Abstract
In flowering plants, the accumulation of small deletions through unequal homologous recombination (UR) and illegitimate recombination (IR) is proposed to be the major process counteracting genome expansion, which is caused primarily by the periodic amplification of long terminal repeat retrotransposons (LTR-RTs). However, the full suite of evolutionary forces that govern the gain or loss of transposable elements (TEs) and their distribution within a genome remains unclear. Here, we investigated the distribution and structural variation of LTR-RTs in relation to the rates of local genetic recombination (GR) and gene densities in the rice (Oryza sativa) genome. Our data revealed a positive correlation between GR rates and gene densities and negative correlations between LTR-RT densities and both GR and gene densities. The data also indicate a tendency for LTR-RT elements and fragments to be shorter in regions with higher GR rates; the size reduction of LTR-RTs appears to be achieved primarily through solo LTR formation by UR. Comparison of indica and japonica rice revealed patterns and frequencies of LTR-RT gain and loss within different evolutionary timeframes. Different LTR-RT families exhibited variable distribution patterns and structural changes, but overall LTR-RT compositions and genes were organized according to the GR gradients of the genome. Further investigation of non-LTR-RTs and DNA transposons revealed a negative correlation between gene densities and the abundance of DNA transposons and a weak correlation between GR rates and the abundance of long interspersed nuclear elements (LINEs)/short interspersed nuclear elements (SINEs). Together, these observations suggest that GR and gene density play important roles in shaping the dynamic structure of the rice genome.
Flowering plants vary tremendously in nuclear genome size. Along with polyploidization, accumulation of repetitive DNA, particularly long terminal repeat retrotransposons (LTR-RTs), is the primary mechanism driving plant genome expansion (Bennetzen et al. 2005). In large-genome species such as maize, barley, and wheat, LTR-RTs make up more than 60%–80% of their genomes, and the majority of these elements have amplified within the last few million years (SanMiguel et al. 1996, 1998; Vicient et al. 1999; Wicker et al. 2001; Bruggmann et al. 2006). A particularly striking study shows that Oryza australiensis, a wild species of rice, has accumulated more than 90,000 copies of LTR-RTs during the last three million years, leading to a twofold increase in genome size without polyploidization (Piegu et al. 2006).
To counteract expansion, plants may generate small deletions that lead to genome shrinkage (Devos et al. 2002; Ma et al. 2004). This process is reflected by the presence of solo LTRs and partially deleted LTR-RTs in all plant genomes investigated to date (Bennetzen et al. 2005). Based on studies in yeast (Roeder et al. 1980), solo LTRs are thought to be formed by unequal intraelement homologous recombination (UR) between two highly identical LTRs. In contrast, partially deleted or truncated elements are thought to be the outcome of illegitimate (nonhomologous) recombination (IR) (Devos et al. 2002; Wicker et al. 2003; Ma et al. 2004). These two processes have been estimated to have removed >190 Mb of LTR-RT DNA from the rice (Oryza sativa) genome within the past four million years, leaving a current genome of ∼400 Mb that contains <100 Mb of detectable LTR-RT elements or fragments (Ma et al. 2004).
The abundance of LTR-RTs in plant genomes is largely determined by the competing activities of retrotransposon amplification and the generation of small deletions (Bennetzen et al. 2005), but the driving forces that shape transposable element (TE) acquisition, elimination, and distribution remain largely unknown. In all plants investigated, LTR-RTs accumulate dramatically in heterochromatin such as pericentromeric and centromeric regions (The Arabidopsis Genome Initiative 2000; International Rice Genome Sequencing Project 2005), where genetic recombination (GR) is suppressed. There is, thus, a negative association between GR suppression and LTR-RT accumulation. However, it is not clear if accumulation in low recombination regions reflects biased amplification in those regions or biased removal in high GR regions.
To shed light on the evolutionary forces that govern the distribution and dynamics of retrotransposons, we comprehensively identified LTR-RTs and analyzed their structural variation along the 12 rice chromosomes. We then investigated the relationships between GR rates and local genomic features, including LTR-RT abundance, gene densities, and the estimated amount of DNA loss through LTR-RT rearrangement. We also analyzed patterns of LTR-RT DNA distributions between the indica and japonica rice genomes, as well as distribution patterns among LTR-RT families and among chromosomal regions. Finally, we compared LTR-RT distribution patterns to those of long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and DNA transposons in the context of GR rates and gene densities. Overall, our goal for these analyses was to infer the forces shaping the distribution of LTR retrotransposons and particularly the balance of UR and IR in mediating removal of LTR-RT genomic DNA.
Results
Characterization and structural analysis of LTR-RTs
Although the majority of LTR-RT families in the rice genome were previously identified and collected into databases (McCarthy et al. 2002; Ma and Bennetzen 2006; Chaparro et al. 2007), accurate characterization of their structure and organization has not been performed across the entire genome. Because LTR-RTs undergo rearrangements (Ma et al. 2004) and can be nested (Ma et al. 2005; Ma and Bennetzen 2006), accurate characterization of LTR-RTs is not straightforward. To ensure a comprehensive and accurate identification of LTR-RTs and their boundaries, we applied and improved the method that was previously developed for analysis of a centromeric region of rice (Ma and Bennetzen 2006) (see Methods). In particular, detailed manual inspection was conducted to confirm each predicted element and to define its structure and boundaries.
Using these methods, the LTR-RTs in the rice genome (International Rice Genome Sequencing Project [IRGSP] Build 4.0 pseudomolecules, cultivar, Nipponbare, http://rgp.dna.affrc.go.jp) were identified and analyzed. We identified three types of LTR-RTs: intact elements, solo LTRs, and truncated elements. The intact elements and solo LTRs were categorized using previously described criteria (Ma et al. 2004). These elements have clear boundaries at both ends and are flanked by target site duplications (TSDs). Truncated elements refer to incomplete elements with only one clearly identified boundary, and each of these elements contains at least one identified LTR and partial internal sequence (Ma et al. 2004). In total, our analyses identified 4937 intact elements, 7981 solo LTRs, and 2006 truncated elements in the 12 chromosomes (Supplemental Table 1). Out of the 4937 intact elements, only 2348 (47.6%) were identified by the very conservative program LTR_STRUC (McCarthy and McDonald 2003). Detailed information about the structure and chromosomal locations of all elements identified in this study is listed in Supplemental Table 2.
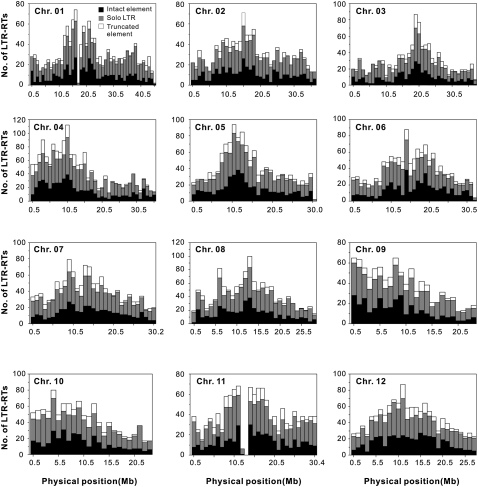
The distributions of intact elements, solo LTRs, and truncated elements, pooled in 1-Mb contiguous subregions along each of the 12 chromosomes, are illustrated in Figure 1. In addition to these three types of elements, a small fraction of “other” elements, as noted in Supplemental Table 1, was identified. These elements, including 14 (0.1%) LTR–internal–LTR–internal–LTR complexes, 741 (4.6%) single LTRs without TSDs, and 334 (2.1%) complete elements without TSDs (Supplemental Table 2), were not analyzed further, because the rearrangement events involved in their formation were not clear. Furthermore, numerous LTR-RT fragments without clearly defined LTRs were excluded from this study, because they were believed to be derived from elements that had undergone multiple independent and often overlapping rearrangements.
Figure 1.
Distribution of LTR-RTs along the 12 rice chromosomes. The exceptionally low proportions of LTR-RTs in chr 01 and chr 11 surrounding respective centromeres are due to the “N” designation present in the assembled rice genomic sequences.
Based on the method described by Wicker et al. (2007), the 16,013 total elements identified in the genome (Supplemental Table 1) were grouped into 393 families, including 298 known families and 95 (32%) previously unknown families identified in this study (Supplemental Table 3). The previously unknown families contained 5496 elements, and their copy numbers varied from 1 to 464. Overall, the intact elements, solo LTRs, and truncated elements listed in Supplemental Table 1 make up 20.4% of the genome.
Distribution of LTR-RTs in the context of GR rates and gene densities
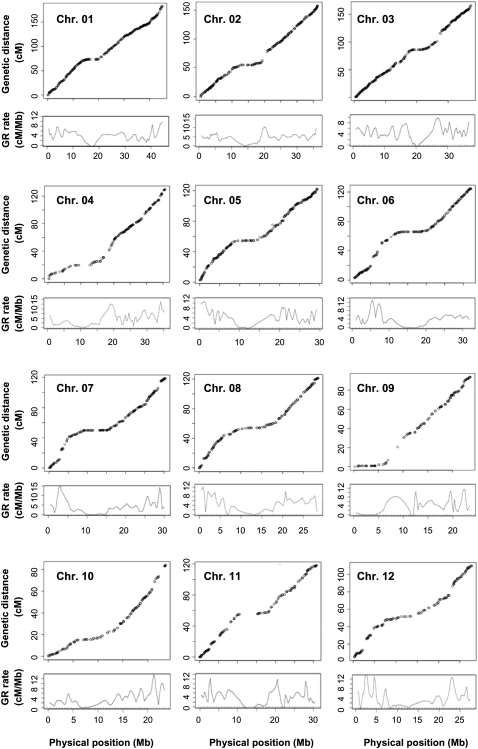
To explore the relationship between GR and structural features, size variation, and distribution of LTR-RTs, we first estimated local GR rates along each of the 12 chromosomes. To do this, we used MareyMap, an R-based tool for estimating recombination rate by comparison of genetic and physical maps (Rezvoy et al. 2007) (see Methods). The cM/Mb plots along each of the rice chromosomes were obtained based on 3982 markers (http://rgp.dna.affrc.go.jp; Supplemental Table 4) that were genetically mapped using a single F2 population of two rice cultivars, Nipponbare and Kasalath (Harushima et al. 1998), and were physically anchored to the Build 4.0 pseudomolecules of Nipponbare rice in this study (Fig. 2). With these data, the local recombination rates along individual chromosomes were estimated, based on the Loess method (a two-degree polynomial fitted in each window) provided by the MareyMap package (Rezvoy et al. 2007) (Fig. 2), resulting in GR rates for nonoverlapping 1-Mb windows along the 12 chromosomes (see Methods). To minimize a potential “centromere effect” (Wu et al. 2003; Zhang and Gaut 2003; Ma and Bennetzen 2006; Rizzon et al. 2006), presumably GR-suppressed pericentromeric regions (Supplemental Table 5) were excluded from correlation analyses.
Figure 2.
Genetic and physical maps of the 12 rice chromosomes and the estimated local GR rates. For each chromosome, circles represent the genetic and physical positions of markers. The curves below the marker plots represent the estimated local GR rates.
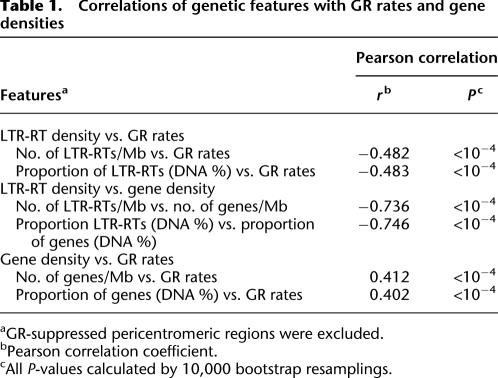
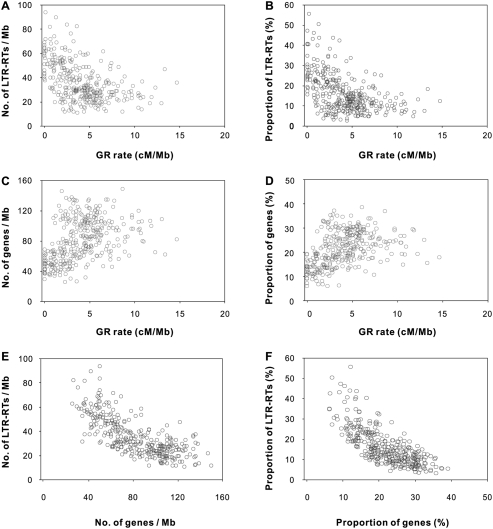
Under these conditions, we investigated the relationship of GR rates with gene densities and LTR-RT densities. Significant negative correlations of GR rates with the numbers of LTR-RTs per Mb were detected in 11 of the 12 chromosomes (Supplemental Table 6). GR rates were also significantly negatively correlated with the proportion of LTR-RT DNA within 1-Mb windows for 10 of the 12 chromosomes (Supplemental Table 6). Overall, significant negative correlations of GR rates with both parameters were detected at the whole genome level (Table 1; Fig. 3A,B). We also calculated gene densities within the contiguous windows on the basis of the latest gene annotation for the rice genome (http://rgp.dna.affrc.go.jp). Nine of the 12 chromosomes exhibit significant positive correlations between GR rates and gene number per Mb; ten of 12 chromosomes exhibit significant positive correlations between GR rates and the proportions of genic DNA; and overall GR rates and gene density were significantly positively correlated at the whole genome level (Table 1; Fig. 3C,D; Supplemental Table 6). Negative correlations between LTR-RT densities and gene densities were observed (Table 1; Fig. 3E,F; Supplemental Table 6). No significant correlation between GR rates and GC content was detected (r = −0.08, P = 0.14).
Table 1.
Correlations of genetic features with GR rates and gene densities
aGR-suppressed pericentromeric regions were excluded.
bPearson correlation coefficient.
cAll P-values calculated by 10,000 bootstrap resamplings.
Figure 3.
Correlations of genomic features with GR rates and gene densities. (A,B) LTR-RT densities plotted against GR rates. (C,D) Gene densities plotted against GR rates. (E,F) LTR-RT densities plotted against gene densities.
To select a linear model to describe the LTR-RT density with a minimum of predictors, a classical stepwise selection procedure (step AIC [Akaike's information criterion]) and a standard alternative testing approach on each predictor were employed (see Methods). Both procedures converged on a final model that suggests GR rates, gene densities, and GC content are all significant predictors of LTR-RT densities (Supplemental Table 7). In this model, GR and gene density are significantly linearly negatively correlated with LTR-RT densities, while GC content is significantly linearly positively correlated with LTR-RT densities.
Correlation of GR rates and gene densities with LTR-RT structural variants
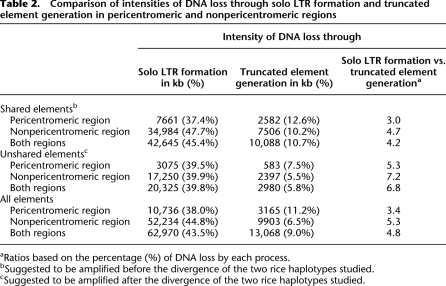
Assuming that each of the solo LTRs and truncated elements was derived from an intact LTR-RT, we estimated that ∼63 Mb and 13 Mb of LTR-RT DNA has been removed from the rice genome through the formation of solo LTRs and truncated elements, respectively, since their initial integration (Table 2). On the basis of the assumptions that solo LTRs are the outcomes of UR and that truncated elements are products of IR, the intensity of DNA loss by UR is therefore about five times higher than by IR (Table 2). This result suggests that UR is initially more active than IR in eliminating LTR-RT DNA in the rice genome. More generally, it is reasonable to assume that a solo LTR has experienced a single UR event, while a truncated element could be an outcome of multiple IR events and thus possibly be older than a solo LTR. If true, the efficiency of UR for initial removal of LTR-RT DNA may be even higher than estimated based on the analysis of LTR-RT structures.
Table 2.
Comparison of intensities of DNA loss through solo LTR formation and truncated element generation in pericentromeric and nonpericentromeric regions
aRatios based on the percentage (%) of DNA loss by each process.
bSuggested to be amplified before the divergence of the two rice haplotypes studied.
cSuggested to be amplified after the divergence of the two rice haplotypes studied.
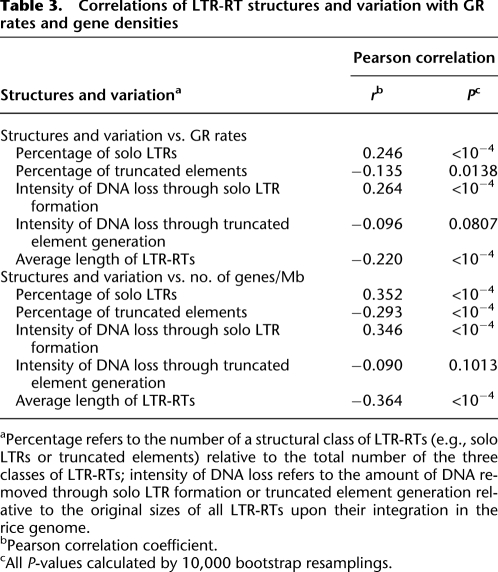
In an attempt to shed light on the effects of GR and gene density on the acquisition and elimination of LTR-RT DNA, we analyzed the relationships between GR rates, gene densities, and LTR-RT structural variants (Table 3; Supplemental Table 8). The relative rates for the formation of solo LTRs and truncated elements were measured by percentages (i.e., the number of solo LTRs and the number of truncated elements relative to the total number of all three categorized LTR-RTs). The percentage of solo LTRs was positively correlated with both GR rates and gene densities across the entire genome. In contrast, truncated elements were negatively correlated with GR rates and with gene densities at the whole genome level (Table 3; Supplemental Table 8).
Table 3.
Correlations of LTR-RT structures and variation with GR rates and gene densities
aPercentage refers to the number of a structural class of LTR-RTs (e.g., solo LTRs or truncated elements) relative to the total number of the three classes of LTR-RTs; intensity of DNA loss refers to the amount of DNA removed through solo LTR formation or truncated element generation relative to the original sizes of all LTR-RTs upon their integration in the rice genome.
bPearson correlation coefficient.
cAll P-values calculated by 10,000 bootstrap resamplings.
As an alternative, we also calculated the relationship between GR rates and the intensities of LTR-RT DNA loss. The intensity of LTR-RT DNA loss was defined as the estimated amount of DNA loss through the formation of solo LTRs or truncated elements in a window relative to the estimated original sizes of all LTR-RTs upon their initial integration (see Methods). The data reveal that, at the whole genome level, GR rates correlate positively with the intensity of DNA loss for solo LTRs but not with truncated elements (Table 3; Supplemental Table 8).
Our intensity calculations also suggest that solo LTR formation is the primary process for LTR-RT DNA removal (Table 2; Supplemental Tables 9, 10). Theoretically, rapid conversion of intact elements to solo LTRs would decrease the chance of creating truncated elements. If this is true, rearrangement of LTR-RTs should follow an age distribution whereby younger elements are more heavily biased than old elements toward solo LTR formation over truncation.
Patterns of recent accumulation and distribution of LTR-RTs
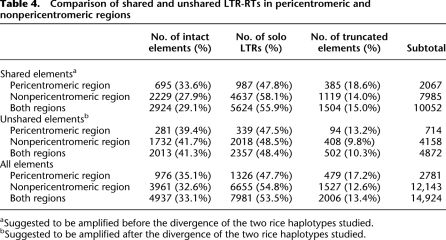
To understand the pattern and dynamics of DNA elimination in the context of the age distribution of elements, we performed comparative analysis of two O. sativa subspecies, japonica (cultivar, Nipponbare) and indica (cultivar, 93-11; Yu et al. 2002) (see Methods). Of the 14,924 solo, truncated, and complete LTR-RTs identified in the japonica genome, 10,052 were shared between japonica and indica, indicating that their presence at these locations in the common ancestor of the two subspecies (Ma and Bennetzen 2004). The remaining 4872 elements were not found in orthologous locations between subspecies, suggesting that these elements inserted in the japonica haplotype after divergence from a shared ancestor with the investigated indica haplotype (Table 4; Supplemental Table 11). This inference is supported by analyses of sequence divergence between LTRs, which suggest that the average ages of the shared and unshared intact elements are 2.28 ± 2.28 (SD) and 0.54 ± 0.75 (SD) million years (Myr), respectively (Supplemental Table 2). This estimate agrees with the previously estimated divergence time (∼0.5 Myr; Ma and Bennetzen 2004) of the investigated japonica haplotype and another indica haplotype (cultivar, GLA4; Han and Xue 2003).
Table 4.
Comparison of shared and unshared LTR-RTs in pericentromeric and nonpericentromeric regions
aSuggested to be amplified before the divergence of the two rice haplotypes studied.
bSuggested to be amplified after the divergence of the two rice haplotypes studied.
The chromosomal distribution of shared and unshared elements is provided in Supplemental Figure 1. Of the 10,052 shared elements, 29.1% are intact elements, 55.9% are solo LTRs, and 15.0% are truncated elements (Table 4; Supplemental Table 11). We estimate ∼42,645 kb of DNA was deleted by solo LTR formation and ∼10,088 kb of DNA was deleted by truncated element generation from shared elements (Table 2; Supplemental Table 9). The intensities of DNA loss through these two processes are 45.4% and 10.7%, respectively (Table 2; Supplemental Table 10). In contrast, of the 4872 nonshared elements, 41.3% are intact, 48.4% are solo, and 10.3% are truncated (Table 4; Supplemental Table 11), corresponding to ∼20,325 kb and ∼2980 kb of DNA lost by solo LTR formation and truncation, respectively (Table 2; Supplemental Table 9). The intensities of DNA loss in unshared elements through the two processes are 39.8% and 5.8%, respectively (Table 2; Supplemental Table 10). These observations support our hypothesis that younger elements are more heavily biased than old elements toward solo LTR formation over truncation and further validate the idea that solo LTR formation is the primary process for initial removal of LTR-RT DNA in the rice genome.
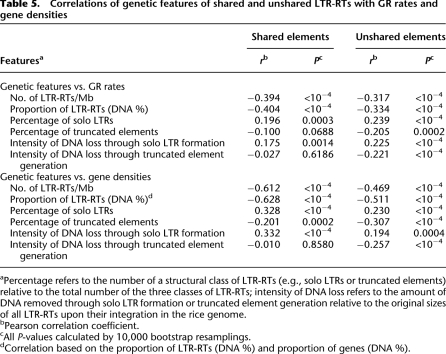
Across the entire genome, the distribution of both shared and unshared elements is negatively correlated with GR (Table 5; Supplemental Table 12). This is true whether we measure LTR-RT distributions as the proportion of DNA within 1-Mb windows or as the number of elements within 1-Mb windows. We also examined structural variants separately. Both shared and unshared solo LTRs were positively correlated with GR, but there was no significant correlation between GR rates and the percentage of shared truncated elements (Table 5).
Table 5.
Correlations of genetic features of shared and unshared LTR-RTs with GR rates and gene densities
aPercentage refers to the number of a structural class of LTR-RTs (e.g., solo LTRs or truncated elements) relative to the total number of the three classes of LTR-RTs; intensity of DNA loss refers to the amount of DNA removed through solo LTR formation or truncated element generation relative to the original sizes of all LTR-RTs upon their integration in the rice genome.
bPearson correlation coefficient.
cAll P-values calculated by 10,000 bootstrap resamplings.
dCorrelation based on the proportion of LTR-RTs (DNA %) and proportion of genes (DNA %).
Analysis of GR-suppressed pericentromeric regions
Thus far, all of our analyses pertain to chromosomal arms, and we have ignored the GR suppressed pericentromeric regions. To examine whether the patterns of LTR-RT accumulation, elimination, and distribution in GR-suppressed pericentromeric regions are consistent with the patterns shown in the chromosome arms, we contrasted results between pericentromeric regions and chromosome arms. The comparative analysis between the two sets of regions on the basis of t-tests, as summarized in Table 6, reveals: (1) the densities of LTR-RTs in pericentromeric regions are significantly higher than those in chromosome arms; (2) gene densities are significantly lower in pericentromeric regions; (3) the percentages of solo LTRs in pericentromeric regions are significantly lower than in chromosome arms, with lower intensities of DNA loss via solo LTR formation; (4) the percentages of truncated elements in pericentromeric regions are significantly higher than in chromosome arms, with commensurately higher intensities of DNA loss; (5) the average sizes of LTR-RTs in pericentromeric regions are significantly larger than those in chromosome arms. Taken together, these observations suggest that pericentromeric regions are fundamentally different from nonpericentromeric regions regarding not only GR rates but also features of LTR-RT structural variation.
Table 6.
Comparison of pericentromeric regions and nonpericentromeric regions
aPercentage refers to the number of a structural class of LTR-RTs (e.g., solo LTRs or truncated elements) relative to the total number of the three classes of LTR-RTs; intensity of DNA loss refers to the amount of DNA removed through solo LTR formation or truncated element generation relative to the original sizes of all LTR-RTs upon their integration in the rice genome.
bP-value by Student's t-test.
Comparison of genomic features and dynamics among different chromosomes
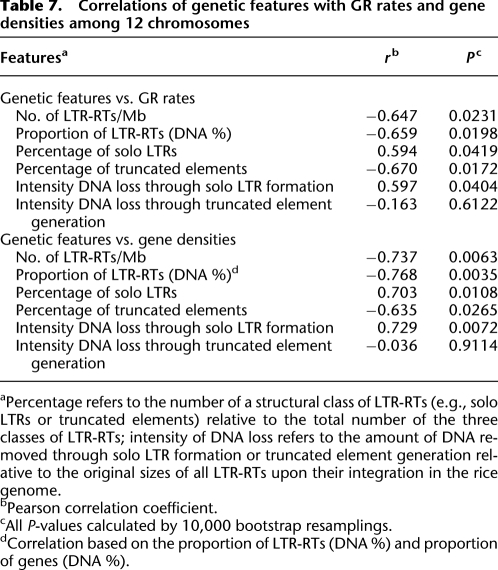
LTR-RT densities vary considerably among chromosomes, ranging from 30 per Mb (chr 3) up to 46 per Mb (chr 4) with an average of 40 per Mb for the whole genome (Supplemental Table 13). Interestingly, these chromosomes also exhibit variation in average GR rates (ranging from 3.54 cM/Mb [chr 10] up to 4.41 cM/Mb [chr 3]) and gene densities (ranging from 58.78 genes per Mb [chr 11] up to 91.23 [chr 3]). We wondered whether the differences in the average GR rates among chromosomes correlates with these genomic features, and therefore, we performed a comparative analysis across the 12 chromosomes (Table 7). The data reveal: (1) significant negative correlations of LTR-RT densities with both average GR rates and gene densities; (2) significant positive correlations of the percentage of solo LTRs with both average GR rates and gene densities; (3) significant negative correlations of the percentages of truncated elements with both average GR rates and gene densities; (4) negative correlations of the intensity of LTR-RT DNA loss in forming solo LTRs with both average GR rates and gene densities; (5) no significant correlation of the intensity of DNA loss in generating truncated elements with either the average GR rates or gene densities. Thus, patterns of LTR-RT accumulation detected along chromosomes also hold among chromosomes.
Table 7.
Correlations of genetic features with GR rates and gene densities among 12 chromosomes
aPercentage refers to the number of a structural class of LTR-RTs (e.g., solo LTRs or truncated elements) relative to the total number of the three classes of LTR-RTs; intensity of DNA loss refers to the amount of DNA removed through solo LTR formation or truncated element generation relative to the original sizes of all LTR-RTs upon their integration in the rice genome.
bPearson correlation coefficient.
cAll P-values calculated by 10,000 bootstrap resamplings.
dCorrelation based on the proportion of LTR-RTs (DNA %) and proportion of genes (DNA %).
Distributions of abundant LTR-RT families
We analyzed the distribution pattern of structural variants for the 10 most numerous LTR-RT families in the rice genome (Supplemental Table 14; Supplemental Fig. 2), in the hope of discerning whether patterns of DNA elimination vary across families. For simplicity, we focused on contrasts between pericentromeric regions and chromosomal arms. Some families show very significant biases in their distribution on all chromosomes, and these biases vary among families. For instance, Park and noaCRR2 elements are exclusively located in the chromosome arms, whereas CRR2 and noaCRR1 are highly enriched in pericentromeric regions (Supplemental Table 15; Supplemental Fig. 3). In addition, the structural types of different families vary greatly (Supplement Fig. 2). For example, rire3 contains ∼24.6% solo LTRs and ∼12.7% truncated elements, whereas osr34 contains ∼63.6% solo LTRs and ∼3.8% truncated elements (Supplemental Table 14). With the current data, it was not possible to establish any relationship between GR rates and the distribution or structural variation of a particular LTR-RT family because of relatively small copy numbers in each of the 12 chromosomes for each family. In general, older families (as estimated by sequence divergence between LTRs) contained higher percentages of solo LTRs and truncated elements (data not shown), but there were some exceptions. For example, the average insertion date of intact elements of the rire3 family is more recent than that for the osr25 family, but the latter is comprised of a higher percentage of solo LTRs (Supplemental Fig. 2).
Distribution of LINEs/SINEs and DNA transposons in the context of GR rates and gene densities
In addition to LTR-RTs, LINEs/SINEs and DNA transposons represent a large fraction (∼14%) of the rice genome (International Rice Genome Sequencing Project 2005). It would be interesting to investigate whether the distribution of these transposable elements was also affected by GR and/or gene density. Thus, we analyzed the correlation of GR rates and gene densities with the distribution of LINEs/SINEs and DNA transposons using the same windows (excluding pericentromeric regions) for the analysis of LTR-RT distribution. The LINEs, SINEs, and DNA transposons in each window were calculated by RepeatMasker-searching against the LINE/SINE and DNA transposon data sets currently available (see Methods). A negative correlation between the abundance of DNA transposons and gene densities was detected at the whole-genome level (r = −0.3703, P < 0.0001), but there was no detectable correlation between the abundance of DNA transposons and GR rates (Supplemental Tables 16, 17). These results, similar to previous observations obtained in Arabidopsis thaliana (Wright et al. 2003), suggest that natural selection against DNA transposon insertion within or near genes plays the major role in shaping the distribution of DNA transposons in both plant species. A weak positive correlation between LINE/SINE abundance and GR rates was observed (r = 0.1682, P = 0.0021), but no clear correlation between LINE/SINE abundance and gene densities was detected at the whole genome level (Supplemental Tables 16, 17). We also compared the distributions of LINEs/SINEs and DNA transposons between chromosome arms and pericentromeric regions and found that both categories of TEs are less abundant in pericentromeric regions than in chromosome arms (Supplemental Table 18).
Discussion
The rice genome is organized along recombinational gradients
The accrual and elimination of LTR-RT DNA have been well documented in Arabidopsis, rice, and several other plant species by analyzing representative samples of LTR-RTs (Devos et al. 2002; Ma et al. 2004; Vitte and Bennetzen 2006). These studies have provided insights into the mechanisms responsible for the size variation among flowering plant genomes. However, the factors that determine specificities in the accumulation, elimination, and distribution of LTR-RTs in local genomic regions have not been comprehensively investigated. Here, we have determined the LTR-RT distribution on the 12 rice chromosomes and compared this distribution with GR rates and gene densities. The observations that GR rates correlate negatively with distributions of LTR-RTs and correlate positively with gene densities indicate that the rice genome is organized along recombinational gradients. The positive correlation of GR with gene density was also observed in the euchromatic regions of maize (Anderson et al. 2006). In contrast, a negative correlation between GR and gene density was detected in the euchromatic regions of Arabidopsis (Wright et al. 2003). Why these genomes differ in this respect remains to be investigated (Gaut et al. 2007). To date, the genome-wide estimates of GR rates have been derived for only two plant (Arabidopsis and rice) genomes by the comparison of genomic sequences and genetic maps (Zhang and Gaut 2003; Rizzon et al. 2006). This study thus provides the first in-depth analysis of the structural variation of LTR-RTs and their distribution in relation to GR rates in any plant genome.
Equal and unequal homologous recombination
Solo LTRs are believed to be the products of UR between the two LTRs of individual elements. Although we cannot rule out the possibility that a solo LTR may be formed at the time of its insertion, the presence of diverged structural forms of some shared LTR-RTs between orthologous regions of indica and japonica genomes—e.g., solo LTRs present in indica versus intact elements present in japonica (Ma and Bennetzen 2006)—favor the hypothesis that solo LTRs are generated by UR. Thus, the positive correlation between the percentages of solo LTRs and GR rates suggests that the components of equal homologous recombination (GR) and UR are shared.
Theoretically, a solo LTR can be generated by intrastrand crossing over or interstrand unequal crossing over (between sister chromatids or between non-sister chromatids). The former causes the removal of an internal segment of an intact element, retaining a solo LTR, while the latter generates both a solo LTR and an LTR–internal–LTR–internal–LTR complex. We identified two LTR–internal–LTR–internal–LTR structures flanked by TSDs (Supplemental Table 2), indicating the occurrence of interstrand unequal crossing over or template switching events (Sabot and Schulman 2007). The relative rarity of LTR–internal–LTR–internal–LTR complexes suggests superficially that intrastrand UR is more frequent than the interstrand events. However, this conclusion needs to be made with the caveat that solo LTRs may be a terminal event, whereas LTR–internal–LTR–internal–LTR structures are likely susceptible to additional UR events that will lead eventually to a solo LTR.
GR and IR
Unlike a solo LTR, which is presumably generated by a single UR event, a truncated element may be the product of several overlapping IR deletions. Hence, the number of IR events, and their possible relationship with GR rates, cannot be precisely determined. Nonetheless, we detected negative correlations between GR and the percentages of truncated elements. Although UR and IR are two independent processes, they both contribute to LTR-RT DNA loss. The genomic regions that allow the persistence of intact LTR-RTs (i.e., those with two LTRs) provide a longer timeframe in which IR can act. Once IR has removed one LTR, then UR is unlikely to act on such an element. Similarly, generation of a solo LTR yields a product that may no longer be able to undergo intraelement UR but can still be subject to further deletion by IR. Taken in total, these observations indicate that UR will most likely be the initial factor to remove LTR-RT DNA (especially from euchromatic regions), but IR will remove most DNA over the long term. This study, for the reasons described above, and because it primarily relies on the identification of LTR-RTs that are recent insertions and thus have at least some intact family members, tends to accentuate the early loss of events and under-represent the later loss events.
Initial frequencies of DNA loss by UR and IR
A previous study revealed rapid elimination of LTR-RT DNA through UR and IR in rice (Ma et al. 2004). On the basis of a survey of randomly chosen LTR-RTs in the rice genome, it was estimated that ∼3.3 Mb and ∼2.5 Mb of LTR-RT DNA was removed through UR and IR, respectively, from the surveyed elements, which made up ∼9.1 Mb of LTR-RT DNA upon their initial integration (Ma et al. 2004). However, a comparative analysis of ∼1.1 Mb of orthologous regions between indica and japonica did not detect much recent DNA loss by IR (<2 kb) from the 17 LTR-RTs present only in japonica. By contrast, eight out of the 17 elements were found to be solo LTRs, indicating a large proportion of LTR-RT DNA loss by UR (Ma and Bennetzen 2004).
To assess the relative degrees of DNA loss by UR and IR that occurred within a recent evolutionary timeframe, only intact elements, solo LTRs, and truncated elements containing at least one LTR were analyzed in this study. Analysis of these elements reveal that LTR-RT DNA loss by UR is about 4.8-fold faster than by IR, and the overall relative degree of DNA loss is ∼53%. When only those elements inserted into japonica after its divergence from indica were counted, the relative degree of DNA loss by UR is about 6.8-fold higher than by IR, and the relative degree of DNA loss by both processes is ∼46%. These calculations indicate that the relative degrees and magnitudes of UR and IR for DNA loss vary over evolutionary time.
Although both UR and IR are responsible for elimination of LTR-RTs in plant genomes, they appear to differ considerably in their relative significance between species. For example, the relative contribution of IR compared with UR was estimated to be approximately twofold higher in Arabidopsis than in rice (Bennetzen et al. 2005; Vitte and Bennetzen 2006). A recent survey of LTR-RTs in wheat, barley, maize, Medicago, and Lotus also revealed that the relative activities of UR compared with IR are highly variable among these species and that there is no apparent correlation of the relative efficiencies of DNA removal by UR or IR with either phylogenetic relatedness or genome size (Vitte and Bennetzen 2006). Although the molecular mechanisms for the amplification and elimination of LTR-RTs are shared by plant genomes, the forces that regulate these mechanisms remain poorly understood. Both population-genetic and comparative approaches are needed to unravel these puzzles (Gaut et al. 2007; Gaut and Ross-Ibarra 2008).
Do GR and gene density affect UR and IR?
Although the positive correlations of the percentages of solo LTRs with GR rates and gene densities detected along several chromosomes are weak or insignificant, a few additional lines of evidence suggest that the formation of solo LTRs by UR was affected by GR. The evidence includes the significantly lower percentages of solo LTRs in GR-suppressed pericentromeric regions than in chromosome arms and the significant positive correlation between the average GR rates of individual chromosomes and the percentages of solo LTRs in corresponding chromosomes. However, we want to point out that the relationships between GR rates and the structural variation of LTR-RTs may still not be able to be fully revealed, because the structural variation is the reflection of many recombination (e.g., UR and IR) events that occurred within different evolutionary timeframes, while the GR rates represent the current status of the local genomic property. Interestingly, the chromosomes that show the strongest correlations between GR rates and the percentages of solo LTRs are among those that contain the highest ratios of unshared elements to shared elements (Supplemental Table 11), suggesting that the effects of GR on the structural variation of LTR-RTs may be better revealed by analyzing younger elements.
The sizes of the windows dissected for the correlation analysis may also influence the accuracy of the calculation of UR frequency. This is particularly true in some euchromatic regions, where limited numbers of LTR-RTs were identified. In such circumstances, larger windows may be able to reduce potential bias in calculating the percentages of solo LTRs and truncated elements caused by relatively small numbers of LTR-RTs in local regions. However, larger-size windows could mask the existence of GR hotspots and coldspots and underemphasize GR-rate heterogeneity on fine scales (Gaut et al. 2007), and thus the “real” effects of local GR on local genomic variation of LTR-RTs would be weakened or even hidden. In addition, LTR-RT families demonstrate distinct distribution patterns along chromosomes, and it appears that different families of intact elements were amplified within distinct evolutionary timeframes. Furthermore, intact elements generally amplified more recently than solo LTRs and truncated elements, and thus the relative abundance of solo LTRs and truncated elements are related to the scales and times of LTR-RT bursts. Establishment of a system to investigate the structural variations of young elements, such as active LTR-RTs, amplified within a recent and narrow timeframe in well-defined coldspots and hotspots of GR, may further validate the relationship between GR and genomic variation.
The correlation between GR rates and intensity of DNA loss by the generation of truncated elements was not found to be statistically significant. This suggests that GR does not strongly affect IR. The gradual accumulation of more truncated elements in lower-GR regions with lower levels of gene density may reflect lower efficiencies of natural selection at removing potential deleterious mutations caused by LTR-RT insertion, in contrast to the higher-GR regions with higher levels of gene densities.
TEs, recombination, and structural variation across eukaryotic genomes
By structural analysis of the LTR-RTs distributed along chromosomes, we observed a negative correlation between GR rates and LTR-RT density. In contrast to this study, such a correlation was not detected in the Drosophila melanogaster genome (Rizzon et al. 2002). In the Drosophila study, the density of DNA transposons was found to negatively correlate with GR rates, but such a correlation was not detected in rice. In Caenorhabditis elegans, a positive correlation was observed between GR rates and the number of DNA transposons, and a lack of correlation was seen between GR rates and the number of LTR-RTs and non-LTR-RTs (Duret et al. 2000). These variable findings suggest that the relative nature of the forces acting on TE distribution and maintenance can vary across organisms. The different accumulation biases of DNA transposons, non-LTR-RTs, and LTR-RTs within or across genomes, as revealed by a number of genome sequencing projects (Adams et al. 2000; The Arabidopsis Genome Initiative 2000; Venter et al. 2001; International Rice Genome Sequencing Project 2005), favor this hypothesis. Nevertheless, the inconsistencies may also be attributable to the type of data used, such as the accuracy of GR rate estimates or the completeness and representation of TE data (Gaut et al. 2007). A recent study revealed significant effects of haplotype polymorphisms in LTR-RTs on genetic recombination in maize (Dooner and He 2008), suggesting that the local genomic compositions in any pair of parental lines can dramatically influence GR rates and hence their subsequent analysis of any relationship with genomic features. However, given that both shared and unshared LTR-RTs between japonica and indica show similar distribution patterns with respect to the GR rates estimated in this study, such effects may be minimal in the rice genome.
This first comprehensive analysis of the relationship between TE distribution, TE evolution, and recombination across entire chromosomes has answered several basic questions regarding the mechanisms that account for chromosome structure. Additional studies are needed to determine how the mechanisms of transposition, GR, UR, and IR generate heterogeneous genome structures among eukaryotes.
Methods
Identification and classification of LTR-RTs
A combination of structural analyses and sequence homology comparisons were used to identify LTR-RTs in the 12 rice chromosomes (IRGSP Build 4.0 pseudomolecules), including two with completely sequenced and comprehensively analyzed centromeres (Wu et al. 2004; Zhang et al. 2004; Ma and Bennetzen 2006; Ma and Jackson 2006). The intact elements were identified by using LTR_STRUC, an LTR-RT mining program (McCarthy and McDonald 2003), and by methods previously described (Ma and Bennetzen 2004, 2006; Ma et al. 2004). Solo LTRs and truncated elements were identified by sequence homology searches against a rice LTR-RT database that was developed by collecting known LTR-RTs (Ma et al. 2004; Nagaki et al. 2005; Ma and Bennetzen 2006; Chaparro et al. 2007), by scanning the rice genome (IRGSP Build 4.0 pseudomolocules) using LTR_STRUC, and by homology-based sequence comparison using BLAST2, CROSS_MATCH, CLUSTALX, and DOTTER programs (Ma and Bennetzen 2004; Ma et al. 2004). The structures and boundaries of all of the identified LTR-RTs were confirmed by manual inspection. The LTR-RTs were classified by sequence homology comparison, and individual families were defined by the criteria described previously (Ma and Bennetzen 2004; Nagaki et al. 2004, Wicker et al. 2007).
Estimation of GR rates
The local GR rates were estimated by using MareyMap (Rezvoy et al. 2007). A total of 3982 markers from the genetic map of rice (http://rgp.dna.affrc.go.jp; http://www.tigr.org) were anchored to the genomic sequence of the rice genome (IRGSP Build 4.0 pseudomolecules), on the basis of their best matches (>95% in identity and >95% in length) and consistent orders in physical and genetic maps. The GR-suppressed pericentromeric regions (Supplemental Table 5) were defined on the basis of the estimated GR rates and the criteria described previously (Rizzon et al. 2006).
The distribution of LTR-RTs and genes, and subsequent statistical analyses
Each chromosome was split into contiguous 1-Mb regions (called windows) from the end of the long arm to the adjacent boundary of the defined pericentromeric region, and from the other boundary of the pericentromeric region to the end of the short arm of the chromosome. GR rates were obtained for each window and plotted on the basis of their midpoints. The distributions and densities of genes were obtained from the latest annotation of IRGSP Build 4.0 pseudomolecules (http://rgp.dna.affrc.go.jp) with modifications. Genes matching TEs and hypothetical genes were excluded. An LTR-RT or gene was assigned to a particular window based on its midpoint. The pericentromeric regions and windows with >0.5 Mb “N” and the windows (<0.5 Mb) adjacent to the pericentromeric regions were not included in the correlation analysis. “N”s, if any, in the 1-Mb contiguous windows were not counted.
The correlations of GR rates with LTR-RT densities, gene densities, proportions of LTR-RT DNA, percentages of solo LTRs, and truncated elements, or intensity of DNA loss were assessed using Pearson's correlation by 10,000 bootstrap resamplings. To select the linear models explaining the LTR-RT densities with the fewest predictors among the GR rates, the gene density and the GC content parameters, a classical stepwise selection procedure based on the Akaike's information criterion (AIC) (Venables and Ripley 2002) was performed with R. The model selected via the stepAIC R procedure was tested using an alternative classical testing approach to confirm the results: The significance of each predictor was tested via a Student's t-test. If the corresponding P-value of the test was low, the predictor was confirmed to participate to the final model. The comparative analysis of the 12 chromosomes was conducted by a Student's t-test.
Analysis of LINEs/SINEs and DNA transposons
The distributions of LINEs/SINEs and DNA transposons were determined based on homology searches against two data sets provided by the laboratories of Thomas Bureau and Ning Jiang, respectively (International Rice Genome Sequencing Project 2005; http://biology.mcgill.ca/faculty/bureau/data.php; N Jiang, pers. comm.), using RepeatMasker (http://www.repeatmasker.org). The percentage of TE DNA in each window was used for correlation analysis.
Comparison of targeted sequences between japonica and indica
The comparative approach for identification of LTR-RT insertions in indica rice was conducted as described previously (Ma and Bennetzen 2006). Two targeted junction segments for each LTR-RT identified in japonica rice cultivar Nipponbare were extracted and used as queries to search against the shotgun sequences from indica rice cultivar 93-11 to identify orthologous segments/sites. An LTR-RT was assumed to be shared by japonica and indica when one or both junction segments exhibited unique matches to 93-11 genome shotgun sequence data.
Estimation of insertion time
The insertion times of LTR–RTs with both LTRs were determined in a manner described previously (Ma et al. 2004). The mutation rate of 1.3 × 10−8 substitutions per base per year proposed for intergenic sequences of rice (Ma and Bennetzen 2004) was employed to convert sequence divergence into dates of insertion.
Acknowledgments
We thank Loic Ponger, Christophe Ambroise, Claudine Devauchelle, and Min Zhang for their assistance on statistical analysis; Ning Jiang for sharing some unpublished transposon data; and three anonymous reviewers for their constructive suggestions. This work was partially supported by National Science Foundation Plant Genome Research Program (grant nos. DBI-0321678, DEB-0426166, DEB-0723860, and DBI-0501814) and Purdue University faculty startup funds.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083899.108.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Lai A, Stack SM, Rizzon C, Gaut BS. Uneven distribution of expressed sequence tag loci on maize pachytene chromosomes. Genome Res. 2006;16:115–122. doi: 10.1101/gr.4249906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, Wei F, Haberer G, Fuks G, Du C, et al. Uneven chromosome contraction and expansion in the maize genome. Genome Res. 2006;16:1241–1251. doi: 10.1101/gr.5338906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro C, Guyot R, Zuccolo A, Piégu B, Panaud O. RetrOryza: A database of the rice LTR-retrotransposons. Nucleic Acids Res. 2007;35:D66–D70. doi: 10.1093/nar/gkl780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, He L. Maize genome structure variation: Interplay between retrotransposon polymorphisms and genic recombination. Plant Cell. 2008;20:249–258. doi: 10.1105/tpc.107.057596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Marais G, Biémont C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Ross-Ibarra J. Selection on major components of angiosperm genomes. Science. 2008;320:484–486. doi: 10.1126/science.1153586. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK. Recombination: An underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 2007;8:77–84. doi: 10.1038/nrg1970. [DOI] [PubMed] [Google Scholar]
- Han B, Xue Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol. 2003;6:134–138. doi: 10.1016/s1369-5266(03)00004-9. [DOI] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci. 2006;103:383–388. doi: 10.1073/pnas.0509810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Jackson SA. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 2006;16:251–259. doi: 10.1101/gr.4583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, SanMiguel P, Lai J, Messing J, Bennetzen JL. DNA rearrangement in orthologous Orp regions of the maize, rice and sorghum genomes. Genetics. 2005;170:1209–1220. doi: 10.1534/genetics.105.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EM, McDonald JF. LTR_STRUC: A novel search and identification program for LTR-retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- McCarthy EM, Liu J, Gao LZ, McDonald JF. Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 2002;3:RESEARCH0053. doi: 10.1186/gb-2002-3-10-research0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Neumann P, Zhang D, Ouyang S, Buell CR, Cheng Z, Jiang J. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol. 2005;22:845–855. doi: 10.1093/molbev/msi069. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvoy C, Charif D, Guéguen L, Marais ABG. MareyMap: An R-based tool with graphical interface for estimating recombination rates. Bioinformatics. 2007;23:2188–2189. doi: 10.1093/bioinformatics/btm315. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Marais G, Gouy M, Biémont C. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 2002;12:400–407. doi: 10.1101/gr.210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS, Farabaugh PJ, Chaleff DT, Fink GR. The origins of gene instability in yeast. Science. 1980;209:1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Sabot F, Schulman AH. Template switching can create complex LTR retrotransposon insertions in Triticeae genomes. BMC Genomics. 2007;8:247. doi: 10.1186/1471-2164-8-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake Berhan A, Springer PS, Edwards KJ, Avramova Z, Bennetzen JL. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. 4th ed. Springer; New York: 2002. [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jónsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu ZD, Dubcovsky J, Keller B. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell. 2003;15:1186–1197. doi: 10.1105/tpc.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wright SI, Agrawal N, Bureau TE. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mizuno H, Hayashi-Tsugane M, Ito Y, Chiden Y, Fujisawa M, Katagiri S, Saji S, Yoshiki S, Karasawa W, et al. Physical maps and recombination frequency of six rice chromosomes. Plant J. 2003;36:720–730. doi: 10.1046/j.1365-313x.2003.01903.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Yamagata H, Hayashi-Tsugane M, Hijishita S, Fujisawa M, Shibata M, Ito Y, Nakamura M, Sakaguchi M, Yoshihara R, et al. Composition and structure of the centromeric region of rice chromosome 8. Plant Cell. 2004;16:967–976. doi: 10.1105/tpc.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gaut BS. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 2003;13:2533–2540. doi: 10.1101/gr.1318503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang Y, Zhang L, Li Y, Lu T, Lu Y, Feng Q, Zhao Q, Cheng Z, Xue Y, et al. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 2004;32:2023–2030. doi: 10.1093/nar/gkh521. [DOI] [PMC free article] [PubMed] [Google Scholar]