Abstract
Human activities have altered the composition of biotas through two fundamental processes: native extinctions and alien introductions. Both processes affect the taxonomic (i.e., species identity) and phylogenetic (i.e., species evolutionary history) structure of species assemblages. However, it is not known what the relative magnitude of these effects is at large spatial scales. Here we analyze the large-scale effects of plant extinctions and introductions on taxonomic and phylogenetic diversity of floras across Europe, using data from 23 regions. Considering both native losses and alien additions in concert reveals that plant invasions since AD 1500 exceeded extinctions, resulting in (i) increased taxonomic diversity (i.e., species richness) but decreased phylogenetic diversity within European regions, and (ii) increased taxonomic and phylogenetic similarity among European regions. Those extinct species were phylogenetically and taxonomically unique and typical of individual regions, and extinctions usually were not continent-wide and therefore led to differentiation. By contrast, because introduced alien species tended to be closely related to native species, the floristic differentiation due to species extinction was lessened by taxonomic and phylogenetic homogenization effects. This was especially due to species that are alien to a region but native to other parts of Europe. As a result, floras of many European regions have partly lost and will continue to lose their uniqueness. The results suggest that biodiversity needs to be assessed in terms of both species taxonomic and phylogenetic identity, but the latter is rarely used as a metric of the biodiversity dynamics.
Keywords: alien species, alpha diversity, biodiversity, phylogenetic beta diversity, phylogeny
Globalization is progressively altering the composition of biotas worldwide (1–3). The interplay of two fundamental processes — extinctions of native species and introductions and successful establishment of alien species (sensu 4; hereafter referred to as invasion) — has been known to reduce the distinctiveness of species communities. Global species extinctions lead to a continuous decrease of overall species richness (i.e., γ-diversity) (5). However, at the scale of continents, regions, and countries, invasions exceed local extinctions and result in an increase in local or regional species richness (i.e., α-diversity) (5–9).
Changes in species composition driven by the combined effects of invasions and extinctions can result in decreasing (i.e., homogenization), increasing (i.e., differentiation), or unchanged compositional turnover of species (i.e., β-diversity) (10, 11) and traits between and within continents (12, 13). The higher magnitude of invasions compared with extinctions is known to lead to decreased β-diversity between regions at continental (14–17) and regional scales (18–21).
Previous evidence of biotic homogenization at continental and regional scales has largely examined impacts on the taxonomic structure of species assemblages, yet extinctions and invasions might also affect phylogenetic structure (22). The phylogenetic structure of a species assemblage represents the evolutionary history of its members and reflects the diversity of genetic and thus morphologic, physiologic, and behavioral characteristics (23). High phylogenetic diversity within and across communities may enable rapid adaptation to changing environmental conditions across both ecologic and evolutionary time scales (24, 25). Research on the relative effects of extinctions and invasions upon the structure (α-diversity) and spatial distribution (β-diversity) of phylogenetic patterns may thus enhance our understanding of how evolutionary and ecologic factors contribute to general diversity patterns (26, 27).
The composition of species invasions and extinctions (28) is not randomly distributed among plant taxa but reflects the response of specific life-history traits to natural and human-induced environmental change (29, 30). Extinctions usually befall specialized endemic or rare species, often from species-poor families, which form distinct parts of biotas (31, 28). Hence, their loss should result in a pronounced decrease of phylogenetic and taxonomic β-diversity within and between regions to which these species are unique (32, 33). Successful invaders are often ecologic generalists with wide distributional ranges (34), often belonging to species-rich families (35). Hence, the gain of such common species should also decrease phylogenetic and taxonomic β-diversity within and between regions.
Recent studies of the effect of alien species on the phylogenetic composition of plant communities revealed ambiguous results; species composition can be modified by the addition of either (i) more distantly related species than expected by chance (phylogenetic overdispersion) (36) or (ii) more closely related species (phylogenetic clustering) (37; and see ref. 38 for review).
Generally, very little is known about the relative effects of extinctions and invasions on phylogenetic relationships among species at large spatial scales (27). On the basis of a comprehensive dataset of original (before extinctions and introductions) and current (after extinctions and introductions) floras across several European regions, here we use phylogenetic information to assess the consequences of species extinctions and introductions on taxonomic and phylogenetic diversity.
Results and Discussion
Since AD 1500, the processes of species extinctions and invasions, acting in concert, resulted in a net increase in overall European plant species richness (γ-diversity) (Table 1). This increase is due to 69 extinctions of European plants and 1,621 invasions of plants from outside Europe (Fig. S1 and Table S1) and is also accompanied by increased phylogenetic γ-diversity. The increased species richness at the European level is reflected by higher species richness in all European regions (α-diversity), where species invasions also exceeded extinctions (Table S1). However, the prevalence of invasions over extinctions decreased rather than increased phylogenetic α-diversity over the same period; an additional decrease was caused by extinctions (both P < 0.001; Table 1, Table S1, and Table S2).
Table 1.
Taxonomic and phylogenetic diversity components across Europe
| Level | Diversity |
||
|---|---|---|---|
| γ | β | α | |
| Taxonomic level | |||
| Original flora | 10,928 | 0.5432 ± 0.1622 | 2,194 ± 1194 |
| Current total flora | 12,624 | 0.5155 ± 0.1490 | 2,664 ± 1215 |
| Phylogenetic level | |||
| Original flora | 1.3797 | 1.4493 ± 0.1049 | 1.4772 ± 0.0202 |
| Current total flora | 1.3942 | 1.4462 ± 0.0951 | 1.4725 ± 0.0189 |
γ applies to overall European diversity, whereas β and α are mean values (± SD) across European regions. Taxonomic diversities are based on species numbers (γ and α) and β tax values. Phylogenetic diversities are based on Δ+ (γ and α) and β phyl values.
Despite increased species richness at the European and regional level, extinctions and invasions in concert resulted in decreased β-diversity among species (βtax) and β-diversity among phylogenetic lineages (βphyl) of European regions (Table 1), indicating that European floras became phylogenetically and taxonomically impoverished. A decrease of phylogenetic richness with increasing species richness was previously reported on a national scale (39).
However, considering extinctions and invasions separately revealed contrasting patterns. Invasions have generally led to taxonomic and phylogenetic homogenization, whereas species extinctions result in differentiation. Species extinctions increased βtax and βphyl (Fig. 1B), although βtax and βphyl of extinct species were higher than those of extant native species (P < 0.001). Species from distantly related taxonomic branches have become extinct in different regions. By contrast, invasions decreased βtax and βphyl of European floras (Fig. 1C), although βtax and βphyl of alien species was higher than those of native species (both P < 0.001). Hence, although alien floras are taxonomically and phylogenetically diverse, the constituent species are either native to other European regions or, if introduced from outside Europe, tend to be closely related to native European species.
Fig. 1.
Changes in βtax vs. changes in βphyl among European regions (n = 253 pairwise comparisons). Current native flora = current natives species, excluding extinct species; current total flora = current natives plus alien species; original flora = native species including extinct species. Red areas indicate significant effect of both measurements (Fisher's paired comparison design test; P < 0.001). (A) Combined effect: extinctions of native species and invasions of alien species lead to taxonomic and phylogenetic homogenization [βtax (current total flora) < βtax (original flora) and βphyl (current total flora) < βphyl (original flora)]. (B) Extinct effect: extinctions of native species lead to taxonomic and phylo-genetic differentiation [βtax (current native flora) > βtax (original flora) and βphyl (current native flora) > βphyl (original flora)]. (C) Alien effect: invasions of alien species lead to taxonomic and phylogenetic homogenization [βtax (current total flora) < βtax (current native flora) and βphyl (current total flora) < βphyl (current native flora)].
We reason that many species that have become extinct or have been introduced in a particular region are components of the current native floras of other European regions, because (i) the loss of taxonomically more diverse native species (extinctions) led to increased βtax, and the gain of more diverse aliens led to decreased βtax; and (ii) invasions of phylogenetically more diverse alien species decreased βphyl, and the loss of more diverse extinct species increased βphyl among floras. This is because extinctions of plant species in Europe are mainly regional rather than continent-wide extinctions. Moreover, ca. 53% of plant invasions in European floras are due to species exchange among European regions (40). Such a high percentage is likely to increase taxonomic and phylogenetic similarities among European floras.
Generally, the negative effect of aliens on βtax and βphyl is approximately seven times the magnitude of the positive effect of species losses (compare βtax values, Fig. 1 B and C). Therefore, the taxonomic and phylogenetic homogenization among European floras due to species invasions masks any regional differentiation due to species extinctions. These patterns are consistent with those found for North America, where species extinctions only played a minor role in defining compositional patterns for state floras, owing to the low numbers of extinct species (41). Although extinction processes are much slower than invasions, it is unlikely that accounting for lag effects in extinctions (8) would change the pattern dramatically.
With compositional changes due to extinctions and invasions, βtax increased linearly with increasing βphyl (Fig. 1A; R2 = 0.86, P < 0.001). However, this relationship is not inevitable (27); the high correlation indicates that species determining this pattern are probably widespread and closely related to extant native species. The weaker relationship between βtax and βphyl derived from the effect of extinctions indicates that there were more phylogenetically unique species among extinct native plants. In contrast to the main patterns, among some regions we did observe occasions when species introductions led to taxonomic and phylogenetic differentiation, and in some circumstances extinctions led to increased homogenization (as species that are unique to only one region are lost). However, we did not find any systematic trend in these patterns, and they were not related to geographic distance, species richness, or other specific attributes of the floras (e.g., level of endemism).
As with α-diversity, two scenarios whereby βtax and βphyl show contrasting patterns (homogenization vs. differentiation) are possible (27): (i) a high βtax (differentiation) and a low βphyl (homogenization) could be observed with a high proportion of resident endemic species or if the different communities consist of close congeners; (ii) a low βtax (homogenization) and a high βphyl (differentiation) is very unlikely, because a high species overlap will always generate a high phylogenetic overlap (27).
Effects of scale dependencies are known for biodiversity patterns in general (42) and invasion processes in particular (43, 44). Phylogenetic structures could also be scale dependent (45). It has been argued that large-scale patterns of phylogenetic clustering reflect biogeographic rather than ecologic processes (38). Moreover, the perception of taxonomic homogenization is dependent on the spatial scale at which samples are gathered and increases with increasing sample area (46). Similarly, phylogenetic homogenization may also be scale dependent because closely related alien and native species may not cooccur in the same plant communities (47, 48). Thus using regions may overestimate the degree of homogenization experienced by local plant communities. Furthermore, political boundaries will often encompass various biogeographic regions, further increasing the similarity across these sample regions (49).
Invasions and extinctions are co-occurring processes, but evidence for a causal relationship is generally scarce (50). Thus, although it is clear that alien plant species have altered European ecosystems (51), there is no example of a native plant species being driven to extinction because of competition with an alien plant species on the European mainland or islands (2, 52). Biotic impoverishment caused by extinctions and invasions are often a symptom of wider degradation of the environment (e.g., by simplification of landscape through the loss of habitat heterogeneity, urbanization, increasing global transport, and nitrogen input into ecosystems) (6, 53).
In conclusion, we have shown that increasing species numbers in European regional floras over the last 5 centuries have been accompanied by a decrease of phylogenetic and taxonomic uniqueness. To restrict indicators of conservation priorities or ecosystem health simply to species richness can thus be misleading and does not capture the important effects of taxonomic or phylogenetic distinctiveness (22, 54, 55). Even if native diversity is considered separately, the decreasing numbers of native species can have more serious consequences for the phylogenetic diversity than can be inferred from species richness alone. We show that by combining taxonomic and phylogenetic information on α- and β-diversity, we gain new insight into changes in biodiversity patterns at different levels, which are likely to be relevant for ecosystem processes (37, 27, 39). Phylogenetic diversity reflects the evolutionary history of a community, which may also reflect its functional diversity (23, 26). Hence, diminished phylogenetic and taxonomic information could decrease the capacity of species assemblages to respond to environmental changes and therefore threaten ecosystem functioning (39). Considering the rate of species invasions into Europe (40, 56), the above trends are likely to continue.
Methods
Species Data.
Species occurrence data and their status (alien, native, or extinct) were collected for 23 European countries or regions (e.g., the Baltic States are represented as one region; Table S1; all countries/regions are hereafter referred to as regions). Because extinction and invasion rates of island ecosystems are much higher than those of mainland regions (52), we excluded island data and worked only with mainland data for which the geographic definition of Flora Europaea (57) allowed us to distinguish between mainland and island data (see details in Fig. S1): Greece (Crete, Karpathos, Kasos, Gavdhos, and those Aegean Islands outside of Europe defined by Flora Europaea), Spain (Balearic Islands), France (Corse), Italy (Sardinia, Sicily), and the Malta Archipelago.
Lists of native species (see ref. 4 for definition) for European regions were derived from the European Science Foundation European Documentation System database of Flora Europaea (57; http://rbg-web2.rbge.org.uk/FE/fe.html). Data on extinct plant species were taken from national Red Lists of individual countries (see full list of references in supporting information).
Lists of alien species per country were compiled from the DAISIE database (58; www.europe-aliens.org). An alien species is defined as one introduced to the region as a result of human activities and successfully naturalized (i.e., forming reproducing populations in the wild) (sensu 4). Only plant species introduced after AD 1500 were considered (4), which are hereafter referred to as “aliens.”
We only considered taxa at the species level. In Europe there are 12,624 species, of which 10,928 are native, 537 extinct, and 3,353 alien. Some species were assigned to different categories in different regions [e.g., 1,726 species are native to one region but are alien to another (see ref. 40), or 468 native species are extinct in one region but occur in other regions (Table S1).
Taxonomic β-Diversity.
We used the Morisita-Horn dissimilarity index (MH) to calculate βtax among floras. The index is computed as:
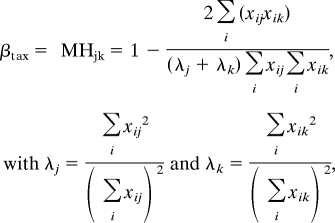 |
where xij and xik represent the frequency of species i in regions j and k, respectively. The index ranges from 0 (total identity between two samples and low β-diversity) to 1 (absolute dissimilarity of these samples and high β-diversity). This index is less sensitive to species richness and sample size than most other indices (59). Because the index needs abundance data, we defined pseudo-abundance of one for each species.
Phylogenetic β-Diversity.
βphyl was defined as β-diversity with a temporal dimension measured as phylogenetic distance between communities according to branch lengths (27). The online software tool Phylomatic (60) was used to construct a supertree using species and genus data of all species considered. In the absence of phylogenetic branch lengths for the whole tree, we calculated pseudobranch lengths to weight the height of the nodes according to their position in the tree (61). We assigned a relative height (with tips at 0 and root at 1) to each node and then calculated branch lengths as the difference between the heights of two nodes. This ensures that the total branch length from root to any tip is constant. Grafen's method sets node height from the tip proportional to the number of descendent terminal nodes (taxa) minus 1 (61). To calculate the branch length we used the function brlen of the R-package ape (62).
The βphyl between two floras was assessed using the PhyloSor index (37). To facilitate the comparison of βphyl and βtax, βphyl was also computed as dissimilarity:
 |
BLjk is the branch length common to communities j and k, and BLj and BLk are the total branch lengths of community j and k, respectively. βphyl ranges from 0 (both communities are composed of the same taxa) to 1 (two communities share no taxa).
To disentangle the effect of the loss and gain of species on the β-diversity between regions at the species (βtax) and phylogenetic (βphy) level, we separated the processes of species extinctions and invasions. Because comprehensive dates of plant extinctions do not exist, we arbitrary defined that all extinctions occurred after AD 1500. Thus, we defined the “original flora” (before AD 1500) as all extant and extinct native species, the “current native flora” as only extant native species, and the “current total flora” as extant natives and alien species. Introducing these three categories, we calculate different effects of different floristic elements on βtax and βphyl: (i) the combined effect as the difference between the β-diversities of original and current flora [β(current total flora) − β(original flora)]; (ii) the effect of extinct native species as the difference between β-diversities of original and current native flora [β(current native flora) − β(original flora)]; and (iii) the effect of alien species as the difference between β-diversities of current total flora and current native flora [β(current total flora) − β(current native flora)]. Furthermore, we calculated separately the βtax and βphyl of extinct, alien, and native species among regions.
Alpha Diversity.
Beside taxonomic α-diversity in terms of species numbers (Table S2), we also assessed phylogenetic α-diversity of the original and current floras by using Warwick's average taxonomic distinctness (Δ+) (63). Δ+ was originally developed on taxonomic relationships but can be adapted to phylogenetic information (64). The index was calculated as:
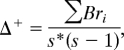 |
where Bri is built from the distance matrix of species based on branch lengths, and s is the number of species. The index is based on the sum of branch lengths between species (as provided by the distance matrix) and can be interpreted as the mean distance between two randomly chosen species independent of their distance from the root of the tree. Δ+ is mathematically unbiased by species richness; that is, it does not automatically increase with sample size and reflects the phylogenetic structure of a subset from a phylogenetic tree best, unlike the majority of other available phylogenetic diversity indices (64). Smaller Δ+ values indicate that, on average, the assemblage of species is phylogenetically more closely related and less distinct. Analogous to βtax and βphyl, we calculated (i) combined effect [Δ+(current total flora) − Δ+(original flora)], (ii) extinct effect [Δ + (current native flora) − Δ+(original flora)], and (iii) alien effect [Δ+(current total flora) − Δ+(current native flora)]. Significant differences between all relationships of β-diversity or Δ+ were assessed using Fisher's paired comparisons design test (65).
Supplementary Material
Acknowledgments.
We thank Stefan Michalski and Emmanuel Paradis for providing phylogenetic and technical expertise; and the Delivering Alien Invasive Species Inventories for Europe (DAISIE) consortium for providing the data. This study was partly funded by the European Union through the FP 6 projects DAISIE (contract number SSPI-CT-2003–511202) and ALARM (GOCE-CT-2003–506675). M.H., J.P., and P.P. were also supported by Grants AV0Z60050516 (from the Academy of Sciences of the Czech Republic), MSM0021620828, and LC06073 (both from the Ministry of Education, Youth and Sports of the Czech Republic).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907088106/DCSupplemental.
References
- 1.Elton CS. The Ecology of Invasions by Animals and Plants. London: Methuen; 1958. [Google Scholar]
- 2.Davis MA. Biotic globalization: Does competition from introduced species threaten biodiversity? Bioscience. 2003;53:481–489. [Google Scholar]
- 3.Hulme PE. Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J Appl Ecol. 2009;46:10–18. [Google Scholar]
- 4.Pyšek P, et al. Alien plants in checklists and floras: Towards better communication between taxonomists and ecologists. Taxon. 2004;53:131–143. [Google Scholar]
- 5.Sax DF, Gaines SD. Species diversity: From global decreases to local increases. Trends Ecol Evol. 2003;18:561–566. [Google Scholar]
- 6.Hobbs RJ, Mooney HA. Broadening the extinction debate: Population deletions and additions in California and Western Australia. Conserv Biol. 1998;12:271–283. [Google Scholar]
- 7.Stohlgren TJ, Barnett DT, Jarnevich CS, Flather C, Kartesz J. The myth of plant species saturation. Ecol Lett. 2008;11:313–322. doi: 10.1111/j.1461-0248.2008.01153.x. [DOI] [PubMed] [Google Scholar]
- 8.Sax DF, Gaines SD, Brown JH. Species invasions exceed extinctions on islands worldwide: A comparative study of plants and birds. Am Nat. 2002;160:766–783. doi: 10.1086/343877. [DOI] [PubMed] [Google Scholar]
- 9.Wilson K-J. Extinct and introduced vertebrate species in New Zealand: A loss of biodistinctiveness and gain in biodiversity. Pac Conserv Biol. 1997;3:301–305. [Google Scholar]
- 10.Olden JD, Poff NL. Toward a mechanistic understanding and prediction of biotic homogenization. Am Nat. 2003;162:442–460. doi: 10.1086/378212. [DOI] [PubMed] [Google Scholar]
- 11.Cassey P, Blackburn TM, Lockwood JL, Sax DF. A stochastic model for integrating changes in species richness and community similarity across spatial scales. Oikos. 2006;115:207–218. [Google Scholar]
- 12.McKinney ML. Species introduced from nearby sources have a more homogenizing effect than species from distant sources: Evidence from plants and fishes in the USA. Divers Distrib. 2005;11:367–374. [Google Scholar]
- 13.Winter M, Kühn I, Nentwig W, Klotz S. Spatial aspects of trait homogenization within the German flora. J Biogeogr. 2008;35:2289–2297. [Google Scholar]
- 14.Rahel FJ. Homogenization of freshwater faunas. Annu Rev Ecol Syst. 2002;33:291–315. [Google Scholar]
- 15.Olden JD, Poff NL, McKinney ML. Forecasting faunal and floral homogenization associated with human population geography in North America. Biol Conserv. 2006;127:261–271. [Google Scholar]
- 16.Spear D, Chown SL. Taxonomic homogenization in ungulates: Patterns and mechanisms at local and global scales. J Biogeogr. 2008;35:1962–1975. [Google Scholar]
- 17.McKinney ML. Do human activities raise species richness? Contrasting patterns in United States plants and fishes. Glob Ecol Biogeogr. 2002;11:343–348. [Google Scholar]
- 18.Duncan JR, Lockwood JL. In: Biotic Homogenization. McKinney ML, Lockwood JL, editors. New York: Kluwer Academic/Plenum; 2001. pp. 247–256. [Google Scholar]
- 19.Marchetti MP, et al. In: Biotic homogenization. Lockwood JL, McKinney ML, editors. New York: Kluwer Academic/Plenum Publishers; 2001. pp. 259–278. [Google Scholar]
- 20.Castro SA, Jaksic FM. How general are global trends in biotic homogenization? Floristic tracking in Chile, South America. Glob Ecol Biogeogr. 2008;17:524–531. [Google Scholar]
- 21.Rooney TP, Wiegmann SM, Rogers DA, Waller DM. Biotic impoverishment and homogenization in unfragmented forest understory communities. Conserv Biol. 2004;18:787–798. [Google Scholar]
- 22.Vane-Wright RI, Humphries CJ, Williams PH. What to protect?—Systematics and the agony of choice. Biol Conserv. 1991;55:235. [Google Scholar]
- 23.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 24.Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- 25.Jump AS, Marchant R, Penuelas J. Environmental change and the option value of genetic diversity. Trends Plants Sci. 2009;14:51–58. doi: 10.1016/j.tplants.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Chave J, Chust G, Thébaud C. In: Scaling Biodiversity. Storch D, Marquet P, Brown JH, editors. Cambridge, UK: Cambridge Univ Press; 2007. pp. 151–167. [Google Scholar]
- 27.Graham CH, Fine PVA. Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol Lett. 2008;11:1265–1277. doi: 10.1111/j.1461-0248.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 28.Vamosi JC, Wilson JRU. Nonrandom extinction leads to elevated loss of angiosperm evolutionary history. Ecol Lett. 2008;11:1047–1053. doi: 10.1111/j.1461-0248.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- 29.Pyšek P, Richardson D. In: Biological Invasions, Ecological Studies. Nentwig W, editor. Vol 193. Berlin: Springer; 2007. pp. 97–126. [Google Scholar]
- 30.Freville H, McConway K, Dodd M, Silvertown J. Prediction of extinction in plants: Interaction of extrinsic threats and life history traits. Ecology. 2007;88:2662–2672. doi: 10.1890/06-1453.1. [DOI] [PubMed] [Google Scholar]
- 31.Gaston KJ. Species-range size distributions: Products of speciation, extinction and transformation. Philos Trans R Soc Lond B Biol Sci. 1998;353:219–230. [Google Scholar]
- 32.Sechrest W, et al. Hotspots and the conservation of evolutionary history. Proc Natl Acad Sci USA. 2002;99:2067–2071. doi: 10.1073/pnas.251680798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mace GM, Gittleman JL, Purvis A. Preserving the tree of life. Science. 2003;300:1707–1709. doi: 10.1126/science.1085510. [DOI] [PubMed] [Google Scholar]
- 34.Pyšek P, et al. The global invasion success of Central European plants is related to distribution characteristics in their native range and species traits. Divers Distrib. 2009;15:891–903. [Google Scholar]
- 35.Pyšek P. Is there a taxonomic pattern to plant invasions? Oikos. 1998;82:282–294. [Google Scholar]
- 36.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian oak communities. Am Nat. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- 37.Bryant JA, et al. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA. 2008;105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proches S, Wilson JRU, Richardson DM, Rejmanek M. Searching for phylogenetic pattern in biological invasions. Glob Ecol Biogeogr. 2008;17:5–10. [Google Scholar]
- 39.Knapp S, Kühn I, Schweiger O, Klotz S. Challenging urban species diversity: Contrasting phylogenetic patterns across plant functional groups in Germany. Ecol Lett. 2008;11:1054–1064. doi: 10.1111/j.1461-0248.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- 40.Lambdon PW, et al. Alien flora of Europe: Species diversity, temporal trends, geographical patterns and research needs. Preslia. 2008;80:101–149. [Google Scholar]
- 41.Qian H, Ricklefs RE. The role of exotic species in homogenizing the North American flora. Ecol Lett. 2006;9:1293–1298. doi: 10.1111/j.1461-0248.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- 42.Whittaker RJ, Willis KJ, Field R. Scale and species richness: Towards a general, hierarchical theory of species diversity. J Biogeogr. 2001;28:453–470. [Google Scholar]
- 43.Rouget M, Richardson DM. In: Plant Invasions: Ecological Threats and Management Solutions. Child LE, et al., editors. Leiden: Backhuys; 2003. pp. 3–15. [Google Scholar]
- 44.Kühn I, Klotz S. In: Biological Invasions, Ecological Studies. Nentwig W, editor. Vol 193. Berlin: Springer; 2007. pp. 181–196. [Google Scholar]
- 45.Cavender-Bares J, Keen A, Miles B. Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:S109–S122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Hulme PE. Contrasting alien and native plant species-area relationships: The importance of spatial grain and extent. Glob Ecol Biogeogr. 2008;17:641–647. [Google Scholar]
- 47.Lambdon PW, Hulme PE. How strongly do interactions with closely-related native species influence plant invasions? Darwin's naturalization hypothesis assessed on Mediterranean islands. J Biogeogr. 2006;33:1116–1125. [Google Scholar]
- 48.Diez JM, Sullivan JJ, Hulme PE, Edwards G, Duncan RP. Darwin's naturalization conundrum: Dissecting taxonomic patterns of species invasions. Ecol Lett. 2008;11:674–681. doi: 10.1111/j.1461-0248.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 49.Olden JD. Biotic homogenization: A new research agenda for conservation biogeography. J Biogeogr. 2006;33:2027–2039. [Google Scholar]
- 50.Gurevitch J, Padilla DK. Are invasive species a major cause of extinctions? Trends Ecol Evol. 2004;19:470–474. doi: 10.1016/j.tree.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Hulme PE. In: Biodiversity Under Threat, Issues in Environmental Science and Technology. Hester R, Harrison RM, editors. Cambridge, UK: Royal Society of Chemistry; 2007. pp. 56–80. [Google Scholar]
- 52.Sax DF, Gaines SD. Species invasions and extinction: The future of native biodiversity on islands. Proc Natl Acad Sci USA. 2008;105:11490–11497. doi: 10.1073/pnas.0802290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKinney ML, Lockwood JL. Biotic homogenization: A few winners replacing many losers in the next mass extinction. Trends Ecol Evol. 1999;14:450–453. doi: 10.1016/s0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- 54.Purvis A, Hector A. Getting the measure of biodiversity. Nature. 2000;405:212–219. doi: 10.1038/35012221. [DOI] [PubMed] [Google Scholar]
- 55.Emerson BC, Gillespie RG. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol Evol. 2008;23:619–630. doi: 10.1016/j.tree.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 56.Hulme PE, Pyšek P, Nentwig W, Vila M. Will threat of biological invasions unite the European Union? Science. 2009;324:40–41. doi: 10.1126/science.1171111. [DOI] [PubMed] [Google Scholar]
- 57.Tutin TG, et al. Flora Europaea. Cambridge, UK: Cambridge Univ Press; 1964–1980. [Google Scholar]
- 58.DAISIE. Handbook of Alien Species in Europe. Dordrecht: Springer; 2009. [Google Scholar]
- 59.Wolda H. Similarity indexes, sample-size and diversity. Oecologia. 1981;50:296–302. doi: 10.1007/BF00344966. [DOI] [PubMed] [Google Scholar]
- 60.Webb CO, Donoghue MJ. Phylomatic: Tree assembly for applied phylogenetics. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 61.Grafen A. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- 62.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 63.Warwick RM, Clarke KR. Taxonomic distinctness and environmental assessment. J Appl Ecol. 1998;35:532–543. [Google Scholar]
- 64.Schweiger S, Klotz S, Durka W, Kühn I. A comparative test of phylogenetic diversity indices. Oecologia. 2008;157:485–495. doi: 10.1007/s00442-008-1082-2. [DOI] [PubMed] [Google Scholar]
- 65.Manly BFJ. Randomization and Monte Carlo Methods in Biology. London: Chapman & Hall; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



