Abstract
DNA methylation is an important epigenetic mechanism, affecting normal development and playing a key role in reprogramming epigenomes during stem cell derivation. Here we report on DNA methylation patterns in native monkey embryonic stem cells (ESCs), fibroblasts, and ESCs generated through somatic cell nuclear transfer (SCNT), identifying and comparing epigenome programming and reprogramming. We characterize hundreds of regions that are hyper- or hypomethylated in fibroblasts compared to native ESCs and show that these are conserved in human cells and tissues. Remarkably, the vast majority of these regions are reprogrammed in SCNT ESCs, leading to almost perfect correlation between the epigenomic profiles of the native and reprogrammed lines. At least 58% of these changes are correlated in cis to transcription changes, Polycomb Repressive Complex-2 occupancy, or binding by the CTCF insulator. We also show that while epigenomic reprogramming is extensive and globally accurate, the efficiency of adding and stripping DNA methylation during reprogramming is regionally variable. In several cases, this variability results in regions that remain methylated in a fibroblast-like pattern even after reprogramming.
DNA methylation is considered a key factor in the formation of cellular memory and identity, but owing to experimental and conceptual limitations, we still do not truly understand how the cell writes and erases DNA methylation marks in the course of normal cellular differentiation, and how these marks revert to their original embryonic stem cell (ESC)-like form following somatic cell nuclear transfer or induced pluripotent stem cell (iPS) reprogramming (Reik 2007). Progress in the field was hampered for years by lack of quality methods for high-throughput DNA methylation profiling, but recently several effective assays for profiling DNA methylation in large fractions of the mammalian genome were developed and applied successfully (Weber et al. 2005; Keshet et al. 2006; Rollins et al. 2006; Cokus et al. 2008; Irizarry et al. 2008; Meissner et al. 2008). Another major source of confusion and difficulty in understanding the role of mammalian DNA methylation is the nonuniform CpG content of the genome, which led most of the experimental attention toward regions with high CpG content (CpG islands). Recent evidence suggests that classical CpG islands of high CpG content are almost never methylated under normal conditions, yet much dynamic DNA methylation (manifested as differentially methylated regions, DMRs) can be found in regions with intermediate CpG content, some of which are classically defined as CpG islands and some of which are not (Irizarry et al. 2009; Straussman et al. 2009). Adding to these difficulties, multiple studies have shown that DNA methylation is stably acquired in culture, forming significant line-to-line variability (Allegrucci et al. 2007) and deterministic tissue culture effects (Mikkelsen et al. 2008; Brunner et al. 2009), all of which make the interpretation of the functional role of DNA methylation difficult to verify. Culture effects and variability are of particular importance when analyzing DNA methylation in stem cells and induced differentiation, since the compatibility of stem cell lines with various clinical applications may greatly depend on their epigenomic state. Taken together, the recent experiments in the field have completely changed the way by which DNA methylation is studied, yet left many challenges unresolved.
Results
We sought to approach these challenges using a suite of rhesus monkey (Macaca mulata) stem cell lines that allowed us to compare epigenetic programming (reorganization of DNA methylation during normal differentiation) and reprogramming (reorganization of DNA methylation patterns following derivation of stem cells from somatic cells) (Fig. 1A). The comparison of these two reciprocal processes and the use of ES cell lines of markedly different developmental origin allowed for better control of the ES cell line and culture effects. Primate somatic nuclear transfer cells are currently unique to rhesus monkeys and represent an opportunity to study DNA methylation patterns in a native reprogramming environment. We used a native in vitro fertilization ES cell line (ORMES-22), a primary XY fibroblast line, and the CRES-2 line generated through somatic cell nuclear transfer from the fibroblast line (Byrne et al. 2007). We supplemented our panel with a homozygous parthenote (unfertilized, spontaneously diploid and active) ES cell line (ORMES-9), which provided an additional control against ES cell-line-specific effects. We obtained gene expression data from each cell type and performed MeDIP-chip (Keshet et al. 2006; Mohn et al. 2009) using tiling arrays designed to encompass rhesus regions orthologous to human ESC H3K4me3–H3K27me3 bivalent domains (Bernstein et al. 2006) with additional extensive control regions. For each cell type, data from three biological replicates were averaged. The array provided us with comprehensive quantitative data on key genomic regions of diverse CpG contents and developmental relevance, something that is still difficult to achieve using alternative technologies (Cokus et al. 2008; Meissner et al. 2008).
Figure 1.
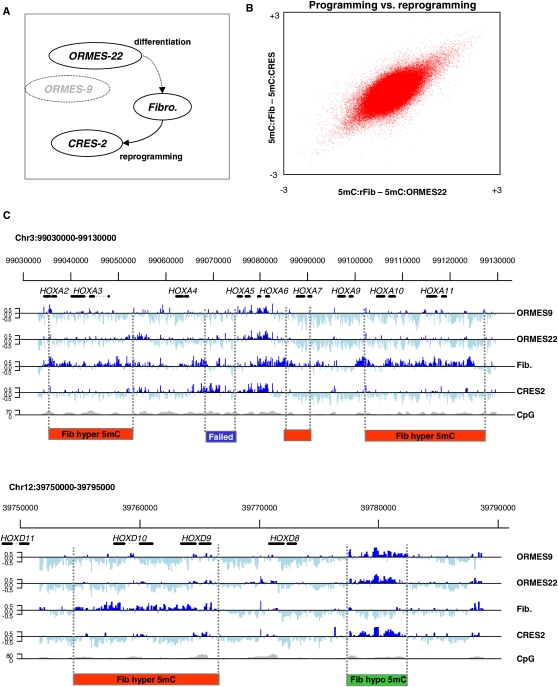
Profiling monkey ES methylation. (A) Experimental design. We studied DNA methylation in native ESCs (ORMES-22), fibroblasts, and ESCs generated by somatic cell nuclear transfer (CRES-2). We also assayed a distinctly different native ES line (the homozygous parthenote ORMES-9) to control for ES line-specific effects. DNA methylation profiles in these four cell types were assayed using MeDIP and tiling arrays. Values of zero correspond to average genomic methylation. (B) Near perfect DNA methylation reprogramming in CRES-2. Shown are the differential methylation values for 380,000 array probes covering orthologous human K4–K27 bivalent domains and selected DNA methylation hotspots. The differences between fibroblasts and the two stem cell lines are highly correlated, showing that at the global level, reprogramming of the fibroblast epigenome during nuclear transfer is near perfect. (C) Conserved and differential methylation in HOX clusters. Shown are the methylation profiles at the (upper) HOXA and (lower) HOXD clusters, which were tiled completely on our array and reflect an excellent overall correlation between the native and reprogrammed ESCs. Regions undergoing fibroblast (red) hypermethylation (hyper-DMRs) or (green) hypomethylation (hypo-DMRs) are highlighted. In contrast to the good overall correspondence between native and reprogrammed ESC methylation, a (blue) small region in the HOXA cluster shows a CRES-2 methylation pattern that is similar to the fibroblast profile, suggesting incomplete reprogramming or independent hypomethylation in OMRES-22 and ORMES-9.
We first assessed the overall degree of reprogramming in the CRES-2 line. Strikingly, although the fibroblasts and native ESCs showed considerable differences in methylation patterns (see below), these differences were almost completely reversed upon reprogramming, generating a CRES-2 DNA methylation pattern that is highly similar to that of the native stem cell (correlation between differential methylation, ρ = 0.53, P ≪ 10−100; Fig. 1B, similar to correlation between biological replicates; Supplemental Fig. S1). As demonstrated in the HOXA and HOXD loci (Fig. 1C), specific genomic regions are subject to different DNA methylation dynamics, including gain or loss of methylation in fibroblasts compared to ESCs (we denote these hyper- and hypomethylated regions, respectively). Interestingly, even though reprogramming is globally accurate, some relatively rare domains remain methylated in a fibroblast-like pattern in the CRES-2 reprogrammed line (labeled “Failed” in Fig. 1C).
A set of DMRs was then extracted using a statistical procedure that searched blindly for significant methylation differences between any two of the three lines (native ESC, somatic, and reprogrammed) (see Methods). The median methylation values of each of the lines at each of the DMRs were clustered to provide an unbiased view of the global methylation dynamics in different groups of loci. As shown in Figure 2A, we observed a similar number of hypo-DMRs (loss of methylation in fibroblasts compared to ESCs, 391 regions) and hyper-DMRs (gain of methylation in fibroblasts compared to ESC, 331 regions). Distributions of differential methylation values in hypo- and hyper-DMRs are shown in Supplemental Figure S2. Importantly, in the vast majority of DMRs, the methylation levels in all three ES lines (including the reprogrammed CRES-2) were similar. A smaller group of 97 DMRs was characterized by high levels of methylation in the native ESCs and lower levels in the reprogrammed line. These DMRs represent either failure to reprogram the CRES-2 line or ESC culture hypermethylation, but importantly, they constitute only a minority of the detected DMRs, which we analyzed separately.
Figure 2.
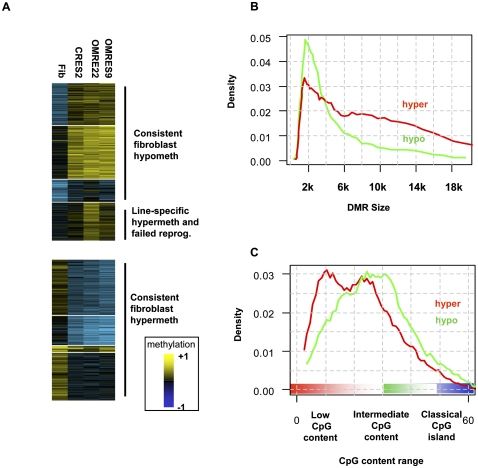
Differentially methylated regions (DMRs). (A) Global patterns of methylation reprogramming. DMRs were statistically extracted from the data by comparing methylation in all pairs of cell types, thereby not pre-assuming any type of organization. Median methylation values for each DMR over all cell types were then clustered (k-means). Shown are the color-coded methylation values of each DMR (rows), organized into clusters showing higher methylation in fibroblasts than in native stem cells (hyper-DMRs) and clusters showing lower methylation in fibroblasts than in native stem cells (hypo-DMRs). Overall, the clusters reflect different basal levels of methylation across the genome, but good correspondence between methylation in the different ES lines. An important exception is a cluster including DMRs with significantly higher methylation in ORMES-22 than in CRES-2. Some of the DMRs in this cluster may reflect ORMES-22 line-specific effects and were excluded from further analysis. Other DMRs in this cluster are also hypermethylated in the ORMES-9 line and were classified as “failed reprogramming” DMRs and analyzed separately. (B) Distribution of DMR sizes. Shown is the distribution of sizes of genomic intervals determined to be (red) hyper- and (green) hypo-DMRs. Hypo-DMRs have a more specific length distribution, peaking around 2 kb. (C) DMR CpG content. The average number of CpGs in 500-bp windows was computed for each DMR (each CpG was counted twice), and the distribution of CpG contents for hyper- and hypo-DMRs was plotted. Hyper-DMRs have a lower overall CpG content. Importantly, both types of DMRs generally occupy regions of low to medium CpG content, and are not observed in classical CpG islands (CpG content >50).
The natural grouping of DMRs into regions undergoing gain or loss of DNA methylation during differentiation is further supported by genomic properties of these loci. Regions gaining DNA methylation are typically larger than regions losing it (P < 6 × 10−9 [KS]; Fig. 2B), the latter having a well-defined distribution of lengths with mean ∼2 kb, suggesting association with more spatially defined genomic elements. Furthermore, regions gaining DNA methylation have lower overall CpG content than regions losing DNA methylation (P < 4 × 10−6 [KS]; Fig. 2C), and both groups are generally of much lower CpG content than classical CpG islands. Our analysis therefore suggests the existence of a group of large regions with low CpG content that are unmethylated in ESCs, gain methylation in fibroblasts, and are capable of losing it upon reprogramming. The data also suggest the existence of well-localized (1–3 kb) regions with intermediate CpG content and high levels of ESC methylation, which lose methylation in fibroblasts and regain it following reprogramming. Additional differences between hyper- and hypo-DMRs are described in Supplemental Figures S3 and S4.
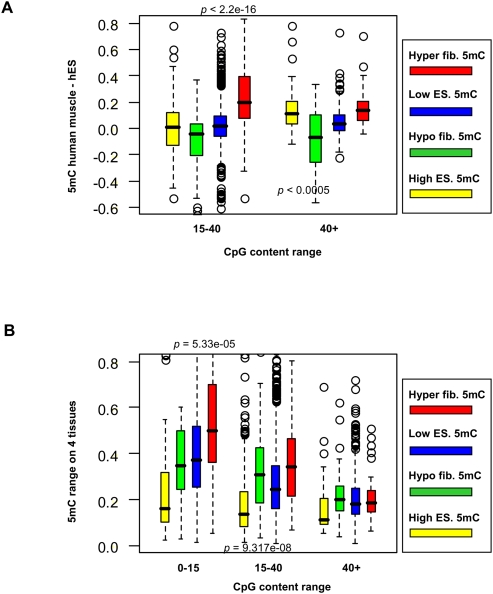
How important are the DMRs we have characterized for programming and reprogramming? One cannot rule out the possibility that some of the hypo-DMRs represent culture effects that are accumulated deterministically and independently in the three ES lines we have analyzed, generating hypermethylation in regions that are normally never methylated. Similarly, it is possible that some of the hyper-DMRs represent accumulation of DNA methylation in the fibroblast culture. To further describe the universality and robustness of the monkey DMRs, we computed the differential DNA methylation between human ESCs and muscle tissues (Straussman et al. 2009) in regions of the human genome that are orthologous to monkey DMRs and regions of high and low methylation. The data (Fig. 3A) demonstrated very good conservation of the monkey DMR methylation patterns, where hyper-DMRs have higher methylation in human muscle tissues (P < 2.2 × 10−16) and hypo-DMRs have higher methylation in human ESCs (P < 0.0005). Furthermore, analysis of the range of methylation of our DMRs across a panel of four human tissues (spleen, liver, colon, and brain) (Irizarry et al. 2009) shows that monkey DMRs, which are defined based on comparisons of fibroblasts and ESCs, are significantly more likely to have variable methylation in the human tissues (P < 10 × 10−8, hypo-DMRs; P < 6 × 10−5, hyper-DMRs; Fig. 3B). These lines of evidence suggest that at least some of our DMRs are real targets of methylation changes during programming and reprogramming, and that culture effects cannot explain all of the epigenomic changes we observe between ESCs and fibroblasts.
Figure 3.
Monkey DMRs are conserved in human ESC and tissues. (A) Muscle-ES differential methylation. Shown are box plots of the DNA methylation differences between human muscle tissues and human ESCs (Straussman et al. 2009), computed for regions of the human genome that are orthologous to monkey (red) hyper- and (green) hypo-DMRs, or to regions with (blue) low or (yellow) high monkey ES methylation. Since the human data span only CpG islands, the statistics only cover regions with intermediate or high CpG content. P-values indicate the significance (using KS test) of difference between hyper-DMRs and low ES methylation regions, and between hypo-DMRs and high ES methylation regions. (B) Range of methylation across a panel of human tissues. Shown are box plots for the differences between the minimum and maximum DNA methylation in human brain, colon, spleen, and liver (Irizarry et al. 2009), for regions that are orthologous to monkey DMRs or regions of high and low monkey ES methylation (same color scheme as in A). P-values indicate the significance of difference between hypo-DMRs and high ES methylation (for CpG range 0–15) and between hyper-DMRs and low ES methylation (for CpG range 15–40).
Based on the observations on robustness and conservation of monkey DMRs in human, we next examined the extent to which hyper- and hypomethylated DMRs are correlated with changes in gene expression or occupancy of epigenomic marks and regulatory factors. Using gene expression array data, we identified rhesus genes with induced or repressed expression in fibroblasts compared to ESCs. We then mapped array probes with gain or loss of DNA methylation to these regulated genes and computed the enrichment of hypo- and hypermethylated probes around transcription start sites (TSSs) of induced and repressed genes. As shown in Supplemental Figure S5, the well-documented (Weber et al. 2007; Gal-Yam et al. 2008) anticorrelation between gene expression changes and DNA methylation changes is observed, where induced TSSs tend to lose DNA methylation and repressed TSSs gain them. Nevertheless, TSS-related changes in DNA methylation account for less than a quarter of the observed DMRs, even when assigning DMRs to TSSs as distant as 5 kb and relaxing our definition of gene induction or repression. We note that the annotation of the monkey genome, which is based on mapping of known human genes, is clearly incomplete, but that such incompleteness is likely to affect only a small fraction of the DMRs. We must conclude that changes in DNA methylation during programming and reprogramming are only partially associated with changes in gene expression in cis, and that other factors also contribute to modify DNA methylation patterns from their ESC pattern to a somatic pattern and back, either affecting transcription through long-range interactions or global epigenomic reorganization, or not affecting transcription at all.
We and others have previously proposed that Polycomb occupancy in embryonic stem cells (ESCs) predisposes genomic regions to retain high CpG content during evolution (Tanay et al. 2007) or to gain DNA methylation in cancer cells (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007; Gal-Yam et al. 2008). We therefore computed the distribution of human ESC SUZ12 occupancy (Lee et al. 2006) in rhesus-mapped regions with low or high ES methylation and in DMRs. In accordance with previous reports, we observe a general deficit of Polycomb occupancy in regions of high methylation (across CpG content classes) (Mikkelsen et al. 2007; Fouse et al. 2008). Moreover, hyper-DMRs in medium and high CpG contents are strongly enriched for high SUZ12 occupancy in human ESCs (Fig. 4A). Surprisingly, SUZ12 enrichment is also detected at hypo-DMRs, suggesting that a significant minority of the Polycomb targets in ESCs sustain significant levels of DNA methylation and are predisposed to lose these upon differentiation. Polycomb complexes are therefore correlated with DNA methylation programming and reprogramming as previously suggested, but their role may be rather heterogenic, either passive (by blocking de novo or housekeeping methylation) or active (by promoting methylation or demethylation upon specific regulatory queues). Alternatively, other underlying uncharacterized epigenetic factors may facilitate DNA methylation changes in epigenetic hotspots, generating indirect correlation with Polycomb occupancy in these regions.
Figure 4.
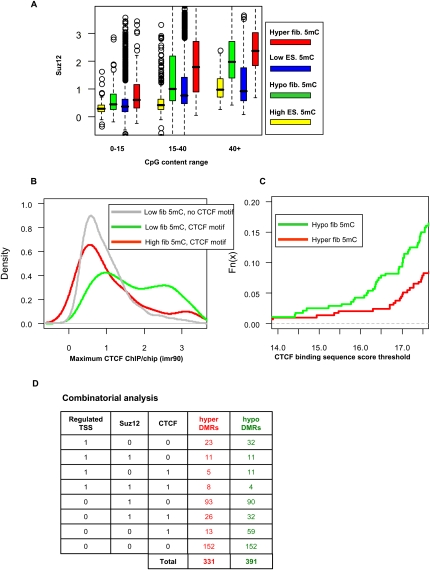
CTCF and Polycomb are correlated with differential methylation. (A) SUZ12 occupancy. Shown are box plots for average human SUZ12 occupancy on mapped monkey DMRs and background regions. We separately plot groups of regions with different levels of CpG content, dissected into (red) hyper-DMRs, (green) hypo-DMRs, (blue) regions with low ES methylation, and (yellow) regions with high ES methylation. In general, regions with high methylation have low SUZ12 levels (e.g., lower than regions with low methylation; see CpG content 15–40). Moreover, regions with higher CpG content (>40) that are hyper- and hypomethylated are enriched in SUZ12 targets. (B) CTCF occupancy at CTCF motifs. Shown are distributions of CTCF binding levels in three groups of genomic loci: (1, gray) background regions lacking CTCF motifs and having low DNA methylation; (2, red) regions of high methylation featuring CTCF binding motifs; (3, green) regions with low DNA methylation featuring CTCF binding motifs. (C) CTCF binding capacity at DMRs. Shown are cumulative probability distributions for the predicted binding energy of the CTCF motif in (red) hyper-DMRs and (green) hypo-DMRs. About 15% of the hypo-DMRs have a strong CTCF binding site, much higher than the percentage for hyper-DMRs. (D) Combinatorial analysis. Shown are counts of DMRs associated with combinations of regulated TSS, SUZ12 hotspot, or CTCF binding site. More than half of the DMRs have at least one factor associated with them.
In search of additional factors involved in DNA methylation reprogramming, we studied the genomic distribution of the CTCF genomic insulator protein using data from human fibroblasts (Kim et al. 2007). CTCF is known to have DNA methylation-dependent activity at several key loci, including the H19 imprinting control DMR. CTCF binding is characterized by a highly specific and informative DNA binding motif, making it one of the mammalian DNA binding proteins with the highest in vivo sequence specificity. Nevertheless, sequence-based prediction of CTCF binding is still <50% accurate (Kim et al. 2007). As shown in Figure 4B, DNA methylation can account for much of this limited specificity, since the distribution of CTCF binding in methylated regions with a CTCF binding site is essentially the same as that of regions without a CTCF binding site. On the other hand, CTCF binding in unmethylated CTCF binding sites is significantly higher (P < 4 × 10−5 [KS]), representing a highly specific binding distribution. This supports the mutual exclusion between CTCF binding activity and DNA methylation and suggests that some of the DMRs we detected may be related to changes in CTCF occupancy. Indeed, as shown in Figure 4C, many hypo-DMRs, but fewer hyper-DMRs, have an underlying CTCF binding site, suggesting a possible link between CTCF recruitment and DNA hypomethylation in fibroblasts, and between the re-methylation of these sites upon reprogramming and CTCF loss. We did not detect significant correlation between DNA methylation changes and the pluripotency factors POU5F1 (also known as OCT4) and NANOG, as shown in Supplemental Figure S6.
We have considered several factors that may be correlated with DNA methylation changes during differentiation and de-differentiation. These include (1) the machinery that drives activity at TSSs, (2) Polycomb complexes, and (3) CTCF factors. In all three cases, occupancy of some specialized protein complexes is generally excluded from regions of high DNA methylation. Changes in DNA methylation may therefore depend (directly or indirectly) on the recruitment or loss of these protein complexes. Using conservative thresholds (Methods), a systematic analysis suggests that 239 out of 391 (61%) hypo-DMRs and 179 out of 331 hyper-DMRs (54%) are correlated with at least one of these factors. Only a minority of the DMRs are associated with regulated TSSs (32/391 hypo-DMRs, 23/331 hyper-DMRs). CTCF sites are associated mostly with hypo-DMRs (106/391) (Fig. 4D). We note that these numbers are based on comparison of monkey methylation data and human CTCF and SUZ12 profiles, so we may be underestimating the overlaps in cases of evolutionary divergence.
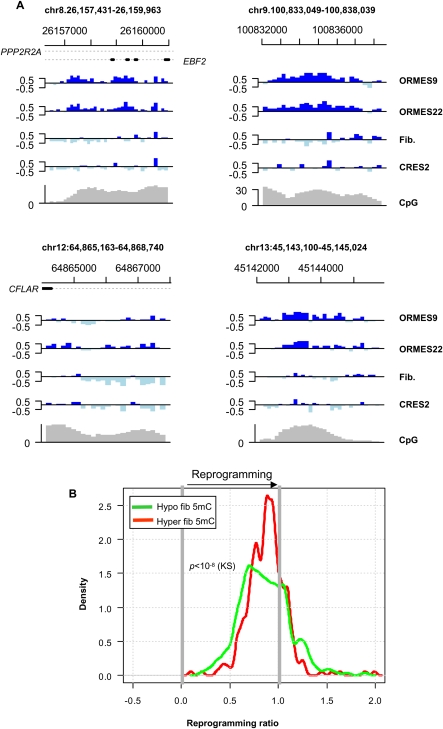
A minority of the DMRs we detected show a consistent native ESC pattern (in two ESC lines) that differs from a pattern common to the fibroblasts and the reprogrammed ESC line (Fig. 5A). These cases potentially represent failure to reprogram the DNA methylation patterns of the somatic cells to their ESC templates. To systematically quantify the efficiency of reprogramming, we computed the ratio between the difference in the methylation median of the reprogrammed ESC and the fibroblast and the difference in the methylation median of the native ESC and the fibroblast (denoted as the DMR reprogramming ratio). A reprogramming ratio of 1 represents perfect reprogramming, and smaller values represent imperfect reprogramming. As shown in Figure 5B, the distribution of reprogramming ratios for hyper-DMRs is centered near 1 (median = 0.84), with a general tendency to values lower than 1, but very few cases near zero. Reprogramming for hyper-DMRs is therefore close to perfect on average. In contrast, the distribution of reprogramming ratios for hypo-DMRs indicate poorer overall reprogramming (median = 0.70, P < 10−8 [KS]), and several cases that partially or completely lack reprogramming (ratio < 0.25, n = 27). One possibility is that the distribution of reprogramming ratio reflects an ongoing process of ESC hypermethylation during reprogramming, which occurs at very different rates for different regions, making slowly reprogrammed regions appear nonreprogrammed and rapidly reprogrammed regions appear perfectly reprogrammed. Another possibility is that reprogramming is terminated, or never occurs, in some of the low-reprogramming-ratio DMRs. Both scenarios result in imperfections of the reprogrammed epigenome, but the implications for stem cell biology remain unclear. We did not detect significant systematic correlations between low reprogramming ratios and other genomic features (Supplemental Fig. S7). Since our methylation profile covers selected parts of the genome, it is possible that additional DMRs are slowly or improperly reprogrammed during somatic cell nuclear transfer, and this may also be the case for the epigenomic state of stem cells derived by induction of pluripotency factors (Takahashi and Yamanaka 2006; Mikkelsen et al. 2008).
Figure 5.
Partial and failed reprogramming. (A) Failure to reprogram DMRs. Shown are examples of DMRs in which the reprogrammed ES DNA methylation pattern follows the fibroblast pattern. These stand in marked contrast to the overall genomic trend (e.g., Fig. 1) and may represent complete lack of reprogramming, partial reprogramming that could not complete, or ongoing reprogramming with much slower kinetics than the genomic trend. (B) Reprogramming ratios. Reprogramming ratios were computed as the ratio of the difference between the reprogrammed ES and fibroblast methylation medians and the difference between the native ES and fibroblast methylation medians. A ratio of 1 indicates perfect reprogramming, and a ratio of 0 represents no reprogramming. Plotted is the distribution of reprogramming ratios of hypo-DMRs and hyper-DMRs. Data are only shown for DMRs that had similar methylation levels in the two native ES lines (ORMES-22 and ORMES-9).
Discussion
Our experiments and analysis, together with other recent measurements of DNA methylation in mouse and human ESCs (Farthing et al. 2008; Fouse et al. 2008; Meissner et al. 2008; Deng et al. 2009), differentiated cell lines (Meissner et al. 2008; Mohn et al. 2008), and somatic tissues (Rakyan et al. 2008; Irizarry et al. 2009; Straussman et al. 2009), outline a rather dynamic picture of the DNA methylation landscape. Comparing fibroblasts to ESCs, a large number of regions are either hyper- or hypomethylated, most of which have a medium level of CpG content. Such changes in DNA methylation are correlated with changes in TSS activity, Polycomb occupancy, or CTCF occupancy for at least 58% of the cases we have profiled. We hypothesize that for the remaining regions, other protein complexes or more accurate information on the current protein complexes may account for the observed methylation dynamics. Interestingly, very little dynamics is observed in the methylation of high CpG content CpG islands, which are generally devoid of methylation in ESCs and differentiated cells. These CpG islands are very frequently located next to developmental regulators and are occupied by Polycomb complexes in ESCs. Many of these CpG islands are aberrantly methylated in cancer, but we have not found significant data suggesting their normal hypermethylation in the present study or any of the other recent high-throughput studies. We therefore believe that future experiments quantifying DNA methylation programming/reprogramming should carefully distinguish between different classes of CpG-rich regions and avoid focusing on promoters or CpG islands alone. Finally, the epigenome of reprogrammed ESCs is shown here to follow closely that of native ESCs, matching the striking similarity of gene expression in native and somatic cell nuclear transfer (SCNT) stem cells (Byrne et al. 2007) or iPS cells (Takahashi and Yamanaka 2006). However, we detect several exceptions to this general trend. Further analysis of these exceptions should clarify whether the failure to reprogram specific genomic domains has functional consequences. Slow- or limited-reprogramming DMRs may also serve as key examples to contrast those many regions that are reprogrammed efficiently, leading to better understanding of the epigenomics of stem cells and the dynamics of DNA methylation and demethylation in general.
Methods
Stem cells culture
Methods for isolation and culture of monkey ESCs from in vitro fertilization, SCNT, and parthenogenetic embryos used in this study were reported previously (Mitalipov et al. 2006; Byrne et al. 2007; Dighe et al. 2008). Briefly, ESCs were grown on feeder layers (mouse embryonic fibroblasts, mEFs) in DMEM/F12 medium with glucose and without sodium pyruvate, supplemented with 1% nonessential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, and 15% FBS at 37°C, 3% CO2, 5% O2, and 92% N2. Culture medium was changed daily; the ESC colonies were typically split every 5–7 d by manual dissociation, and the collected clumps were replated onto fresh mEFs.
MeDIP
MeDIP was performed as previously described (Gal-Yam et al. 2008) with the following alterations: 10 μg of sonicated genomic DNA (300–1000 bp in length) was denatured, incubated O/N at 4°C with 10 μg of anti-methyl cytosine antibody (Diagenode), and subsequently with 40 μL of Dynabeads (M-280 Sheep anti-Mouse IgG; 6.7 × 108 beads/mL; Invitrogen) for 2 h at 4°C. The beads were washed and incubated with digestion buffer and proteinase K for 3 h at 50°C, and the DNA was extracted by phenol chloroform and EtOH precipitation. For array experiments, the output from three MeDIP reactions was combined (total of 30 μg of starting DNA) to constitute one replicate. The sonicated DNA served as input. MeDIP arrays were performed in biological triplicates.
Array design
We collected a set of human ESC bivalent domains (Bernstein et al. 2006) and combined them with additional methylation-related domains and control regions. We mapped these regions from the human genome to the rhesus genome using the UCSC liftOver program and tiled them with probes at 100-bp resolution (Roche-NimbleGen).
Sample preparation and array hybridization
The MeDIP DNA was amplified with a Sigma GenomePlex Complete Whole Genome Amplification (WGA) kit using a protocol developed in the Farnham lab (O'Geen et al. 2006). The amplified samples were column-cleaned with the GenElute PCR Clean-Up Kit. DNA quality and quantity were assessed with a Nanodrop device, and the size distribution was estimated on a 1% agarose gel. The IP samples were labeled with Cy5 dye-labeled 9 mers (blue) and the reference samples with Cy3 dye-labeled 9 mers (pink) from Trilink Biotech, and after EtOH washing, drying, and rehydrating, each sample was requantified with NanoDrop. Adhering to the NimbleGen protocol, we pooled 6 μg of each sample and 6 μg of appropriate reference into the same tube prior to hybridization. The hybridizations were conducted with the NimbleGen Hybridization kit and X1 mixers, and placed on a four-bay station for 18 h. The slides were then washed, and the spot intensity in the two channels was recorded with a Pix 4000B scanner. The data were synchronized with the NimbleScan software and exported for analyses. MeDIP data were normalized as described (Supplemental Fig. S6; Gal-Yam et al. 2008), with the exception of subtraction of M.SssI data, which was omitted since the main application of the data was the analysis of DMRs. When averaging triplicates we excluded two arrays (one fibroblast and one CRES sample) because of lower technical quality. When screening for DMRs, we used the triplicates as is.
Detection of DMRs
To detect DMRs, we screened genomic windows of size 500 bp to 20 kb. For each cell line (ORMES-22, fibroblast, CRES-2, ORMES-9), we computed the distribution of methylation values for all probes in the window (using triplicates as independent observations). We then tested the difference between any two distributions using Kolmogorov-Smirnov (KS) statistics and scored the window using the lowest P-value thus derived. Given the P-values for all windows, we selected the lowest P-value windows while excluding window overlap. This resulted in a set of nonoverlapping DMRs with locally optimal P-value. We used a P-value threshold of 10−5 to generate the set of DMRs analyzed, and a set of nonoverlapping 2-kb regions with a P-value larger than 10−3 and median methylation higher than 0.3 or lower than 0 for background regions with high and low methylation, respectively. We note that the P-values we computed are not corrected for the correlation between adjacent probes, but that such correction would affect all genomic loci uniformly, effectively only changing the thresholds we used (Figs. 3 and 4).
Comparison to human data
We renormalized the MeDIP data of Straussman et al. (2009) as described above and computed the mean MeDIP signal for regions in the human genome that were orthologous to DMRs or background monkey regions. We disregarded regions that were not covered on the human array (which focused only on CpG islands). We used the data from Irizarry et al. (2009) as provided by the authors, computing the minus average comprehensive high-throughput arrays for relative methylation (CHARM) levels for genomic regions that were orthologous to the monkey DMRs and background regions.
Genomic and epigenomic analysis
To define rhesus TSS, we used human genes mapped onto the monkey genome (UCSC). Gene expression data (GSE7748) were mapped onto these genes based on an overlap between the gene expression probe and the mapped gene body. Induced and repressed genes were defined as having at least one mapped gene expression probe with log2(fibroblast/ESC) > 1.5. We considered DMRs as associated with a regulated TSS (Fig. 4) if they overlapped the region 1 kb around the TSS. Comparison of rhesus methylation and human ESC SUZ12 or human fibroblast CTCF was carried out by mapping monkey data onto the human genome. SUZ12 ChIP (Lee et al. 2006) and CTCF ChIP (Kim et al. 2007) data were renormalized as previously described. To generate the counts in Figure 4, we associated a DMR with CTCF or SUZ12 if at least one of the probes in the DMR had a normalized CTCF or SUZ12 ChIP value >1.5.
Acknowledgments
We thank E. Segal, E. Meshorer, and I. Simon for critical reading of the manuscript and members of the Tanay laboratory for discussion. Research in the laboratory of A.T. was supported by ISF grant 1372/08 and by the ISF converging technologies program. A.T. is an Alon fellow. Funding in the S.M. laboratory was provided by start-up funds from the Oregon National Primate Research Center and the Oregon Stem Cell Center, grants from the Stem Cell Research Foundation and NIH NS044330, HD18185, and RR00163. The A.P. laboratory is supported by the University of Iceland Research Council, the Icelandic Centre for Research (Project: 070260022), and a Marie Curie International reintegration grant (MIRG-CT-2007-46510). We thank Steingrimur Einarsson and his colleagues at NimbleGen for technical advice.
Footnotes
[Supplemental material is available online at http://www.genome.org. DNA methylation profiles from this study have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE17981.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.096685.109.
References
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe V, Clepper L, Pedersen D, Byrne J, Ferguson B, Gokhale S, Penedo MC, Wolf D, Mitalipov S. Heterozygous embryonic stem cell lines derived from nonhuman primate parthenotes. Stem Cells. 2008;26:756–766. doi: 10.1634/stemcells.2007-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific Polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]