Abstract
The Human Microbiome Project (HMP), funded as an initiative of the NIH Roadmap for Biomedical Research (http://nihroadmap.nih.gov), is a multi-component community resource. The goals of the HMP are: (1) to take advantage of new, high-throughput technologies to characterize the human microbiome more fully by studying samples from multiple body sites from each of at least 250 “normal” volunteers; (2) to determine whether there are associations between changes in the microbiome and health/disease by studying several different medical conditions; and (3) to provide both a standardized data resource and new technological approaches to enable such studies to be undertaken broadly in the scientific community. The ethical, legal, and social implications of such research are being systematically studied as well. The ultimate objective of the HMP is to demonstrate that there are opportunities to improve human health through monitoring or manipulation of the human microbiome. The history and implementation of this new program are described here.
It has been known for some time that the human body is inhabited by at least 10 times more bacteria than the number of human cells in the body, and that the majority of those bacteria are found in the human gastrointestinal tract (Savage 1977). Throughout the history of microbiology, most human studies have focused on the disease-causing organisms found on or in people; fewer studies have examined the benefits of the resident bacteria. As noted in reviews by Relman and Falkow (2001) and Relman (2002), the endogenous flora of the human body are poorly understood. Following the publication of the human genome sequence in 2001 (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), Julian Davies argued that although completing the human genome sequence was a “crowning achievement” in biology, it would be incomplete until the synergistic activities between humans and microbes living in and on them are understood (Davies 2001). Relman and Falkow (2001) called for a “second human genome project” that “would entail a comprehensive inventory of microbial genes and genomes at the four major sites of microbial colonization in the human body: mouth, gut, vagina, and skin.” Relman (2002) envisioned that the “characterization of the microbiome would be accomplished through random shotgun sequencing procedures, targeted large-insert clone sequencing, and assessments of intra- and inter-individual variation by using high-density microarrays.” This approach, coupled with a “study of host genome-wide expression analysis,” would yield major “insights into the role of the endogenous flora in health and disease.”
Scientific background
The concept of the human microbiome was first suggested by Joshua Lederberg, who coined the term “microbiome, to signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space” (Lederberg and McCray 2001). Initial efforts to determine the numbers of microbes in a community and their phylogenetic relationships comprised analyzing the relatively well-conserved 16S rRNA genes in mixtures of organisms (Woese and Fox 1977; Stahl et al. 1984; Woese and Olsen 1986; Giovannoni et al. 1990; Schmidt et al. 1991; Dymock et al. 1996). Much of our understanding of the human microbiome comes from culture-based approaches using the 16S rRNA technology. However, it is estimated that as much as 20% to 60% of the human-associated microbiome, depending on body site, is uncultivable (Pei et al. 2004; Verhelst et al. 2004; Zhou et al. 2004; Aas et al. 2005; Bik et al. 2006), which has likely resulted in an underestimation of its diversity.
More recently, studies have been published that describe the human microbiome in different biological states using the 16S rRNA gene sequencing technique. For example, studies of the gut microbiome at the 16S rRNA gene level have revealed a significant diversity in the flora of individuals (Eckburg et al. 2005), have shown differences in the flora of obese versus lean donors (Ley et al. 2006), and have followed the evolution of the microbiome in infants (Palmer et al. 2007). Studies also have used the 16S rRNA gene as a metagenomic marker of the microbiome in the oral cavity (Faveri et al. 2008), vagina (Hyman et al. 2005), and skin (Gao et al. 2007).
Although enormously important in helping scientists define evolutionary relationships among bacteria, there are limitations to the 16S rRNA gene sequencing approach that have prompted more recent studies to examine the complexity of environmental samples by sequencing genomic libraries made from DNA extracted directly from the mixed sample (Handelsman et al. 1998). This approach is called “metagenomics” and was initially applied in several studies of environmental microbial communities (Handelsman 2004; Tyson et al. 2004; Tringe and Rubin 2005; Nealson and Venter 2007).
Initiation of the HMP
The early studies examining the microbiome stimulated interest in undertaking a large-scale investigation of the human intestinal microbiome. An international meeting was held in Paris in November 2005 to discuss such an effort. This meeting, hosted by the French National Institute for Agricultural Research (INRA) and chaired by Dusko Ehrlich, led to the recommendation that a Human Intestinal Metagenome Initiative (HIMI) be undertaken to define more completely the human intestinal microbiome in health and disease. The meeting attendees also recommended that an International Metagenome Consortium be formed to bring together common efforts from around the world to accomplish the goals of the HIMI (http://human-microbiome.org).
Directly following the Paris meeting, the NIH held discussions about the merits of initiating an NIH-sponsored Human Microbiome Project (HMP) to study the human microbiome broadly by examining at least four body sites (the gastrointestinal tract, the mouth, the vagina, and the skin). Eventually, the HMP was organized and funded as a cutting edge, high-priority, transformative initiative of the NIH Roadmap for Biomedical Research.
This paper is a “marker paper” for the HMP. The “marker paper” is a concept originally developed at a 2003 meeting (the “Ft. Lauderdale Meeting”) on data release (see Sharing Data from Large-Scale Biological Research Projects: A System of Tripartite Responsibility; http://www.wellcome.ac.uk/About-us/Publications/Reports/Biomedical-science/WTD003208.htm [2003]) and recently reconfirmed at an international meeting in Toronto, Canada (Toronto International Data Release Workshop Authors 2009). The attendees of both meetings recommended that large, genome-scale “community research projects” be described in a manuscript at the outset that discusses the scope and vision of the project to inform the research community about the experimental design, plans to make data available, expected data standards, quality control, and Consortium publication plans.
The goal of the HMP
The goals of the HMP are to demonstrate the feasibility of characterizing the human microbiome well enough to enable study of the variation in the human microbiome (with population, genotype, disease, age, nutrition, medication, and environment) and its influence on disease, while providing both a standardized data resource and technological developments to enable such studies to be undertaken broadly in the scientific community. The HMP is a limited effort per se, but has the ultimate objective of creating broad opportunities to improve human health through monitoring or manipulating the human microbiome.
The HMP is a 5-yr project that will be funded at a level of $150 million over that period. It is envisioned that, by the conclusion of the HMP, the project will have created the needed resources (data, technology, computational tools, and preliminary analyses of the ethical, legal, and social implications of the research) and will have documented enough examples of the microbiome's importance to human health that support of microbiome research will become a priority in the human disease portfolio of many of the NIH Institutes and Centers and of many other funding agencies. If the importance of considering the impact that changes in the normal microbiome can have on health and disease can be demonstrated, it will have the potential to transform medicine. The websites http://nihroadmap.nih.gov/hmp/ and http://hmpdacc.org/, and links therein, provide additional information about the HMP and access to HMP data.
Implementation of the NIH HMP
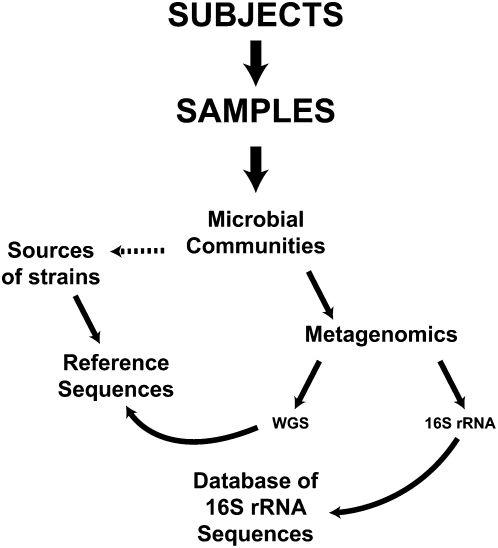
The guiding model for the HMP (Fig. 1) has several inter-related parts. First, the HMP will add at least 900 additional reference bacterial genome sequences to the public database. At the same time, samples will be collected from “normal” (see below) volunteers to characterize the microbiome at several important body sites. Initially, 16S rRNA gene sequencing will be used to identify the microbiome community structure at each site. Comparison of the resulting 16S data to the data already available in databases will be used to relate the sources to known species. Metagenomic shotgun sequencing also will be performed on the same samples for deeper analysis. For example, the results will be BLASTed against the data in GenBank, KEGG, and so on, to identify genes and potential functions. The data from many individuals will be analyzed to determine whether there is a core microbiome at each body site.
Figure 1.
Scheme for studying the “normal” human microbiome. Subjects are recruited, and samples are collected. The microbial communities in the samples are analyzed using metagenomic sequencing approaches. 16S rRNA sequences are compared to databases of 16S rRNA sequences, while whole-genome shotgun (WGS) sequences are compared to existing reference strains, many of which will have been sequenced under the HMP. The microbial communities from the donors can also serve as sources of strains that will be sequenced to increase the number of known reference sequences.
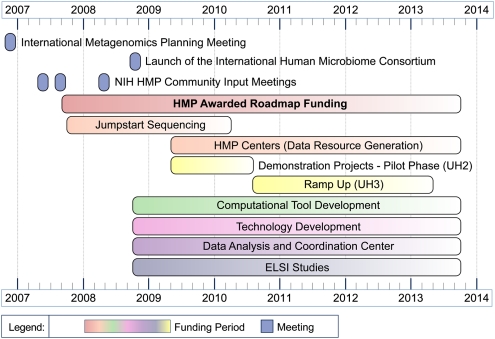
The NIH HMP is being implemented in a phased manner (for a list of funded projects, see timeline in Fig. 2 and http://nihroadmap.nih.gov/hmp/fundedresearch.asp).
Figure 2.
HMP timeline.
The Jumpstart phase
The initial phase of the project, called the “Jumpstart” phase, was initiated in 2007. The participants were four large-scale sequencing centers supported by NIH, The Baylor College of Medicine, The Broad Institute, The J. Craig Venter Institute, and the Washington University School of Medicine. The Jumpstart funding supported:
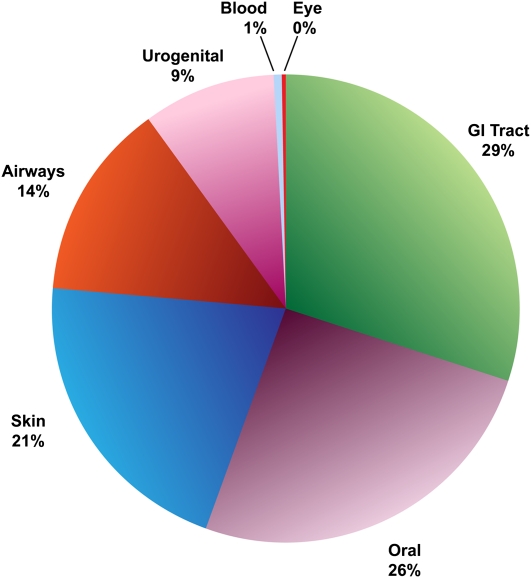
Sequencing of 500 new reference bacterial genomes (see Fig. 3 for distribution by body site).
Developing a protocol for recruiting and sampling at five body sites (the gastrointestinal tract, the mouth, the vagina, the skin, and the nasal cavity) in a pilot with 250 “normal” volunteers made up of equal numbers of men and women recruited from a diverse population.
Performing 16S rRNA gene sequencing on samples from five body sites from female and four body sites from male volunteers (with subsampling from different anatomical regions at each of those sites, for a total of 15 samples from males and 18 from females) (http://www.hmpdacc.org/sampling.php).
As of July 2009, more than 500 bacterial genomes are being pursued by a sequencing center. Roughly 375 are in draft sequencing pipelines or have been completed and deposited in GenBank. A number of these reference bacterial genomes were initially proposed by members of the research community as part of white papers submitted to NHGRI's process for selecting sequencing targets. These, mainly found in the human gastrointestinal tract and to a lesser extent the vagina, were subsequently made part of the HMP reference bacterial genome sequencing effort. As the NIH HMP got under way, it developed a process to solicit recommendations from the research community for bacterial genomes to be sequenced. Four working groups to address the five body sites (the gastrointestinal tract, the mouth, the vagina, the skin, and the nasal cavity) were established with members from the microbiological research community, the NIH, and the HMP Sequencing Centers, to identify and recommend bacterial genomes to be sequenced. The list of sequencing projects under way and completed can be found at http://www.hmpdacc.org/.
Figure 3.
Bacterial distribution by body site. This figure shows the distribution by body site of bacteria that have been sequenced under the HMP or are in the sequencing pipelines.
The umbrella protocol for recruitment of normal subjects was agreed to through consultation with groups of experts in the five body sites to be sampled (the GI tract, the mouth, the vagina, the skin, and the nasal cavity), working along with experts in research ethics and informed consent. The issue of whose samples to include in the “reference” microbiome resource was debated, and it was acknowledged that it would not be realistic to obtain a sample large enough or diverse enough to be truly representative of the entire U.S. population. However, efforts were made to recruit a sample that was reasonably diverse in terms of race, ethnicity, and other demographic features.
The term “normal” rather than “healthy” is used in this study to denote that in order to achieve an umbrella protocol, body site-specific experts revised some of the exclusion criteria so that they would no longer define the biology at the site as “healthy.” This was done in order to reduce the number of exclusion criteria to make it, in the clinicians' opinion, possible to recruit volunteers. There was concern that recruitment using a protocol calling for volunteers who were “healthy” at each site (as defined by the sample site experts) would have so many exclusion criteria that recruitment would be very slow or impossible.
Special attention was paid to the informed consent process, so that potential sample donors were adequately informed about the benefits and risks associated with participation in a “community resource” project. A template for an informed consent form was developed and then adapted for use at the two centers where sampling took place (Baylor College of Medicine and Washington University; see http://hmpdacc.org/clinical.html for consent forms). Particular attention was given in the consent process to informing donors about how their privacy would be protected and the limitations of the available protections. Donors were informed that the microbiome data from the study of their samples would be deposited in an open access database on the Internet, while any human DNA data, and any personal medical information collected from them, would be in a controlled access database (dbGaP), available only to human microbiome researchers approved by an NIH Data Access Committee.
Progress to date includes recruitment and sampling of nearly all of the 250 volunteers (approximately equal numbers of men and women) with collection of a second sampling time point started. The 16S rRNA gene sequencing of those samples is well under way with completion of the samples from the first 18 volunteers. The sequence data, from which contaminating human DNA has been removed, have been submitted to the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/home/); phenotypic data for these individuals will be deposited at dbGAP (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap) in the near future once the data structures are in place. In accomplishing these goals, the Jumpstart investigators have developed a common set of sampling and sequencing protocols, plus a set of rigorous standards and quality-control guidelines, to ensure that the data from different laboratories and from all sequencing platforms (including “next-gen”) are comparable and reliable (http://hmpdacc.org/sops.php).
One of the key efforts has been a set of experiments using a reconstructed (“mock”) community to test the reliability of the data from multiple sites and set standards for analyzing the microbiome; the sequence data from the mock community have been deposited in GenBank, and the results of these experiments will be published elsewhere. A significant finding from the mock community experiment has been the importance of benchmarking data sets to each other to ensure data reliability and comparability. For example, the initial sequence data from the mock community experiments predicted a complexity far greater than the true set of bacterial genomes present in the mock community mixture. A series of follow-up experiments, for example, using new and better tools to eliminate chimeras, removed the artifacts so that the centers are now confident that their sequence data are comparable and can be expected to reliably reflect community complexity at a body site. A major part of these efforts was development of specialized software such as for chimera checking. Many of these programs are now available as free downloads through the individual Jumpstart sequencing center websites accessible through http://hmpdacc.org/.
Second phase
In the next phase of the HMP, the participants will continue the work started in the Jumpstart phase to generate reference databases and to develop new technology, and then extend the effort with a set of demonstration projects to try to determine changes in the microbiome correlated to specific disease states.
HMP sequencing centers
The four centers that participated in the Jumpstart phase will continue to develop and use next-gen methods in the next phase to sequence at least 400 more bacterial genomes to complete the goal of adding 900 new bacterial reference genomes to the sequence databases. (An additional 100 new bacterial genomes are being sequenced by the European MetaHit Project; thus, a total of at least 1000 new genome sequences from human commensal microorganisms will be added to the public repository.) The centers will also sequence the genomes of viruses and eukaryotic microbes found in the human microbiome and perform 16S rRNA gene and whole-genome shotgun sequencing to characterize the microbiomes of the 250 donors sampled in the Jumpstart phase at additional time points. All of these data will be used to study the diversity of the microbiome at each body site and to attempt to define a core microbiome at each site, at the highest phylogenetic resolution that the data allow (most likely at or below the family level). Conservation of gene function across the sites also will be examined. The HMP centers plan to publish manuscripts describing broad analyses of the set of sequenced bacterial genomes and microbial metagenomic data generated from the collection of normal samples from 18 body sites in more than 250 individuals.
HMP Demonstration Projects
The Demonstration Projects aim to tackle the most important question of the HMP: whether changes in the microbiome can be related to human health and disease. Because of the short time frame of the HMP, the primary goal of these projects is to establish a correlation between microbiome changes and health/disease, rather than demonstrate causation. If a project can successfully demonstrate correlation early in the timeline, work to begin to establish causation may be undertaken. But the HMP recognizes that such studies may take years of work to complete and go beyond the initial goals of the HMP.
Fifteen investigator-initiated projects have recently been funded for an initial 1-yr pilot phase, during which each investigator will have the opportunity to demonstrate the feasibility of his/her project (http://nihroadmap.nih.gov/hmp/fundedresearch.asp). At the end of the pilot phase, the subset of the projects that demonstrate the most promise, as judged by peer review, will be scaled up and continued for three additional years. Collectively, the 15 pilot projects will study bacterial, fungal, and viral changes related to various health conditions in microbiome samples from consented human participants, including those that affect the skin, the nasopharynx, the oral cavity, the gastrointestinal tract, the genitourinary tract, and the blood. Individuals who provide the samples to be studied in the demonstration projects will provide consent analogous to those used for the Jumpstart project, again being informed of the special privacy issues associated with the deposition of data in a public database. High-throughput sequencing will be used to produce microbiome (bacterial, viral, and fungal) sequence data (both bacterial and fungal rRNA genes and bacterial, viral, and fungal whole genomes) of samples from normal and diseased individuals. At the same time, in many of these studies, complementary molecular approaches will be used to measure microbial gene expression and host genotypes. Data analysis will be performed using existing or new computational methodologies, to define the potential relationships between changes in the human microbiomes and the health conditions under study. Benchmarking to compare technical and analytical pipelines from the demonstration projects is currently underway, using the “mock” community approach mentioned above.
Examples of HMP Demonstration Projects
The HMP is supporting a very diverse set of research projects among the 15 funded grants. Rather than attempt to summarize each research project, we give a few examples of projects illustrating approaches being taken in different health conditions:
Skin: Martin Blaser, PI. “Evaluation of the Cutaneous Microbiome in Psoriasis.” Psoriasis, a chronic disease involving the immune system, affecting more than 7.5 million people in the United States, appears on the skin, usually in the form of thick, red, scaly patches. Its cause is unknown. The goal of this study is to examine how changes in the normal cutaneous microbiome may contribute to the disease. The skin microbiome of 75 donors with and without psoriasis will be examined at several taxonomic levels. Additionally, the research will seek to examine whether the immunosuppressive agents used to treat psoriasis alter the microbiome.
Virome: Gregory Storch, PI. “The Human Virome in Children and Its Relationship to Febrile Illness.” An estimated 20 million visits per year to hospital emergency departments are because of fever in children. The causes of a vast majority of these fevers remain undiagnosed. This project seeks to describe the human virome in children and to investigate its relevance to febrile disease. It will also study the relationship of the patients' immune systems to the composition of their viromes. Next-gen sequencing technologies will be used to examine the virome of blood, respiratory, and gastrointestinal samples from healthy, from febrile, and from immunosuppressed children.
GI Tract: Claire Fraser-Liggett and Alan Shuldiner, PIs. “The Thrifty Microbiome: The Role of the Gut Microbiome in Obesity in the Amish.” Obesity is a major health problem in the United States. This project directly addresses the causes of obesity by testing the “Thrifty Microbiome Hypothesis,” which poses that the gut microbiome plays a key role in human energy homeostasis. Previous studies have indicated that a difference in the gut microbiome can be found in obese and lean adults. This study will perform a functional and genomic assessment of the gut microbiome in donors whose genetics and phenotypic traits (weight, fat deposition, etc.) are carefully recorded. The Old Order Amish population was chosen for this study because it is genetically homogeneous and has already been characterized for many of the traits being studied.
Vagina: Jacques Ravel and Larry Forney, PIs. “The Microbial Ecology of Bacterial Vaginosis: A High-Resolution Longitudinal Metagenomic Analysis.” Bacterial vaginosis (BV) arises in women when the vaginal microbiome is disrupted. It is a common condition that is very difficult to control. This project will test the hypothesis that vaginal microbiome dynamics and activities are indicators of risk of BV. The study will examine daily changes, over two menstrual cycles, in the vaginal microbiome of 200 women and correlate them with occurrence of BV to better define the syndrome and identify patterns that are predictive of BV. It will use 16S rRNA genes, metagenome and metatranscriptome sequencing utilizing next-gen sequencing technologies, in combination with metabolomics, to assess the diversity of microbial species, genes, and functions of the microbiome associated with these samples.
Cancer of the GI Tract: Zhiheng Pei, PI. “Foregut Microbiome in Development of Esophageal Adenocarcinoma.” Esophageal adenocarcinoma (EA), the type of cancer linked to heartburn due to gastroesophageal reflux diseases (GERD), is the fastest rising malignancy in the United States. The recent increases cannot be explained by environmental or host factors. Initial research by the PI's laboratory has shown that patients carrying particular types of microbiome are more likely to have the early stages of EA than those that do not. The group will examine the 16S rRNA genes and whole-genome sequence composition of the microbiome in each stage of development of EA. If a significant association of the changes in the microbiome during development of EA can be shown, early diagnosis and treatment of the disease may be possible through strategies that convert the disease-related microbiome to the healthy microbiome.
Data Analysis and Coordination Center (DACC)
The HMP Data Analysis and Coordination Center is an informatics resource that will make information about the HMP, including results and conclusions, available to the scientific community. The HMP DACC will also coordinate the development of data standards and facilitate the analysis and deposition of data to the appropriate public repositories (http://hmpdacc.org/). The HMP DACC website contains the HMP Project Catalog of the reference strains. This searchable and sortable Project Catalog contains various types of information about each of the reference strains, including body site of isolation, sequencing status, and which center is sequencing the strain. Links from the Project Catalog lead to more biological information and references for the strains.
Technology development
Producing a reference set of complete genome sequences is central to the HMP because such information is needed to interpret metagenomic sequence data. To sequence individual microbial genomes, it has generally been necessary to grow cultures of those microbes; however, much of the microbiome cannot currently be grown in culture. Therefore, the HMP is also supporting the development of new technologies that will allow the isolation of many more purified microbial species so their genomes can be sequenced, thereby expanding the reference collection. Among the approaches supported are the development of methods to culture previously uncultivable bacteria; to isolate single microbial cells; to isolate, amplify, or clone unamplified or amplified DNA of whole genomes from individual cells at high fidelity and coverage; and to use tagging strategies to facilitate the enrichment of cells of a given species to essential purity. Individual projects seek, for example, to collect the microbiome from distinct sites in the gut that would not routinely be sampled (e.g., tightly adherent to the intestinal mucosa) and then enrich individual species by flow-sorting from those reduced-complexity samples; to reproduce in vitro growth conditions that mimic the microoxic environment in the gut and that have not previously been applied to cultivation of human microbes; to trap large genomic DNA fragments by virtue of small amounts of known sequence within them; and to produce microfluidic devices to capture large numbers of individual microbes and then amplify and sequence those that are recognized by various criteria as being unknown or rare.
Computational tools
Vast amounts of data will be generated by the HMP using next-gen sequencing technologies and other high-throughput methods. Computational methods to process and analyze such data are in their infancy, and, in particular, objective measures and benchmarks of their effectiveness have been lacking. Projects supported by the HMP will address issues related to: genome assembly and gene-finding software for metagenomics data sets generated from new sequencing technologies; characterization of biodiversity in samples; statistical models and simulations to compare different measures of microbial diversity; and gene annotation tools for classification of protein families and functional prediction of HMP data.
Ethical legal and social implications (ELSI) of HMP research
Since its inception, the field of genomics has been characterized by a component designed to anticipate and address the ethical, legal, and social implications of genomics research at the same time that the genomics research itself is being conducted. The findings of these studies have in many cases been very influential in helping to inform the way genomics research is carried out, while also increasing the knowledge base regarding more distal applications of the science, including its broader societal impact. Although ELSI research in the field of microbiology has a less well-established tradition, its importance in the field is increasingly being recognized. Thus, an integral part of the HMP involves consideration of the potential ELSI issues that might be associated with microbiome research (Box 1). To date, five projects have been initiated as part of this component of the HMP. One of these projects will involve interviews with the individuals who donated the samples to be studied to develop the reference microbiome resource, as well as with individuals who were asked to donate samples but declined, in order to explore general perceptions and attitudes in the public about this new area of research. Other funded projects in this area include studies to analyze how risk and benefit are conceptualized in human microbiome research; to investigate patient perceptions of bioengineered probiotics and clinical metagenomics; to analyze existing regulatory frameworks for the federal regulation of probiotics; and to investigate the implications of research on the ancient and contemporary human microbiome for the social and ancestral identities of indigenous people.
Box 1.
Ethical, legal, and social implications
Data release
The NIH has designated the HMP as a “community resource.” As such, NIH has designed data release policies for the HMP (http://nihroadmap.nih.gov/hmp/datareleaseguidelines.asp) according to the principles of rapid data release as outlined in the Fort Lauderdale accords (http://www.genome.gov/10506376), and the participants in the data production components of the Project have agreed to abide by those policies, for example, data generated will be released rapidly after it is produced, before publication. The Demonstration Projects have agreed to a 12-mo publication moratorium on their data—that is, other researchers may use the data once they are released, but the producers have a 12-mo window in which to publish on the data. Users of data from the large-scale sequencing centers (which have no publication moratorium) are asked to respect the principles put forth in the Ft. Lauderdale agreement, for example, respect the data producers' rights to publish a first global analysis of the data, credit the data producers in any publication, and confer with them regarding any use of the data in a publication prior to their first publication on the data.
As earlier noted, human participants who provide samples for the HMP have consented to this broad data release, and procedures have been established to safeguard the identity of individual sample donors.
The HMP Research Network
The production components of the HMP and the demonstration projects are organized as a Research Network to enhance coordination among, and synergy from, the individually funded efforts. The HMP grants supporting technology development, bioinformatics tool development, and ELSI projects are not formally members of the Research Network, but are regularly invited to attend its meetings. Similarly, the HMP Research Network is open to other U.S. investigators conducting research projects that have similar goals. Specifically, researchers who are using 16S rRNA gene or whole-genome shotgun metagenomic analyses to study changes in the human microbiome in health and disease or who are generating a significant number of reference microbial genome sequences are welcome to join the research consortium as full members. The condition for membership is agreement with the principles of rapid (prepublication) data release and intellectual property management as set out in the HMP Policy for Membership (http://nihroadmap.nih.gov/hmp/datareleaseguidelines.asp).
The International Human Microbiome Consortium (IHMC)
In December 2007, the NIH hosted a meeting of interested researchers from around the world to discuss the formation of an International Human Microbiome Consortium (IHMC) (see http://www.human-microbiome.org/) that would coordinate human microbiome research around the world to eliminate unnecessary duplication of effort, ensure rapid data release of human microbiome data sets (molecular and clinical) with appropriate informed consent, agree to common data quality standards, and share tools and research results of human microbiome research. The IHMC was officially launched in September 2008 with a membership of 10 countries. The IHMC is open to membership by any research project that is willing to agree to the IHMC principles (http://www.human-microbiome.org).
Conclusion
The HMP is designed to push the frontiers of human microbiology by providing the data, tools, and resources to inform medical science more fully about the role of changes in the resident human microbiome in disease and health. It will provide a database of information that will serve as a reference for future studies. The HMP is being launched at a time of tremendous technological change, when the cost of DNA sequencing is dropping exponentially and throughput is increasing many-fold. HMP's focused effort to set standards for the use of the next-gen technologies in metagenomic studies and generate accurate, quality controlled data will serve as a foundation for future human microbiome research.
Acknowledgments
This work was supported by funding from the NIH Roadmap for Medical Research and the National Human Genome Research Institute. The authors thank the PIs and staff at the HMP large-scale sequencing centers at Baylor College of Medicine (especially Richard Gibbs, Joe Petrosino, Sarah Highlander, and Jim Versalovic), The Broad Institute (especially Bruce Birren and Doyle Ward), The J. Craig Venter Institute (especially Robert Strausberg, Karen Nelson, and Barb Methe), the Washington University School of Medicine (especially George Weinstock and Erica Sodergren), and the Data Analysis Coordination Center at the University of Maryland (especially Owen White and Jennifer Wortman) for their permission to cite the data, results, and plans discussed in this marker paper as well as for their help with the manuscript. We thank the HMP Working Group Co-chairs, Lawrence Tabak, Griffith Rodgers, and Anthony Fauci, and the NIH Roadmap staff, Mary Perry and Elizabeth Wilder, for their guidance through the inception and launch of this project.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.096651.109.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. In a map for human life, count the microbes, too. Science. 2001;291:2316. doi: 10.1126/science.291.5512.2316b. [DOI] [PubMed] [Google Scholar]
- Dymock D, Weightman AJ, Scully C, Wade WG. Molecular analysis of microflora associated with dentoalveolar abscesses. J Clin Microbiol. 1996;34:537–542. doi: 10.1128/jcm.34.3.537-542.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23:112–118. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lederberg J, McCray AT. ’Ome Sweet ’Omics—a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Venter JC. Metagenomics and the global ocean survey: What's in it for us, and why should we care? ISME J. 2007;1:185–187. doi: 10.1038/ismej.2007.43. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. New technologies, human–microbe interactions, and the search for previously unrecognized pathogens. J Infect Dis. 2002;186:S254–S258. doi: 10.1086/344935. [DOI] [PubMed] [Google Scholar]
- Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001;9:206–208. doi: 10.1016/s0966-842x(01)02041-8. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Lane DJ, Olsen GJ, Pace NR. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984;224:409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- Toronto International Data Release Workshop Authors. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L, De Ganck C, Temmerman M, Vaneechoutte M. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ. Archaebacterial phylogeny: Perspectives on the urkingdoms. Syst Appl Microbiol. 1986;7:161–177. doi: 10.1016/s0723-2020(86)80001-7. [DOI] [PubMed] [Google Scholar]
- Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]