit A.E., VanderWerf F., Bakker C.E., et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Dictenberg J.B., Swanger S.A., Antar L.N., Singer R.H., Bassell G.J. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irwin S.A., Galvez R., Greenough W.T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 53.McKinney B.C., Grossman A.W., Elisseou N.M., Greenough W.T. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;136B:98–102. doi: 10.1002/ajmg.b.30183. [DOI] [PubMed] [Google Scholar]
- 54.de Vrij F.M., Levenga J., van der Linde H.C., Koekkoek S.K., De Zeeuw C.I., Nelson D.L., Oostra B.A., Willemsen R. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol. Dis. 2008;31:127–132. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard C.A., Butts D.A., Shatz C.J. Regulation of CNS synapses by neuronal MHC class I. Proc. Natl Acad. Sci. USA. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vazquez L.E., Chen H.J., Sokolova I., Knuesel I., Kennedy M.B. SynGAP regulates spine formation. J. Neurosci. 2004;24:8862–8872. doi: 10.1523/JNEUROSCI.3213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumari D., Usdin K. Chromatin remodeling in the noncoding repeat expansion diseases. J. Biol. Chem. 2009;284:7413–7417. doi: 10.1074/jbc.R800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res. 2008;18:1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leehey M.A., Berry-Kravis E., Min S.J., Hall D.A., Rice C.D., Zhang L., Grigsby J., Greco C.M., Reynolds A., Lara R., et al. Progression of tremor and ataxia in male carriers of the FMR1 premutation. Mov. Disord. 2007;22:203–206. doi: 10.1002/mds.21252. [DOI] [PubMed] [Google Scholar]
- 60.Brunberg J.A., Jacquemont S., Hagerman R.J., Berry-Kravis E.M., Grigsby J., Leehey M.A., Tassone F., Brown W.T., Greco C.M., Hagerman P.J. Fragile X premutation carriers: characteristic MR imaging findings of adult male patients with progressive cerebellar and cognitive dysfunction. AJNR Am. J. Neuroradiol. 2002;23:1757–1766. [PMC free article] [PubMed] [Google Scholar]
- 61.Jacquemont S., Hagerman R.J., Leehey M., Grigsby J., Zhang L., Brunberg J.A., Greco C., Des Portes V., Jardini T., Levine R., et al. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gai W.P., Power J.H., Blumbergs P.C., Culvenor J.G., Jensen P.H. Alpha-synuclein immunoisolation of glial inclusions from multiple system atrophy brain tissue reveals multiprotein components. J. Neurochem. 1999;73:2093–2100. [PubMed] [Google Scholar]
- 63.Gordon N. Alexander disease. Eur. J. Paediatr. Neurol. 2003;7:395–399. doi: 10.1016/j.ejpn.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Hagerman R.J., Berry-Kravis E., Kaufmann W.E., Ono M.Y., Tartaglia N., Lachiewicz A., Kronk R., Delahunty C., Hessl D., Visootsak J., et al. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123:378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bontekoe C.J., Bakker C.E., Nieuwenhuizen I.M., van der Linde H., Lans H., de Lange D., Hirst M.C., Oostra B.A. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 66.Lim I.A., Merrill M.A., Chen Y., Hell J.W. Disruption of the NMDA receptor-PSD-95 interaction in hippocampal neurons with no obvious physiological short-term effect. Neuropharmacology. 2003;45:738–754. doi: 10.1016/s0028-3908(03)00276-4. [DOI] [PubMed] [Google Scholar]
- 67.Iwahashi C., Tassone F., Hagerman R.J., Yasui D., Parrott G., Nguyen D., Mayeur G., Hagerman P.J. A quantitative ELISA assay for the fragile X mental retardation 1 protein. J. Mol. Diagn. 2009;11:281–289. doi: 10.2353/jmoldx.2009.080118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]