Abstract
Although a highly accurate sequence of the Caenorhabditis elegans genome has been available for 10 years, the exact transcript structures of many of its protein-coding genes remain unsettled. Approximately two-thirds of the ORFeome has been verified reactively by amplifying and cloning computationally predicted transcript models; still a full third of the ORFeome remains experimentally unverified. To fully identify the protein-coding potential of the worm genome including transcripts that may not satisfy existing heuristics for gene prediction, we developed a computational and experimental platform adapting rapid amplification of cDNA ends (RACE) for large-scale structural transcript annotation. We interrogated 2000 unverified protein-coding genes using this platform. We obtained RACE data for approximately two-thirds of the examined transcripts and reconstructed ORF and transcript models for close to 1000 of these. We defined untranslated regions, identified new exons, and redefined previously annotated exons. Our results show that as much as 20% of the C. elegans genome may be incorrectly annotated. Many annotation errors could be corrected proactively with our large-scale RACE platform.
One of the goals of obtaining full genome sequences in the 1990s was to precisely identify the full complement of proteins, or proteome, used by model organisms and humans. The Caenorhabditis elegans genome was the first metazoan genome to be sequenced (The C. elegans Sequencing Consortium 1998) and remains today the only complete and contiguous animal genome sequence ever established. The sequence is of excellent quality, with less than one mismatch per 30,000 nucleotides in the original release 10 yr ago and an error rate currently of less than one mismatch per 40,000 nucleotides (Hillier et al. 2005).
Despite this high-quality genome sequence, the complexity of gene transcription initiation and termination as well as differential splicing has made it extremely challenging to experimentally verify the exact sequence of the full complement of predicted protein-coding open reading frames (ORFs), or ORFeome (Walhout et al. 2000b). More than 10 yr after the first release of the genome sequence, the ORFeome remains imprecisely defined.
Our first attempt to experimentally verify the C. elegans ORFeome used PCR-based cloning of ORFs using the Gateway system (Walhout et al. 2000a) carried out from a high-quality cDNA library (Walhout et al. 2000b) with ORF-specific primers based on the WS9 release of WormPep (August 1999). In this first attempt, we demonstrated that the number of protein-coding genes in C. elegans exceeds 17,300, a remarkably high number given that the Drosophila genome had been annotated with just 13,600 genes (Reboul et al. 2001). Our first genome-wide effort at cloning the C. elegans ORFeome experimentally verified ∼55% of all ∼19,000 predicted ORFs, with 4000 ORFs that had remained strictly computationally defined until that point (Reboul et al. 2003). In a second effort, using redesigned ORF-specific PCR primers for 4232 repredicted ORFs based on WS100 release of WormPep (May 2003), we successfully cloned 1378 repredicted ORFs and 937 newly predicted ORFs (55%) (Lamesch et al. 2004). In both, the success rate was higher (63%) for ORFs supported by expressed sequence tags (ESTs) or other cDNA sequence evidence (“touched” ORFs), than for “untouched” ORFs lacking experimental support (42%). The remaining 45% of predicted ORFs could not be amplified even with redesigned ORF-specific primers, owing most likely to mispredicted ORF boundaries (Reboul et al. 2001, 2003; Lamesch et al. 2004). Because these verification experiments were done reactively, that is, they were limited to PCR amplification of the predicted computational models, the possibility of extended exon structure beyond the verified portion cannot be overlooked.
Other approaches have provided insight into worm transcript structure. These include reconstruction of mRNA structure based on available EST and cDNA sequences (Thierry-Mieg and Thierry-Mieg 2006); TEC-RED (Hwang et al. 2004), which makes use of trans-spliced leaders to capture short reads from the 5′-end of transcripts; and tiling arrays (He et al. 2007) to investigate which parts of the genome are transcribed. Two recent “next-generation” sequencing efforts of the C. elegans transcriptome (Shin et al. 2008; Hillier et al. 2009) provided information on transcribed genome regions and on splice junctions. These efforts have provided further insights into the transcriptional landscape and transcript structure of the worm. However, complete transcript structures, including cis-connectivity of exons, remain to be fully defined for all encoded genes.
As for any organism, a complete description of the C. elegans full complement of gene products—noncoding RNAs and proteins—including all variants obtained from alternative transcription and splicing is necessary for a comprehensive systems description of its biology. Experimental verifications of computationally derived structural transcript models suggest that current understanding of the rules of transcription initiation and termination, as well as splicing, remain approximate and incomplete. A proactive strategy that can define ORF and transcript structures without relying completely on computational predictions is urgently needed. Here, we describe a large-scale RACE platform and demonstrate its value by applying it to approximately 2000 unverified ORF models. Using existing ORF annotations as a point of departure, we defined approximately 1000 alternative transcript and ORF models. With the strategy described here, we are now in a position to proactively obtain models for entire ORFeomes for C. elegans and other organisms.
Results
The RACE approach
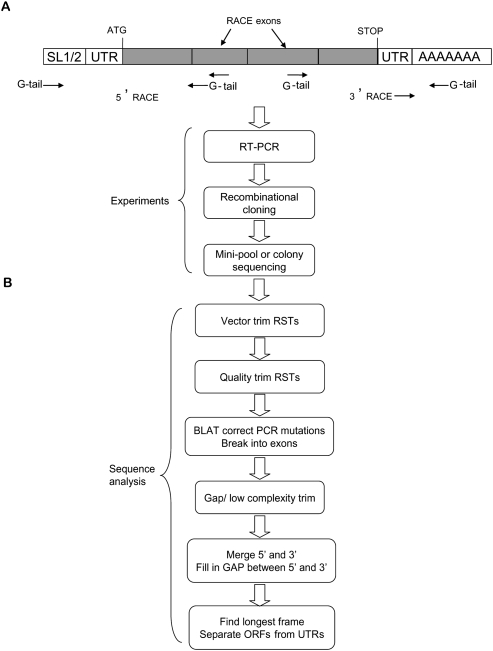
Since almost half of predicted ORFs in C. elegans remain only partially supported by experimental data from cloning, new experimental strategies are needed to improve transcript and ORF annotation. RACE (rapid amplification of cDNA ends) (Bao and Hull 1993) can proactively explore protein-coding transcript models. Large-scale RACE has been hampered by low throughput, low specificity and sensitivity, and occasionally false capture of transcript ends. We adapted RACE for genome-wide studies in C. elegans by (1) improving throughput by combined use of Gateway cloning technology (Walhout et al. 2000b) and minipool sequencing (Reboul et al. 2001, 2003; Rual et al. 2004b); (2) increasing sensitivity and specificity by carrying out nested PCR; and (3) making use of C. elegans trans-spliced leader sequences to ensure capture of true 5′ transcript ends (Fig. 1). About 70% of all C. elegans mRNAs have a trans-spliced leader sequence (Krause and Hirsh 1987; Blumenthal 2005); mostly the 22-base-long SL1 sequence (Conrad et al. 1995), with SL2 ranking as the next most frequently used trans-spliced leader (Huang and Hirsh 1989; Blumenthal et al. 2002). For 5′-RACE, the use of SL sequences, as opposed to the ligation of an arbitrary sequence to the 5′-ends of transcripts in conventional RACE, provides two distinct advantages: no additional manipulation of RNA is needed, and the presence of SL1/SL2 ensures that the mRNA has an intact 5′-end. To increase sensitivity and specificity for both 5′- and 3′-RACE, we carried out nested PCRs, with secondary primers matching to regions inside of the primary amplification regions. The secondary primers were tailed with Gateway sequences (G-tail) to permit recombinational cloning. Established protocols (Walhout et al. 2000b) were used for cloning, and for sequencing of products as minipools (Reboul et al. 2003). Sequencing from minipools, besides improving throughput, advantageously provides sequence information on the dominant transcript species, while still allowing detection of major alternatively spliced variants (showing up as discrete regions of “mixed called” bases in the sequence traces) when they are present in the minipools.
Figure 1.
Experimental and computational RACE pipelines. (A) Experimental strategy begins with reverse transcription of messenger RNA (with a tailed dT24 for 3′-RACE) to generate a complementary DNA strand used as template to generate the first set of RACE products. These amplicons are then reamplified with nested internal primers to generate the final RACE products, which are cloned, then sequenced. (B) Computational pipeline begins with vector and quality trimming of the reads. RACE sequences are aligned by BLAT against the C. elegans genome then parsed as genome (exon) blocks. RACE sequences are then replaced by matching genomic sequences to correct sporadic sequencing or PCR errors. If an exon is of low complexity or has regions that cannot be aligned to the genome, the exon, together with subsequent exons (if present), is trimmed. For short transcripts, 5′ RACE Sequence Tags (RSTs) and 3′ RSTs can be readily assembled to generate full transcripts. For longer transcripts, the gaps of nonoverlapping 5′ RSTs and 3′ RSTs are filled by sequences from WormBase transcript models.
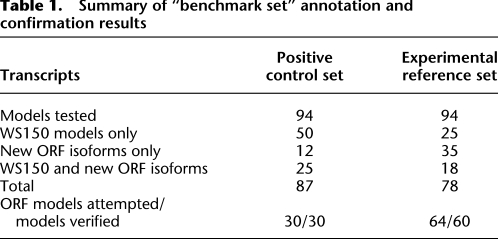
To establish both a benchmark for RACE and an automated annotation pipeline, we gathered a positive control set (PCS) of 94 well-annotated protein-coding transcripts, as well as an experimental reference set (ERS) of 94 transcripts whose ORF annotations had not been experimentally defined previously. The PCS consisted of transcripts for which the corresponding ORFs are (1) previously cloned, (2) shown to contain the SL1 leader sequence, and (3) known to have poly(A) sites. The ERS was picked randomly from transcripts for which either (1) ORF cloning was unsuccessful in the first two worm ORFeome project efforts (Reboul et al. 2003; Lamesch et al. 2004), or (2) cloning was successful but for which exon structure had been mispredicted such that the actual coding region was cloned out of frame. Of the 94 transcripts of the ERS, 50 have an SL1 trans-splice acceptor based on WormBase WS150 annotation. The only experimental data supporting the remaining 44 transcripts were one or more ESTs.
Primer design and generation of RACE fragments were as described (Methods). We arranged the primers according to the sizes of the expected RACE fragments in each row. Examination of our PCS RACE products (Supplemental Fig. 1) shows a general increase of size in each row, in line with the design of the primers. The ERS RACE products also show an increase in size in each row; however, consistent with being inaccurately modeled sometimes, the sizes are less regular than with the PCS (Supplemental Fig. 1).
Both 5′- and 3′-RACE products were cloned as described (Methods). Following transformation, PCR amplimers were either generated from minipools made of multiple transformants, or from individual colony isolates. RACE sequence tags (RSTs) were obtained for each 5′- and 3′-RACE product (5′ and 3′ reads for the 5′-RACE and only 5′ reads for the 3′-RACE). Each RST was initially aligned to its corresponding annotated CDS using bl2seq. We only retained RSTs with an alignment length >100 bp and with high sequence quality (having 200 or more consecutive bases with phred ≥ 20) for further analysis.
When minipools of transformants were analyzed, we observed an ∼98% success rate (for the combined 5′ and 3′ reads) for our PCS, and an 82% success rate (combined 5′ and 3′ reads) for the ERS. When individual colonies were sequenced, these numbers increased to 100% and 90%, respectively. For 3′-RACE when minipools were analyzed, we observed an ∼86% success rate for the PCS and a 77% success rate for the ERS. When individual colonies were sequenced, these numbers increased to 95% and 86%, respectively. The increase in success rate going from minipools to colony sequencing was contributed to by the reduction of (1) failed minipool reads that were redone as multiple single colonies, and (2) mixed sequence traces due to the presence of alternative splicing, which interferes with base-calling of the sequences.
We carried out manual evaluation of the PCS and ERS sequence data followed by automated evaluation (see next section and Methods). For manual analysis, after clipping vector sequences as well as SL and poly(A) sequences from the original traces, RSTs were aligned to the C. elegans genomic sequence version WS150 using the Acembly program of AceDB (“A C. elegans DataBase”). For each set of RSTs that aligned to the expected genomic region, a transcript and ORF model was generated and compared with the existing WS150 models (complete listings in Supplemental Files 2 and 3). Examination of the remodeled ORFs enabled detection of alternatively spliced variants for both the ERS and the PCS (Tables 1, 2). Several of these variants had different internal exon structures, while others had alternative 5′ and 3′ exons. Many in the experimental set had mispredicted start/stop codon positions, which could explain the previous failed attempts at cloning these ORFs. These results confirm previous suggestions (Reboul et al. 2001) that many of the genes that failed to amplify have mispredicted models.
Table 1.
Summary of “benchmark set” annotation and confirmation results
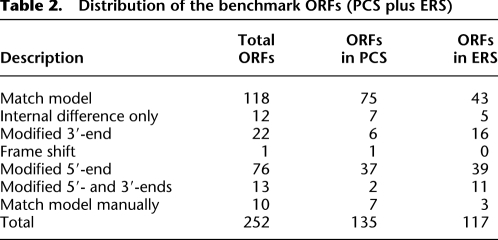
Table 2.
Distribution of the benchmark ORFs (PCS plus ERS)
To compare the obtained 5′-RACE results to the known trans-splice acceptor sites of PCS and ERS transcripts, the clipped sequences of 5′ RSTs were also aligned to the C. elegans genome with the BLAT program. RSTs containing sequences that could not be aligned to the genome were discarded. Out of 144 known trans-splice acceptor sites, 129 were verified by the RSTs; the other 15 were different but located within 25 kb of the RST starting site. BLAT results also located trans-splice acceptor sites for 35 ORFs among 44 ORFs without known trans-splice sites. In addition, 22% of genes have more than one trans-splice acceptor site.
Computational pipeline for generating RACE-defined transcript and ORF models
To process and assemble RSTs and to ultimately construct transcript and ORF models, we established a computational pipeline with multiple quality-control filters (Fig. 1). We obtained both 5′-RACE and 3′-RACE products for all 94 PCS transcripts. Our automated pipeline generated one or more ORF models for 87 of the 94 PCS transcripts, whereas we were able to generate ORF models for all 94 manually (Supplemental Files 1–3). For the ERS, we were able to generate models for 78 transcripts using our automated algorithm and for 81 transcripts manually. Out of all the transcripts with at least one ORF model, there were 12 PCS and 35 ERS transcripts with only new ORF models (i.e., no confirmatory WormBase models were found); and 25 PCS transcripts and 18 ERS transcripts containing both new models and WormBase matching models (Table 1). In total, 252 ORF models were generated for the PCS and ERS sets (details in Table 2). The RACE data also defined new ORF structures for ∼13% of the PCS ORFs, indicating that alternative models remain to be discovered even for “well-annotated” worm transcripts.
To experimentally verify the ORF models derived from the RACE results, we generated RACE-defined transcript models for PCS and ERS sets, and then used these to generate primer pairs to PCR-amplify 94 full-length ORFs. This set included 34 ORFs from the PCS and 60 ORFs from the ERS. Following RT-PCR on RNA prepared from N2 worms (Supplemental Fig. 1C), we cloned the generated PCR products. Sequencing of these revealed a 96% (90/94) success rate in amplifying the targeted ORFs. Hence, our RACE strategy efficiently delineates transcript boundaries and can guide more accurate ORF cloning.
Large-scale RACE on approximately 2000 unverified gene models
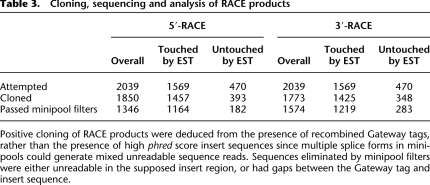
With the necessary experimental and computational pipelines in place, we next scaled up the RACE experiments to interrogate 2039 ORF models for which previous attempts at ORFeome cloning had failed (Reboul et al. 2003), representing ∼25% of the unverified worm ORFeome (Supplemental Fig. 2). Of these, 1569 ORF models were touched, and 470 were untouched. Each RACE product was cloned and then sequenced from an individual minipool, unidirectionally from the 5′-end. Of the 1569 touched ORF models, 74% of the 5′ RSTs and 82% of the 3′ RSTs passed all quality-control filters. Of the untouched genes, 39% of the 5′ RSTs and 60% of the 3′ RSTs passed the filters (Table 3; Supplemental File 4).
Table 3.
Cloning, sequencing and analysis of RACE products
Positive cloning of RACE products were deduced from the presence of recombined Gateway tags, rather than the presence of high phred score insert sequences since multiple splice forms in minipools could generate mixed unreadable sequence reads. Sequences eliminated by minipool filters were either unreadable in the supposed insert region, or had gaps between the Gateway tag and insert sequence.
We note that the lower observed success rate for these RACE reactions relative to the benchmarking experimental sets (ERS) is not unexpected. All ERS transcript models were supported by EST evidence, so that their expression level is likely to be higher and their internal exons correspondingly more likely to be accurately annotated. Also, individual colonies were sequenced from each minipool in the ERS set. Minipool sequencing provides several advantages such as rapidity, cost effectiveness, and evidence for the presence of major isoforms (if present). When alternative forms are present in more or less equal ratios, base-calling becomes difficult, and the apparent success rate decreases. Individual colony sequencing deconvolutes the mixture and provides readable sequence, increasing the success rate. Future primer walking experiments, guided by available tiling-array data and transcriptome sequencing data, would help obtain RACE information on such transcripts.
Large-scale RACE-derived structural annotation
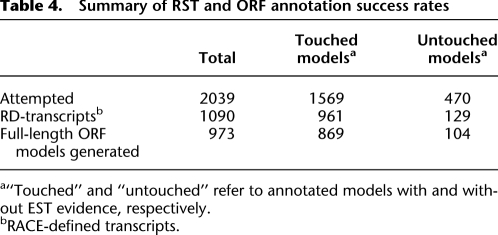
To construct transcript and ORF models using RSTs, we considered only the 5′ and 3′ RSTs that passed our quality-control filters, from which we constructed 1090 RACE-defined (RD) transcripts (Table 4; Supplemental File 4). Out of the 1090 RD-transcripts, 973 constituted a full-length ORF, that is, an ORF with a recognizable ATG start codon and a stop codon (Table 4; Supplemental File 1; see Supplemental File 5 for complete listing with full-length sequences). Among these 973 generated ORF models, 627 (64%) confirmed WormBase release WS150 ORF models (or splice variants), while 346 (36%) were new, not present in WS150. Of the 346 new ORF models, 328 (or ∼95%) were with redefined ends: 225 (or ∼65%) had redefined 5′-ends, 53 (or ∼15%) had redefined 3′-ends, and 50 (or ∼15%) had redefined 5′- and 3′-ends (Fig. 2A). The remaining 18 (or 5%) represented internal alternative splice variants of models with unchanged ends. Among the 328 ORF models with redefined ends, 93 (28%) showed internal exon changes as well.
Table 4.
Summary of RST and ORF annotation success rates
a“Touched” and “untouched” refer to annotated models with and without EST evidence, respectively.
bRACE-defined transcripts.
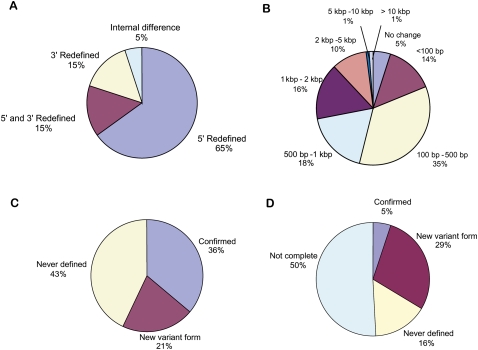
Figure 2.
Generated transcript and ORF models compared with the corresponding WormBase annotation. (A) The percentage of new ORF models that differ from the corresponding WormBase models. If an ORF model had internal changes coexisting with changes in boundary (redefined 5′-, 3′-, or both ends), the ORF model was counted as having a change in boundary. (B) Chromosomal span change of novel ORF models in kilobases (kb). A change can be either an increase or a decrease in span. (C,D) Changes in annotated UTRs. UTRs in the RD-transcript models are divided into several categories: confirmed (WormBase), new variant (different from WormBase), and never previously defined (no UTR indicated in WormBase). Many transcript models in WormBase do not have UTRs defined, and a model can be annotated as having multiple UTRs. “Not complete” UTRs are those 3′ UTRs that could not be defined with certainty.
We further investigated the 80% (262/328) of new ORF models with 5′ boundary changes, discovering that 30% (98/328) of these represented in-frame extensions or reductions of the 5′-end. In 50% (164/328) of these, the change was more complex, with the 5′-end eliminated and replaced by a new end. For some, the redefinition involved significant changes in chromosomal span. While most newly redefined ORFs showed <1 kb change in chromosomal span (Fig. 2B), we observed three ORFs (C10E2.3, T02C5.5c, and C37F5.1) with large changes in chromosomal span (over 10 kb). The C10E2.3 ORF model was 2.6 kb with a chromosomal span that was 10 kb longer than that of the original ORF model, while the C37F5.1 ORF model was 1.2 kb with a chromosomal span 21 kb shorter than the original model. A shorter transcript for C37F5.1 is also annotated in AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) (Thierry-Mieg and Thierry-Mieg 2006), and the confirmation of the shorter form does not necessarily indicate that the longer annotation is incorrect. ORF model T02C5.5c was 6 kb long with a predicted span of 18 kb. A new 5′ start site ∼18 kb upstream of the annotated site was found for T02C5.5c, but the ORF size changed only slightly. We tested C10E2.3 and C37F5.1 by RT-PCR (T02C5.5c was not tested because of its long ORF length). We confirmed the complete ORF of C37F5.1 and the ends of C10E2.3 by cloning and sequencing.
Comparison of the 973 newly generated ORF models with those in WS150 found 84 new exons in 69 of the ORFs (Supplemental File 6). These exons were entirely novel, in that no part of the corresponding genomic sequences had previously been defined as exonic. We additionally identified 313 exons in 280 ORFs that modified the annotated exons by extending or truncating previously predicted exons (Supplemental File 7). We examined the splice sites of introns for both the new and modified exons. In WS150 transcript models, 99,592 out of 100,506 (or ∼99%) exons corresponded to the most common GT/AG splicing signals. Among all the introns we examined, 99.5% (391/393) have GT/AG (386) or GC/AG (5) splicing signals (Supplemental File 8), the latter being the second most abundant splice site in C. elegans (Sheth et al. 2006).
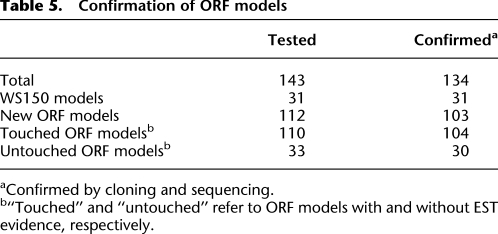
To validate the generated ORF models, we probed 143 of the models by RT-PCR followed by cloning and sequencing. We only attempted validation for ORFs less than 3 kb because of low processivity of the reverse transcriptase. The generated clones were sequenced and analyzed as minipools, or sometimes as single colony isolates. For ORF models shorter than 1.2 kb, we verified the entire ORF length by sequencing from both ends. For ORF models longer than 1.2 kb, we confirmed both ends by single pass sequencing. We confirmed 134 out of 143 (or ∼94%) of the tested ORF models (Table 5). Of the tested cases, 112 represented new ORF models, and we confirmed 103 of these (or ∼92%). Among the tested models we did not observe a statistically significant difference between the confirmation rate of touched and untouched models (104 of 112 [95%] vs. 30 of 33 [91%], P = 0.164). In contrast, we had previously observed a significant difference between the confirmation rate of touched and untouched (Reboul et al. 2003; Lamesch et al. 2004), indicating that once an ORF model is confirmed by RACE, the two classes behave similarly in verification experiments.
Table 5.
Confirmation of ORF models
aConfirmed by cloning and sequencing.
b“Touched” and “untouched” refer to ORF models with and without EST evidence, respectively.
Our RACE platform captures the full-length 5′-untranslated region (UTR) elements, but does not ensure capture of all full-length 3′-UTR elements owing to low sequence complexity or extended length of the 3′ UTR. Among the 973 generated RD-transcripts with full-length ORFs, 366 (36%) confirmed the 5′ UTR in WormBase, 205 (21%) added new variants, and 402 (43%) had no 5′ UTR information available in WormBase (Fig. 2C). Nine percent of our ORFs had no associated 5′ UTRs. In contrast, among genes with defined 5′ transcript boundaries in WormBase, 6% have no 5′ UTR. This difference is statistically significant (P = 0.000042). The higher percentage of genes without a 5′ UTR found by experimental RACE is consistent with trans-splicing of many C. elegans mRNAs, resulting in short 5′ UTRs with the SL located near the start of the ORF (Page et al. 1997). We carried out a similar analysis for the 3′ UTRs that we could completely define (∼49% [479/973] of generated ORF models). Among these, only 10% (49 out of 479) confirmed previously annotated 3′ UTRs. The other 90% (430 out of 479) had new 3′ UTRs that either redefined previously annotated 3′ UTRs or had not been described previously (Fig. 2D).
Alternative trans-splice leader usage
For ∼15% of the 5′ RSTs, the quality score of the SL sequence was poor, while the surrounding sequences (both the Gateway tails and downstream transcript sequences) had high-quality scores. These differences indicate possible alternatively spliced transcripts in the sequenced minipools. Because we had used a mixture of SL1 and SL2 for 5′-RACE and because competitive (or alternative) SL1-SL2 trans-splicing in operons has been reported (Graber et al. 2007), these RACE products likely contained a mixture of transcripts derived from the same gene but linked either to SL1 or SL2, in other words, alternatively trans-spliced. To investigate the extent to which such putative competitive splicing is associated with alternative splicing of downstream transcript sequences, we examined 28 such 5′ RSTs, sequencing 12 colonies isolated from each respective minipool (Supplemental File 9). We found that 16 of the 28 5′-RST minipools investigated contained mixtures of homologous genes due to mispriming. Two categories of mispriming are observed: (1) mispriming of gene-specific primer to a homologous gene, and (2) internal priming by SL1/SL2 in place of the gene-specific primer. These mispriming events were not further investigated.
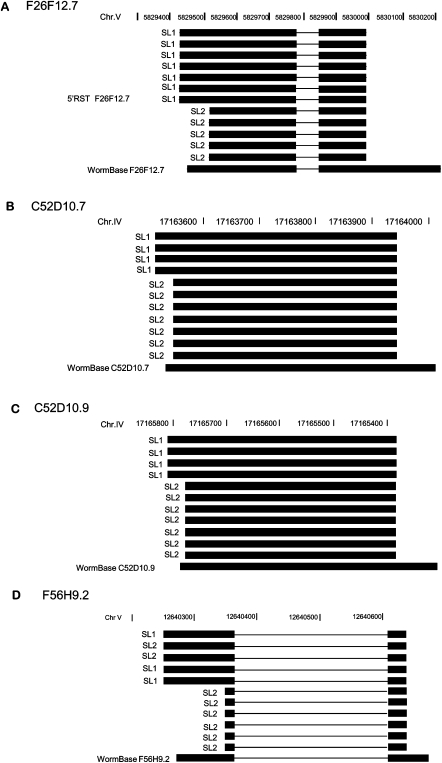
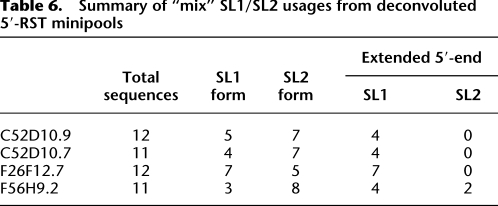
Clones from the remaining 12 minipools did contain transcripts of the same gene linked to either SL1 or SL2, confirming alternative trans-splicing. Among the isolated clones, 60 contained the SL1 sequence, and 62 had the SL2. We manually searched upstream sequences for the most common 3′-splicing signal sequences (UUCAG, UUUCA, AUUU, and UUUUC) within a window from −10 to 0, and the U-rich elements (UAUUUU, UACUU, UAUCU, UAUUU, CUUUU, and UUUCU) from −100 to 0 associated with trans-splicing (Graber et al. 2007). Of the 60 SL1 transcripts, 57 (95%) have both splicing signal and U-rich elements (Supplemental File 9). Of 62 SL2 transcripts, 58 (94%) have U-rich elements, and 39 (63%) have both (Supplemental File 9). In eight of these 12 minipools, sequences linked to SL1 and SL2 showed no difference in the downstream regions. For three of these eight minipools, the corresponding transcript models were reportedly present in cotranscribed operons. R11D1.9 and C42C1.13 are the first genes in their respective operons, while Y56A3A.13b is downstream in its operon. In the remaining four of the 12 minipools, we captured alternative transcript variants. For C52D10.7, C52D10.9, and F26F12.7, SL1 was linked exclusively to isoforms extended at the 5′-end (Fig. 3; Table 6). In F56H9.2, an extended 5′ form was linked preferentially to SL1. C52D10.7 and C52D10.9 are highly homologous tandem repeat genes that displayed a similar SL selection pattern (Fig. 3); neither is reported to be in an operon. While alternative promoter usage can explain alternative SL acceptor sites, alternative trans-splicing can be a competing mechanism leading to formation of these transcripts. In sum, alternative trans-spliced leaders are found in ∼6% of the transcript models tested, and alternative trans-spliced leaders are found in some cases to be preferentially associated with different transcript variants.
Figure 3.
Examples of alternative trans-spliced leader usage. Single colony sequencing was done on RSTs that showed evidence of mixed trans-splicing. (A–C) Alignment of reads obtained from individual 5′-RACE clones shows association of SL1 and SL2 to different transcript variants. (D) Alternative usages of SL1 and SL2 were observed for the longer transcript variant, while SL2 trans-splicing is observed for the shorter variant.
Table 6.
Summary of “mix” SL1/SL2 usages from deconvoluted 5′-RST minipools
Discussion
Proactively defining transcript or ORF structure with the large-scale RACE platform described here offers the opportunity to discover transcript features not conforming to rules used in current ab initio gene predictions. Proactivity is critical where model-based reactive ORF verification (Reboul et al. 2003; Lamesch et al. 2004) fails. The high-throughput RACE approach relies only modestly on predicted transcript models. We have also demonstrated that our approach has excellent sensitivity and specificity. Most C. elegans transcripts have SL1 or SL2 leader sequences (∼55% SL1, 15% SL2) (Blumenthal et al. 2002). By including both SL1 and SL2 primers in the 5′-RACE reactions, we could capture both SL1- and SL2-containing transcripts, targeting ∼70% of protein-coding transcript space. The SL-mediated 5′-RACE will clearly not work for transcripts without SL1 and SL2 sequences. The remaining transcripts, not having SL1 or SL2 leaders, can be captured by the ligation method (Schaefer 1995; Chenchik et al. 1996; Manichaikul et al. 2009), although we expect the efficiency of RNA ligation will be less than the SL1/SL2 approach. Transcripts with SL3-5 splice leaders, though, form a minority of trans-spliced transcripts, and could be captured in future experiments by using primers corresponding to these leader sequences in the RACE PCR reaction.
Of the ORF models that we generated, ∼31% of the touched but unverified models differed from the WormBase models. Strikingly, for untouched models whose cloning was attempted and failed previously, more than ∼73% of generated ORF models were novel relative to WS150 models. We also found alternative ORF structures for 13% of “well-annotated” positive control genes that we examined. Although a novel ORF model does not necessarily imply that the previous model is wrong, our results show that more than one-fifth of the C. elegans ORFeome can be alternatively annotated. Decisions to update models are left to annotation curators, who do so on the basis of their particular criteria for accepting alternative annotations or retiring existing ones. Genome-wide validation of these annotations is urgently needed to gain a systems-level understanding of the protein-coding potential of the worm.
While technologies for whole-genome sequencing have rapidly evolved, the difficulties inherent in defining transcript and ORF structures within metazoan genomes have persisted. Several available large-scale “survey” methodologies, including CAGE (Shiraki et al. 2003), TEC-RED (Hwang et al. 2004), GIS-PET (Chiu et al. 2007; Ng et al. 2007), and MS-PET (Ng et al. 2006), may be used to aid defining transcript structures and boundaries. These methods provide short tags that define transcript end(s), but, importantly, do not probe the internal exon structures. Transcriptome sequencing using parallel sequencing platforms (RNA-seq) can define precise exon boundaries (Cloonan et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Shin et al. 2008; Wilhelm et al. 2008). However, the wide dynamic range of gene expression in higher eukaryotes severely limits the ability of RNA-seq approaches to determine transcript structure of rarely expressed genes. Recent transcriptome sequencing of L1 stage worms with the 454 sequencing platform, while informative, provided reads for only about 6100 protein-coding genes and partially verified only 200 untouched genes (Shin et al. 2008). Platforms with higher coverage might provide greater depth, but determining cis-connectivity of exons across the entire length of transcripts would still not be possible given the short read lengths of these platforms. The recent worm RNA-seq effort using the Illumina Genome Analyzer (Life Sciences [Roche]) platform successfully touched most transcript models (Hillier et al. 2009). However, the “Genelet” models generated from assembly of the obtained sequences are often truncated because of uneven coverage. In addition to cis-connectivity limitations, some transcriptome sequencing approaches lose strand information of the transcripts, limiting use for transcript annotation (Mortazavi et al. 2008).
How does targeted RACE compare with non-targeted, but highly parallel, transcriptome sequencing? We carried out detailed analysis of the SL and exon structures determined by RACE with the mid-L2 transcriptome sequencing reported by Hillier et al. (2009). The SL1 and SL2 trans-splice acceptor sites identified in our RACE were compared to the SL1 and SL2 trans-splice acceptors (based on release WS180) and Hillier et al. (2009) (Supplemental File 1). Out of 1067 SL1 sites we identified, 310 sites overlap with Hillier et al., 564 sites matched to WS180 annotation, and 249 sites appear in all three data sets. For 207 SL2 sites we found, 61 were identified in all three approaches, while 68 and 123 matched to Hillier et al. (Supplemental Fig. 3; Supplemental Table 1; Supplemental File 1) and WS180 annotation, respectively. In defining the position of SL1 trans-splicing, there is only 37% overlap within the tested space between findings reported here and the L2 stage reported by Hillier et al. Likewise, the overlap for SL2 sequence is ∼33%. In summary, while there is overlap between the two data sets, the RACE approach provides new and alternative SL sites absent in the transcriptome sequencing.
We compared the 85 newly detected exons with exons found in Hillier et al. (2009) (Supplemental Table 2; Supplemental File 1). Approximately 42% (36) of exons do not overlap with Hillier et al. altogether; only ∼29% (25) match exactly. We see similar trends for exons that we redefined experimentally (Supplemental Table 3; Supplemental File 1). Of the 313 exons that we modified, 88 (∼28%) show no overlap, i.e., are not annotated by Hillier et al. (2009); 172 (or ∼55%) show different 5′ and/or 3′ boundaries; and only 46 (∼15%) show exact matches.
The comparison of our RACE results with current RNA-seq results shows that transcriptome sequencing generates distinct and potentially complementary results. Improvements in cis-connectivity determination of splicing events are expected with further technological innovations in RNA-seq approaches, such as paired-end sequencing (Fullwood et al. 2009). Ambiguities are still likely to persist, as paired-end sequencing gives connectivity for transcript ends but not for splicing events in the middle of an ORF. The high-throughput RACE platform can correctly annotate protein-coding genes of C. elegans. RACE, transcriptome sequencing, tiling array, PET, and others provide distinctive solutions to defining transcript structure, each with its own strengths and limitations. The final elucidation of genome-wide transcript annotation, in worm or any other eukaryote, rests on integration of multiple high-throughput approaches, benefiting from strengths that each platform offers.
Methods
RACE experiments
General RACE primers
The following primers were used for the first PCR of the 5′-RACE experiments (sequences given 5′ to 3′): SL1, GGTTTAATTACCCAAGTTTGAG; SL2, GGTTTTAACCCAGTTACTCAAG. The second 5′-RACE PCR reactions used: GFSL1, GGGGACAACTTTGTACAAAAAAGTTGGCGGTTTAATTACCCAAGTTTGAG; GFSL2, GGGGACAACTTTGTACAAAAAAGTTGGCGGTTTTAACCCAGTTACTCAAG.
For the 3′-RACE experiments, the following primers, derived from the Invitrogen GeneRACER kit, were used: reverse transcription priming with the GR3 primer, GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTT; the first PCR was done with GR3, GCTGTCAACGATACGCTACGTAACG; and the final amplification was done using GGRn3, GGGGACAACTTTGTACAAGAAAGTTGGGCGCTACGTAACGGCATGACAGTG.
Gene-specific RACE primer design
For the 5′-RACE experiments, we designed two nested reverse primers antisense to the putative ORF region of the gene of interest. The distal primer was placed 100–500 bases 3′ to the putative start of the ORF, while the more proximal reverse primer, typically positioned in tandem to the distal primer, was tailed with the Gateway B2 sequence to allow recombinational cloning (Walhout et al. 2000b). For the forward primer of the 5′-RACE, we used a pool of SL1/SL2 sequences, each tailed with the B1.1 Gateway sequence at its 5′-end. The nested 3′-RACE primers had the same general design as the 5′-RACE primers, except that they were in the forward orientation (sense relative to the mRNA), and the primer proximal to the poly(A) tail also contained a Gateway B1 tail. The distal primer was placed 100 to 400 bases upstream of the putative stop codon. The dT24 primer contained the Gateway B2.2 tail. All gene-specific primers were located in annotated exons and were designed to have a Tm between 55°C and 65°C.
Generation of RACE amplicons
To generate RACE fragments we reverse-transcribed total C. elegans RNA, isolated from mixed-stage, asynchronously growing worm populations, using either dT16 (for 5′-RACE) or the tailed oligo(dT) GR3 primer for 3′-RACE. For the first of the two nested PCRs, we performed touchdown PCR (the annealing temperature of the first 10 cycles was 65°C, on average 5°–10° above the Tm of the gene-specific primers) using the distal gene-specific primers along with the appropriate universal primer (a 1:1 mixture of SL1 and SL2 primers for the 5′-RACE reactions and a tailed dT16 GR3 Primer, for the 3′-RACE reactions). For these PCR reactions, we adjusted the amount of reverse-transcribed material such that ∼150 ng of total RNA was present per reaction. We used less than 0.5 μL of the first PCR reaction for the second-stage PCR with the nested and tailed proximal gene-specific primers. Nested PCR step increases sensitivity and specificity of the experiment while providing Gateway tails for cloning.
Gateway cloning of amplicons and sequencing
PCR products generated in RACE or in ORF verification experiments were recombinationally cloned by a BP reaction into pDONR223 to generate Gateway Entry clones (Rual et al. 2004a). The products from the BP reactions were used to transform chemically competent DH5α Escherichia coli, in 96-well microtiter plates containing spectinomycin, for growth and selection of cells bearing Entry clones. Following growth in liquid media, the transformed bacteria were used as a source of template in PCR reactions, using vector primers to generate the final DNA template for sequencing. PCR products were sequenced using conventional automated cycle sequencing to generate RSTs or OSTs (Reboul et al. 2001). Sequencing was carried out by Agencourt Bioscience Corp.
For the benchmark sets, forward and reverse reads were obtained for the cloned 5′-RACE products. For the rest of the RACE experiments, including all 3′-RACE, only a 5′ forward read was generated. For ORF verification, 5′ and 3′ reads were obtained.
For ORF verification experiments, Gateway-tailed primers were designed to amplify complete ORFs as described (Reboul et al. 2003). PCR products were Gateway-cloned and sequenced from both ends to generate OSTs. Vector and quality trimmed OSTs from both ends were assembled. If there was overlap between the 5′ and 3′ OSTs, a contig was assembled and further analyzed to find the reading frame.
Reconstruction of transcripts from RACE sequence
Manual analysis of the benchmark sets
All sequence traces were assigned a unique ID and stored in a MySQL database. Each RST was initially aligned to its corresponding annotated WS150 CDS using the bl2seq program. Only RSTs with an alignment length >100 bp and with high sequence quality (having 200 or more consecutive bases with a phred ≥ 20) were retained for further analysis. After clipping vector sequences, low-quality sequences, SL1/SL2, and poly(A) sequences from the original traces, RSTs were aligned to the C. elegans genome, WormBase sequence version WS150, using the assembly program of the AceDB. For each set of RSTs that aligned to the expected genomic region, an ORF model was generated and compared with the existing WS150 model.
Large-scale RACE analysis
We generated Perl scripts to process and analyze the RACE data. The computational pipeline: RST sequences were base-called and vector-trimmed with phred. Quality trimming excluded sequences with average phred scores below 15 in a sliding window of 20 nt. Vector- and quality-trimmed RSTs were aligned by BLAT against WormBase WS150 genomic sequence. We enforced two filters. First, for the 5′ RSTs, no gap on RST was allowed between the 5′-SL primer and the continuing sequences. Similarly, in the 3′-RACE, no gap was allowed between the gene-specific primer and the rest of the RST. Such gaps were occasionally observed as short stretches of sequences on the RST that could not be aligned to the genome. Second, for any RST with a gap between RST-exons, the exons following the gap were not further processed. Those RSTs passing the imposed filters were broken into hit blocks (RST-exons) based on their best BLAT hits, to avoid inclusion of homologous gene segments. To eliminate errors (PCR-induced or otherwise), genomic sequences corresponding to RST-exons were identified and used in place of the actual RST-exons in all subsequent steps.
Processed and filtered 5′ and 3′ RSTs of the same targeted gene were merged to generate a transcript model. Usually the RSTs did not span the entire transcript model; for these (∼90% of the transcripts), we used the existing WS150 transcript model sequences to fill in the gap between the 5′ and 3′ RSTs, generating RST-model-hybrids.
ORF and UTR prediction from RD-transcripts
The first ATG that gave rise to the longest open reading frame in each RD- transcript was used to define the start of the open reading frame. The ORF is considered “complete” if a stop codon was found (only rarely was a stop codon not found). Once the ORF region was defined, 5′ and 3′ UTRs were assigned. The first base of 5′ UTR was defined by the end of the SL sequence, and the end of the 5′ UTR was defined by the first ATG of the ORF. For 3′ UTR assignation, the ORF stop codon delineated the start of the UTR, and the poly(A) stretch marked the end. We aligned the sequence to the genome, and a complete 3′ UTR was characterized as exons with no gaps in between and followed by a poly(A) tract. Sometimes, owing to low complexity or extended length, the complete 3′ UTR could not be defined.
Databases
We used WormBase WS150 (released Oct 2005) for primer design, sequence analysis, and comparison. The UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat) was used to analyze and display individual sequences, with the March 2004 assembly of the C. elegans genome sequence as the reference genome sequence.
Acknowledgments
This work was funded by a grant from the Ellison Foundation (M.V.) and by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative in support of the Center for Cancer Systems Biology (CCSB). M.V. is a “Chercheur Qualifié Honoraire” of the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium). F.P.R. was supported in part by NIH grant HG003224 and by the Canadian Institute for Advanced Research. We thank Adnan Derti for helpful discussions and comments on the manuscript.
Authors' contributions: M.V., K.S.A, D.E.H., and F.P.R. conceived the project. K.S.A., D.E.H., and M.V. directed the execution of the project. K.S.A, D.Z., H.J.L., R.R.M, X.Y, S.M., and N.S. carried out the RACE and cloning. C.L., T.H., C.F., and K.S.A. established the computational RACE pipeline. C.L., T.H., L.G, Y.S., and K.S.A. carried out the analyses. K.S.A, C.L., D.E.H., M.E.C., and M.V. wrote the manuscript.
Footnotes
[Supplemental material is available online at http://www.genome.org. 5′- and 3′-RACE sequences are available at http://www.wormbase.org and http://worfdb.dfci.harvard.edu/index.php?page=race.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.098640.109.
References
- Bao Y, Hull R. Mapping the 5′-terminus of rice tungro bacilliform viral genomic RNA. Virology. 1993;197:445–448. doi: 10.1006/viro.1993.1609. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. The C. elegans Research Community. WormBook. 2005. Trans-splicing and operons. http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chenchik A, Diachenko L, Moqadam F, Tarabykin V, Lukyanov S, Siebert PD. Full-length cDNA cloning and determination of mRNA 5′ and 3′ ends by amplification of adaptor-ligated cDNA. Biotechniques. 1996;21:526–534. doi: 10.2144/96213pf02. [DOI] [PubMed] [Google Scholar]
- Chiu KP, Ariyaratne P, Xu H, Tan A, Ng P, Liu ET, Ruan Y, Wei CL, Sung WK. Pathway aberrations of murine melanoma cells observed in Paired-End diTag transcriptomes. BMC Cancer. 2007;7:109. doi: 10.1186/1471-2407-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Conrad R, Lea K, Blumenthal T. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Salisbury J, Hutchins LN, Blumenthal T. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA. 2007;13:1409–1426. doi: 10.1261/rna.596707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang J, Liu T, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, et al. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17:1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Coulson A, Murray JI, Bao Z, Sulston JE, Waterston RH. Genomics in C. elegans: So many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Hirsh D. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BJ, Muller HM, Sternberg PW. Genome annotation by high-throughput 5′ RNA end determination. Proc Natl Acad Sci. 2004;101:1650–1655. doi: 10.1073/pnas.0308384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill D, et al. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14:2064–2069. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Ghamsari L, Hom EFY, Lin C, Murray RR, Chang RL, Balaji S, Hao T, Shen Y, Chavali AK, et al. Metabolic network analysis integrated with transcript verification for sequenced genomes. Nat Methods. 2009;6:589–592. doi: 10.1038/nmeth.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, Sung WK, et al. Multiplex sequencing of paired-end ditags (MS-PET): A strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Wei CL, Ruan Y. Paired-end ditagging for transcriptome and genome analysis. Curr Protoc Mol Biol. 2007;79:21.12.1–21.12.42. doi: 10.1002/0471142727.mb2112s79. [DOI] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes & Dev. 1997;11:1651–1661. doi: 10.1101/gad.11.13.1651. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Tzellas N, Thierry-Mieg N, Moore T, Jackson C, Shin-i T, Kohara Y, Thierry-Mieg D, Thierry-Mieg J, et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, et al. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Rual JF, Hill DE, Vidal M. ORFeome projects: Gateway between genomics and omics. Curr Opin Chem Biol. 2004a;8:20–25. doi: 10.1016/j.cbpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, et al. Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 2004b;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer BC. Revolutions in rapid amplification of cDNA ends: New strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Hirst M, Bainbridge MN, Magrini V, Mardis E, Moerman DG, Marra MA, Baillie DL, Jones SJ. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC Biol. 2008;6:30. doi: 10.1186/1741-7007-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000a;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000b;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]