Abstract
The organization of mammalian DNA replication is poorly understood. We have produced high-resolution dynamic maps of the timing of replication in human erythroid, mesenchymal, and embryonic stem (ES) cells using TimEX, a method that relies on gaussian convolution of massive, highly redundant determinations of DNA copy-number variations during S phase to produce replication timing profiles. We first obtained timing maps of 3% of the genome using high-density oligonucleotide tiling arrays and then extended the TimEX method genome-wide using massively parallel sequencing. We show that in untransformed human cells, timing of replication is highly regulated and highly synchronous, and that many genomic segments are replicated in temporal transition regions devoid of initiation, where replication forks progress unidirectionally from origins that can be hundreds of kilobases away. Absence of initiation in one transition region is shown at the molecular level by single molecule analysis of replicated DNA (SMARD). Comparison of ES and erythroid cells replication patterns revealed that these cells replicate about 20% of their genome in different quarters of S phase. Importantly, we detected a strong inverse relationship between timing of replication and distance to the closest expressed gene. This relationship can be used to predict tissue-specific timing of replication profiles from expression data and genomic annotations. We also provide evidence that early origins of replication are preferentially located near highly expressed genes, that mid-firing origins are located near moderately expressed genes, and that late-firing origins are located far from genes.
Genomes are organized in replicons, defined as chromosomal regions replicated from a single origin. In bacteria, a single bidirectional origin drives replication of the entire multimegabase genome. In eukaryotes, duplication of the entire genome requires the precisely coordinated activation of up to 10,000 replicons.
In yeast and eukaryotic viruses, replication is initiated by interaction between trans-acting factors and a cis-acting element (the replicator) located near or at the initiation region defined as the actual site of initiation of bidirectional replication (Aladjem 2007). Yeast origins of replication contain a recognizable sequence, the ARS motif, present on average every 30 kb in the genome; these sequences are necessary and sufficient to lead to autonomous replication (Gilbert 2001; Aladjem 2007). In mammalian cells, the situation is more complex. Initiation region fragments have been identified using assays based on either nascent strand quantification or leading strand analysis, but electrophoretic techniques based on physical separation of replication intermediates do not generally lead to the identification of precise regions of initiation, but rather of zones where initiation of replication has a greater probability of occurring (Masukata et al. 1993; DePamphilis 1997; Norio 2006; Hamlin et al. 2008). Therefore, it has been proposed (Aladjem 2007) that the locations of initiation events within initiation regions vary, show sequence disparity, and are affected by interaction with distal elements.
It is well established that timing of replication is significantly changed during differentiation, but the molecular mechanisms that control these changes are unknown. Most tissue-specific genes replicate early in the lineage in which they are expressed, while heterochromatic regions, some regions on the inactivated X, and some inactive genes tend to replicate late in S phase. Imprinted alleles often have asynchronous replication patterns with the expressed allele replicated early, and the silent allele replicated late (Goren and Cedar 2003; Schwaiger and Schubeler 2006).
The global organization of the replicons in the mammalian genomes is not well understood, although a few regions have been well characterized. For instance, the transition between an early and late-replicating region has been studied by the Schildkraut laboratory in the murine locus for the immunoglobulin heavy chains (Igh) (Ermakova et al. 1999; Norio et al. 2005). It was found that in most cell types examined, megabase-long early and late-replicating regions were joined by a 600-kb-long temporal transition region (TTR), in which the replication was unidirectional and in which no initiation of replication could be detected. This suggested that large zones of the genome contain multiple origins of replication firing in a coordinated manner, early or late, whereas in the transition regions, the timing of replication is determined by chromosomal location and the absence of active origins of replication.
Two general approaches have been used to measure timing of replication (MacAlpine et al. 2004; White et al. 2004; Woodfine et al. 2004; Jeon et al. 2005; Karnani et al. 2007; Hiratani et al. 2008). The first relies on pulse labeling of newly synthesized DNA using BrdU, separation of the labeled cells in two or more fractions according to their position in the cell cycle, and immunoprecipitation of the labeled DNA. This approach has been used successfully in low and high throughput formats, but it is technically complex and requires an amplification step to detect the immunoprecipitated DNA.
White et al. (2004) studied chromosome 22 in Drosophila cell lines and reported that replication timing was correlated with gene expression, novel transcribed regions of unknown function, sequence composition, and cytological features. Hiratani et al. (2008), using the same approach have recently produced a genome-wide map of timing of replication in mouse embryonic stem (ES) cells and in neurospheres and found that the timing of replication was reorganized during differentiation, and that timing correlated more strongly with promoters expressed at low levels than at high levels. Jeon et al. (2005), using oligonucleotide arrays reported that in transformed cells, early replication was correlated with high gene density, and that at least 60% of the interrogated chromosomal segments replicate equally in all quarters of S phase, suggesting that large stretches of chromosomes are replicated by inefficient, variably located, and asynchronous origins and forks, producing a pan-S phase pattern of replication. Using higher resolution arrays containing 1% of the genome, the same group reported that 20% of the tested regions had a pan S replication profile, again in transformed HeLa cells. Farkash-Amar et al. (2008) using a novel synchronization method, combined with BrdU produced a genome-wide map of the timing of replication in a mouse lymphocytic leukemia cell line and found that a large fraction of the genome replicates asynchronously, and that early replication is frequently correlated with the transcription potential of a gene and not necessarily with its actual transcriptional activity. Finally, using a lower throughput fluorescence in situ hybridization-based assay Dutta et al. (2009) have shown that allele-specific replication of X-linked genes and random monoallelic autosomal genes occur in human embryonic stem cells (hESC), and concluded that epigenetic mechanisms that randomly distinguish between two parental alleles are emerging in the cells of the inner cell mass, the source of hESC.
The second approach relies on detecting variation in copy number during S phase as an indicator for the timing of DNA replication. It has the advantage of simplicity and requires minimal cell manipulation, but it involves the detection of very small differences in copy number that can be difficult to precisely quantify. Woodfine et al. (2004) have pioneered the use of tiling arrays to measure copy-number difference to measure replication timing. Using an array containing about 3500 bacterial artificial chromosomes (BAC), these authors provided a genome-wide map of the timing of replication at a 1-megabase (Mb) resolution in a transformed human cell line and detected a positive correlation between replication timing and a range of genome parameters including GC content, gene density, and transcriptional activity.
Here, we report the development of TimEX (Timing Express), a method that is similar in principle to the BAC array method of Woodfine et al. (2004), but which provides higher resolution timing maps with fine spatial resolution. It relies on precise estimates of copy-number variation based on Gaussian convolution to integrate a highly redundant massive number of individual measurements obtained either by hybridization to high-density arrays containing 400,000 tiled oligonucleotides or by massively parallel next-generation sequencing of genomic DNA libraries.
Results and Discussion
Principle of the TimEX method
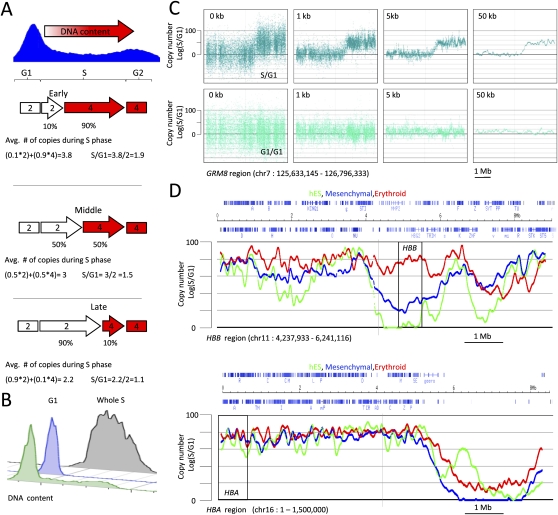
The TimEX method relies on the fact that in nonsynchronized dividing cells, DNA segments that replicate near the beginning of S phase are present in higher copy numbers than DNA segments that replicate near the end of S phase (Fig. 1A). Therefore, determination of DNA copy number in sorted populations of S-phase cells, relative to the corresponding sorted populations of the same cells in the G1 phase of the cell cycle, provides the time in S phase at which replication occurs.
Figure 1.
TimEX. (A) Principle of the technique. Copy number of DNA in sorted S-phase cells compared with sorted G1 cells can be used as a surrogate measurement for the timing of replication (see text). (B) Typical pre- and post-sort DNA content profiles of cycling basophilic erythroblasts detected by staining with propidium iodide. Green profile, pre-sort DNA content profiles; blue and gray, respectively, G1 and S post-sort profiles. (C) Scatterplots illustrating smoothing by Gaussian convolution. Top and bottom panels are, respectively, scatterplots of S/G1 and control G1/G1 ratios for the 8-mb GRM8 region on chr 7 (see Supplemental Table S1). (X-axis) Genomic position; (y-axis) normalized S/G1 or G1/G1 ratio. The left panels illustrate the results without any smoothing; the three panels on the right show the same data smoothed by Gaussian convolution of sigma equal to 1, 5, or 50 kb. As expected, the G1/G1 ratio is flat, while the S/G1 ratio varies. High S/G1 ratios indicate regions that replicate early in S phase, low S/G1 ratios regions that replicate late in S phase. (D) Comparison of the timing of replication in hESCs and in mesenchymal and erythroid cells derived from hESCs. The scatterplots are as above. The red, green, and blue curves, respectively, represent the TimEX profiles of the three cell types. A total of 8-Mb regions containing the beta hemoglobin (HBB) and the alpha hemoglobin (HBA) are shown. All of the other regions present in the arrays are shown in Supplemental Figure S2. The profiles in the three cell types are different, but the overall shape of the curves and the slopes of the transition regions are similar, suggesting that the underlying molecular mechanisms are the same. Differences between cell types are particularly evident in gene-poor regions.
To assess copy numbers in a high-throughput manner, we first used custom Roche NimbleGen tiling arrays containing about 400,000 probes and spanning 18 genomic segments 4–8 Mb long, representing about 3% of the genome, including some of the ENCODE regions (Supplemental Table S1).
Exponentially growing human undifferentiated H1 ESC, mesenchymal stem cells, and basophilic erythroblasts both derived from H1 hESCs (Olivier et al. 2006; Qiu et al. 2008) were sorted into a G1 and an S fraction using propidium iodide staining to assess DNA content (Fig. 1B). After sorting, DNA was extracted, sheared, and labeled using Cy5 or Cy3 fluorescent 7-mer random primers. The S phase and the G1 control DNAs were then mixed in equal amounts and hybridized to the arrays. As an additional control, G1 phase DNA labeled with either Cy3 or Cy5 were mixed in equal amounts and hybridized to the same arrays.
The unprocessed S/G1 and G1/G1 ratios showed that copy-number variations were detectable, but that the signal to noise ratio was low (Fig. 1C). The maximum theoretical S/G1 ratio is 2, but in practice the observed ratios did not exceed 1.6 because of contamination during the sorting of the S-phase fraction by cells in G1 or G2. To improve the resolution, the microarray signals were analyzed in detail as a function of probe sequences and normalized using a novel method (Supplemental Fig. S1). Four types of smoothing algorithms were tested: naïve binning, averaging over a sliding window, sliding median, and finally Gaussian convolution, which in our hands proved to be the most productive algorithm to analyze our data. Gaussian convolution is a technique classically used in image analysis that can be used to filter the noise out of continuously variable signals (see Supplemental Methods). Application of this algorithm to our TimEX data greatly improved the resolution of copy-number differences in the S and G1 signals (Fig. 1C; Supplemental Fig. S1). As expected, the control G1/G1 ratio is nearly flat (Fig. 1C; Supplemental Figs. S1, S2) and the remaining undulations of the G1/G1 ratio provide a direct measure of the limit of the spatial resolution of the TimEX procedure, which is about 50 kb for our chip.
Importantly, the TimEX results were reproducible between biological replicas (Supplemental Fig. S2) and in agreement even when two distinct Roche NimbleGen chips with different probe designs were compared (Supplemental Figs. S3, S4).
General organization and tissue specificity of timing of replication
Comparison of the TimEX profiles in undifferentiated hESC, and derived mesenchymal stem cells and erythroid cells showed, as expected, that the three cell types tested had distinct profiles of timing of replication (Fig. 1D; Supplemental Figs. S2, S4). The coefficient of correlation between the three cell types were comprised between 0.68 and 0.76 (Supplemental Fig. S4).
Tissue-specific changes were particularly evident around some highly regulated genes. For instance, the cluster of the β-like hemoglobin genes and flanking sequences is replicated earlier in the erythroid cells than in the two other cell types (Fig. 1D). However, as reported previously (Vyas et al. 1992), not all tissue-specific genes have such a dramatic effect. For instance, the alpha hemoglobin gene cluster, which is located in a gene-dense region, replicates early in all three cell types tested (Fig. 1D), despite being active only in erythroid cells.
TimEX-seq
To extend our results genome wide, we then developed TimEX-seq, a method similar to TimEX, except that copy-number variations are estimated by sequencing using next-generation massively parallel methods. To measure timing genome wide, we produced rapidly dividing basophilic erythroblasts in vitro from culture of bone marrow CD34+ cells (Qiu et al. 2008), sorted the cells into S and G1 fractions, and extracted genomic DNA. Whole-genome libraries were then prepared and sequenced on an Applied Biosystems SOLiD System sequencer to a depth of about 25 million reads. For each of the two libraries, more than 10 million reads were uniquely mapped to the genome. The TimEX-seq profiles were then obtained by counting the number of reads that mapped to arbitrarily defined 5-kb genomic windows for the G1 and S libraries, filtering regions of low tag densities, calculating the S/G1 ratio, and smoothing the S to G1 ratio by Gaussian convolution as described above. As a control, the genomic DNA from the basophilic erythroblasts was also hybridized to Roche NimbleGen tiling arrays.
The TimEX profiles obtained using tiling arrays and massively parallel sequencing were remarkably similar, providing an important cross-validation for both approaches (Fig. 2A). The correlation coefficient between TimEX and TimEX-seq profiles was above 0.96 with 10.5 million reads (Fig. 2B). To obtain the genome-wide timing of replication profile in hESC, we then created libraries of S and G1 DNA from sorted hESC and sequenced the libraries using the Illumina platform. Again, the profiles obtained were highly similar to the profiles obtained on the Roche NimbleGen arrays (Fig. 2C), with a coefficient of correlation between the two platforms above 0.95.
Figure 2.
TimEX-seq. (A) Comparison of TimEX results obtained using tiling arrays or massively parallel sequencing (basophilic erythroblasts). The S/G1 ratio of the frequency of uniquely matched reads in 5-kb windows was calculated and smoothed as in Figure 1. (X-axis) Genomic distances; (y-axis) S/G1 ratio for sequencing, and log2 (S/G1) for tiling arrays. Sequencing and tiling arrays produce very similar profiles. (B) Coefficient of correlation between tiling array and sequencing results. (C) Comparison of TimEX results obtained using tiling arrays or massively parallel sequencing (hESC). (D, top) Examples of chromosome-wide TimEX-seq profiles obtained based on 10.5 million (basophilic erythroblasts) and 13 million reads (hESC). A sigma of 100 kb was used for the Gaussian convolution. (Bottom) Differential timing curve for hESC and erythroid cells obtained by subtracting the hESC profile from the erythroid profile.
To determine the minimal number of reads necessary to obtain timing profiles, we repeated the above calculations using decreasing amounts of reads. Timing profiles that were well correlated with the profiles obtained with 10 million reads (r = 0.95) could be obtained with as little as 5 million reads (Supplemental Fig.S6).
Tissue specificity of replication timing
Comparison of the genome-wide timing of replication profiles in erythroid and in hESC yielded a coefficient of correlation of 0.71, similar to what we observed with the Roche NimbleGen platform.
To further characterize the tissue-specific differences between DNA replication in hESC and erythroid cells, we subtracted the two TimEX-seq profiles (Fig. 2D). This revealed that about 18% of the genome replicates at least 2 h apart (assuming an 8-h S phase) in ES and in erythroid cells.
Subfractionation experiments
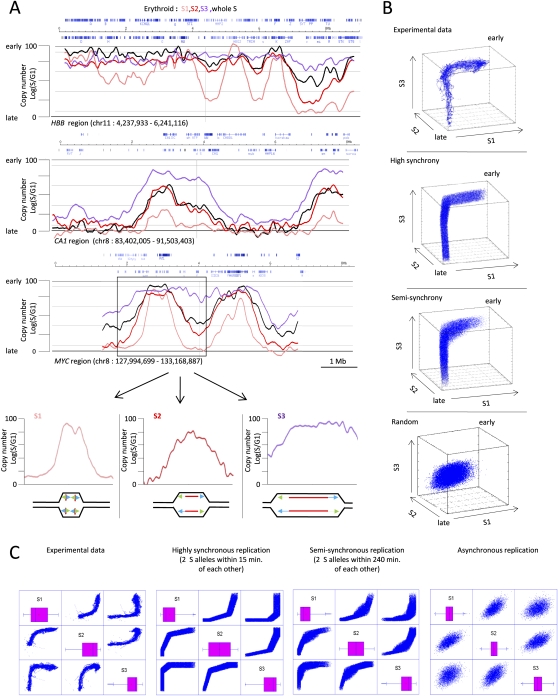
Regions that replicate early or late in S phase are by necessity regions in which all of the origins fire nearly synchronously. But in our experimental system, regions that have S/G1 ratios indicative of mid-S replication are consistent with at least three mechanisms: random firing of origins throughout S phase, regulated synchronous origins firing in the middle of S phase, or asynchronous replication of the two alleles. To discriminate between these mechanisms, we sorted S-phase basophilic erythroblasts into three fractions, (early [S1], middle [S2], and late [S3]) (Supplemental Fig. S7) and hybridized their DNA to Roche NimbleGen arrays as above.
Results of these experiments provided a unique dynamic view of replication in these cells (Fig. 3A; Supplemental Fig. S7). Of particular interest are the peaks and plateaus that are absent in the early fraction (S1) but present in the later fractions (S2, S3). A modelization of this experiment suggests that these patterns can only be explained by origins that fire in mid S phase (Supplemental Fig. S8). Since we found these dynamic peaks and plateaus in both the mid and the late fractions, and since they reach different heights in different regions, we conclude that origins or zones of initiation are programmed to fire in narrow defined temporal windows during S phase and that individual temporal windows can start at any point in S phase.
Figure 3.
S-phase subfractionation experiments. (A) Scatterplots illustrating TimEX profiles for three genomic regions 5–8 Mb in size. The black curve represents the results for the entire sorted S phase; the pink, red, and purple curves the profiles for the early, middle, and late fractions, respectively. As expected, the profile of the whole S fraction resembles the average of the three fractions. The timing of replication varies over large domains. Analysis of these curves suggests that replication is highly regulated. (Bottom) Our molecular interpretation of one of the two major peaks observed in the myc region. Blue and green arrows represent progressing forks of replication; (red lines) newly replicated DNA. (B, top) Three-dimensional scatterplots illustrating the experimental S/G1 ratio of the early, middle, and late (S1/S2/S3) fractions plotted for all 10-kb genomic windows represented in the array. The three panels below illustrate the S1/S2/S3 scatterplots obtained from simulations in which the replication is assumed to be perfectly synchronous, semisynchronous, or asynchronous (see text, movies M1–M4, and Supplemental Fig. S9 for more plots and the algorithms used for the simulations). The experimental data are most similar to the synchronous replication model, suggesting that the order in which DNA is replicated during S phase is highly regulated. (C) Two-dimensional projections of the three-dimensional plots of B.
Synchrony
To assess the synchrony of initiation of replication genome wide, we devised a novel analysis technique based on plotting the S/G1 ratio observed in the S1, S2, and S3 fractions in three-dimensional scatterplots.
In this representation, the regions replicating at the very end of the S phase have coordinates (0, 0, 0), while the earliest replicating regions have coordinates (100, 100, 100). If replication was completely synchronous, and if the S1, S2, and S3 windows did not overlap, all of the genomic windows replicating between these two points would form a thin line starting in the early corner, moving first along the S1 axis, then along the S2 axis, then S3, to end in the late corner. The more the sorting windows overlap, the more the line cuts the corners. In contrast, if the replication was asynchronous in different cells, a cloud would be observed.
To illustrate this concept, we plot in Figure 3, B and C, Supplemental Figure S9, and in movies M1–M4, the distributions of the (early, middle, late) triplets for the experimental data and for simulated diploid cells with an 8-h S phase. High synchrony was simulated by assuming that the two allelic copies replicate in every cell of a population within 15 min of each other. Partial synchrony was simulated by assuming that the allelic copies replicated within 240 min of each other, and complete asynchrony by assuming that replication of both allelic copies occurs randomly in time in each cell of the population. The models incorporate the effects of the overlap of the three fractions during sorting and of the measurement errors of the TimEX procedure.
In the case of perfectly synchronous replication, the simulation yields, as expected, an elongated snake that starts in the early corner of the cube and ends in the diametrically opposite late corner. In contrast, in the partially synchronized simulation, we observe a snake with a “belly” (a protrusion) and in the completely asynchronous simulation a spheroid (Fig. 3B,C; movies M1–M4). The scatterplot of our experimental data is remarkably similar to the plot expected when replication is perfectly synchronous. Thus, we conclude that the timing of DNA replication in cultured basophilic erythroblasts is highly synchronized. To assess the sensitivity of this approach, we then refined the models and simulated genomes in which the two alleles replicated within 2 h of each other and genomes which were partly synchronous and partly asynchronous (Supplemental Fig. S9). These additional simulations suggested that the three-dimensional plots should be very sensitive for the detection of genomic regions where the replication of the two alleles is highly asynchronous, and that for most of the genome, the two alleles replicate within 2 h or less of each others. Since we only analyzed 3% of the genome with this approach, further studies will be necessary to determine whether this conclusion is true genome-wide.
Temporal transition regions
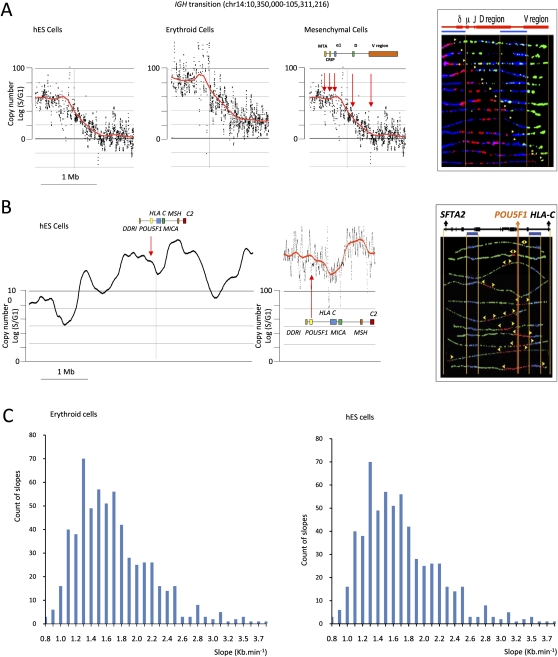
Analysis of the TimEX profiles suggests that the spatial distribution and the timing of the firing origins of replication in the genome are very uneven. In parts of the genome, large regions replicate in a coordinated manner early or late in S phase, suggesting that in these regions the density of early or late firing origins is high. Other genomic regions seem to be replicated in temporal transition regions (TTR) similar to the one described in the mouse Igh locus. Figure 4A illustrates the temporal transition region in the human IGH locus (IGH@), as detected by TimEX in the three cell types analyzed. Other transition regions are illustrated in Figures 1 and 2.
Figure 4.
SMARD analysis. (A) The IGH@ region. (Three left panels) Scatterplots of the results of TimEX analysis in the IGH locus in basophilic erythroblasts, mesenchymal cells, and undifferentiated hESC. The black dots represent the S/G1 ratio using Gaussian convolution windows of 5 kb. The red curve shows the same data smoothed using windows of 200 kb. (Right panel) The SMARD analysis in human mesenchymal cells for a 161-kb PmeI segment of the IGH@ locus that is within the predicted transition region. All of the molecules are stained red at the left end (3′) and green at the right end (5′), indicating that in these mesenchymal stem cells a single replication fork proceeds from 3′ to 5′ (from early to late in S) continuously through the PmeI fragment analyzed at the IGH locus. A genomic map is included above the segment. The blue bars indicate the positions of the two blue biotinylated probes used to identify the segment by FISH (see Supplemental Fig. S8). The vertical orange lines delineate the location of the gene on the segment. The other vertical orange lines indicate the boundaries of the FISH probes. The yellow arrowheads indicate the direction in which the replication fork moves. These results validate the TimEX analysis and demonstrate a long transition region in the human IGH@ locus similar to the one previously reported in the mouse Igh region. (B) The POU5F1 region. (Two left panels) Scatterplots of the results of TimEX analysis in the POU5F1 region in hES cells. The panel to the left represents 6 Mb, the middle panel, 400 kb. Smoothing is as above. This TimEX profile suggests that this region is rich in origins, since replication seems to occur within the first hour of S over a 1-Mb segment. The panel to the right illustrates a SMARD analysis of a 350-kb segment containing the POU5F1 gene. A map of the 350-kb POU5F1 segment is shown above the image. As expected, (yellow) forks going in both directions can be detected in many molecules, suggesting that the region is indeed rich in origins. (C) Histograms illustrating the slope of the transition regions larger than 250 kb calculated genome-wide for hESC and erythroid cells. The method to calculate the slope is described in the Methods section.
To evaluate the rate of replication fork progression in these putative TTR, we have measured the slopes of all the transition regions that were larger than 250 kb in hESC and in erythroid cells (Fig. 4C; see Methods). About 5% of the genome was encompassed in such transition in hESC, and 8% in erythroid cells. The average slope of these transition regions corresponded to a rate of fork progression of 1.8 kb/min +/− 0.5 in hESC and 1.7 kb/ min +/− 0.5 in erythroid cells. These numbers are in good agreement with previous estimates (Jackson and Pombo 1998; Norio et al. 2005; Takebayashi et al. 2005).
To start addressing at the molecular level the question of whether the transition regions observed by TimEX are regions where the forks progress unidirectionally, we analyzed the replication patterns of the human IGH region in hESC by single molecule analysis of replicated DNA (SMARD), (Norio and Schildkraut 2001). As shown in Figure 4A and Supplemental Figure S10, the results supported our hypothesis, since most of the forks progressed unidirectionally in the transition region. These results are also supported by the observation of Hiratani et al. (2008) that in mouse ESC replication domains might be separated by originless transition regions.
We then analyzed by SMARD the region around the POU5F1 gene in hESC, a region that is predicted by TimEX analysis to be rich in origins of replication. Again, SMARD analysis was fully compatible with our interpretation of the TimEX results, since we observed many molecules containing forks progressing in both directions from an initiation site (Fig. 4B; Supplemental Fig. S10), which is characteristic of a region rich in initiation events.
Timing of replication and gene transcription
In accordance with earlier reports, we observed a correlation between gene density and timing of replication with early replicating regions generally gene rich and late regions generally gene poor (data not shown). To assess the correlation between expression and replication, we measured gene expression in our in vitro-derived basophilic erythroblasts and in hESC using Affymetrix expression microarrays. As expected, we observed a correlation between early replication and expression levels, and between late replication and lack of expression. The coefficient of correlation between the timing of replication of all 5-kb windows containing an Affymetrix probeset and expression was around 0.3 in erythroid cells and in hESC. More detailed analysis revealed that differentially expressed genes are preferentially located in regions where the timing differs between ES and erythroid cells (Fig. 5B), although the majority of differentially expressed genes are in the invariant fraction of the genome. Because replication is processive, we hypothesized that timing of replication might correlate most tightly with the distance to genes and to their level of expression, rather than simply with expression levels. To test this hypothesis, we divided the Affymetrix probesets in four quartiles (E1, E2, E3, and E4) according to their expression levels, with E1 containing the most highly expressed genes, and we computed the distance of all 5-kb genomic windows to the closest 5-kb window containing a probeset in each of the four quartiles. Remarkably, computing and plotting the averaged TimEX values against their distance to the closest highly expressed probeset (E1) revealed a very strong inverse correlation (Fig. 5C), over considerable genomic distances (>2 Mb). Regression analysis revealed that the correlation between timing and distance to highly expressed genes can be modeled by reciprocal curves of equation S/G1 = 1/(ax + b), where x equals the distance to the closest expressed gene and “a” and “b” are constants.
Figure 5.
Timing of DNA replication and gene transcription. (A) Scatterplot illustrating the relationship between gene transcription and timing of replication in basophilic erythroblasts and in hESC cells. (X-axis) Mean mRNA expression (determined using Affymetrix U133plus arrays) grouped into 10 bins of equal number of probesets and of increasing expression signals. (Y-axis) Average TimEX values (and standard errors) for all 5-kb genomic windows containing an Affymetrix U133plus probeset are plotted. On average, expressed genes are replicating earlier than unexpressed genes. (B) Histograms illustrating that differentially expressed genes are preferentially located in regions where timing differs between hESC and erythroid cells. (X-axis) Fold differential expression; (y-axis) percent differentially expressed genes. (C) Scatterplots illustrating relationship between timing and distance to expressed genes: Distances of all 5-kb genomic windows to the closest 5-kb window containing either a highly expressed Affymetrix Probeset (top 0–25 quartile), a moderately expressed (25–50 quartile), a poorly expressed (50–75 quartile), or a silent (75–100 quartile) probeset were calculated (see text). The average TimEX value for all windows at the same distance to a probeset was then plotted against their distances to the closest probeset for each of the four quartiles. This plot reveals that the timing of replication is highly dependent on distance to highly expressed probesets since (in the case of erythroid cells) the averaged TimEX value was about 76 for windows containing an actively expressed probeset (distance = 0), and decreased to <30 for windows more than 2-Mb away from a highly expressed probeset. Analysis of the other quartiles shows that the relationship between timing and distance to probeset decreases for less expressed genes, and almost completely disappears for silent genes, suggesting that gen-expression levels directly correlate with timing of origin firing. (D) TimEX values (not averaged) for 5-kb windows covering chromosome 14 (blue line) are plotted against the distance of each window to the closest highly expressed gene (top 0–10 percentile) to illustrate the variability of the TimEX values (which cannot be appreciated in C because of averaging). The red line illustrated the best fitting reciprocal equation (r = 0.81). Supplemental Figure S12 shows similar analysis for all chromosomes. (D) Profiles of predicted timing of replication obtained by calculating the inverse of the distance of each genomic window to the closest Affymetrix Probeset and multiplying it by a coefficient equal to the normalized expression signals of the same probeset (see Methods). The red boxes highlight a peak that is present in hESC but not in erythroid cells, both in the experimental and in the predicted values. (E) Scatterplot illustrating correlation between experimental and predicted timing profiles for chromosome 14.
To determine whether distance to less-expressed genes also correlated with timing of replication, we also plotted the averaged TimEX values against their distances to the closest probeset in the second expression quartile. To discriminate the influence of the genes in the second quartile from that of the genes in the first quartile, we eliminated from this computation all of the windows that were located <250 kb from a gene in the first quartile. Similar computations were performed for the third and fourth quartiles. These plots suggested that all expressed genes have an influence on timing of replication, but that this influence is proportional to levels of expression, since the slope of the curves gradually decreased for the different quartiles (Fig. 5C). In the case of the E4 quartile (least-expressed genes that are far from expressed genes), the TimEX results were almost the same for all windows regardless of their distance to the closest genes (Fig. 5C), suggesting that silent genes have no effect on timing of replication. Scatter and box-and-whisker plots in which the same data is presented before averaging the TimEX values are presented in Figure 5D and Supplemental Figure S11.
This analysis reveals an important novel correlation between timing of replication and expression. We infer from this result that early and middle origins of replication are located very close to genes, and that the timing of the firing of these origins of replication is proportional to the level of expression of the neighboring genes. These results are supported by an earlier report that suggested that origins of replication might be located near promoters (Delgado et al. 1998).
Replication timing predictions
Since the correlation between timing of replication and distance to expressed probesets was very strong, we attempted to predict genome-wide replication timing using the reciprocal equation defined above, genome annotations, and expression data obtained with mRNA from hESC or erythroid cells obtained using the Affymetrix U133 plus2 arrays. Figure 5E illustrates timing profiles generated by an algorithm that predicts timing by calculating the inverse of the distance of each genomic window to the closest highly expressed Affymetrix Probeset and multiplying the results by the normalized expression signal of the same probeset, therefore weighting the prediction by the expression levels of each gene. Constants a and b in the reciprocal equation were adjusted by trial and errors to maximize the coefficients of correlation between predicted and experimental timing values and were respectively equal to a = 13.3 and b = 0.8.
Coefficients of correlation between predicted and experimental values were >0.8 for some chromosomes and, respectively, 0.73 and 0.74 for the whole genome (Fig. 5F) for hESC and erythroid cells.
Importantly, much higher correlation coefficients between experimental and predicted values were obtained if all expressed genes were taken into consideration in the simulation than if genes with lower hybridization signals were eliminated. Weighting of the predicted values by the normalized expression results was also essential to maximize the correlation, again suggesting that all expressed genes are located near origins, and confirming our earlier inference that timing of origin firing is related to level of expression of nearby genes.
The good correlation between predicted and experimental timing values that we observed is particularly remarkable because the Affymetrix arrays we used do not cover all of the known transcripts (Thierry-Mieg and Thierry-Mieg 2006), do not encompass many of the intergenic transcripts that have recently been detected (Katayama et al. 2005; Efroni et al. 2008), and because we measured steady-state levels of mRNA rather than primary transcription levels, which might be expected to correlate more tightly with timing of firing of origins of replication. This suggests that more highly accurate predictions might be obtained with more precise expression data. Of course, we cannot exclude that other factors beyond transcription might also contribute to the regulation of the timing of replication.
To determine whether similar results could be obtained with a microarray of a different design, we hybridized hESC mRNA to the newer Affymetrix Human Gene Array 1.0, which differs from the U133plus2 array by the fact that the individual oligoprobes defining each probeset are localized throughout the genes rather than in the 3′ untranslated region. Very similar results were obtained with this newer chip (genome-wide coefficient of correlation between experimental and predicted values equal to 0.73), suggesting that our observations are robust and do not reflect the idiosyncrasies of a particular chip design.
To determine whether the predictions were tissue specific, we then calculated the coefficient of correlation between experimental and predicted values using timing values from hESC and expression data from erythroid or vice-versa. As expected, higher coefficients of correlation were obtained when mRNA expression and timing data were from the same cell types than when they came from different cell types (Table 1). The predicted values that we obtain are therefore tissue specific and do not simply capture the 80% of the genome that has an invariant timing of replication in ES and erythroid cells.
Table 1.
TimEX predictions are tissue specific
Expression data for hESC and erythroid cells were used to predict Timing values. Predicted TimEX for each cell line was then correlated to the experimental values for the same cell type as well as for the other cell type.
Conclusion
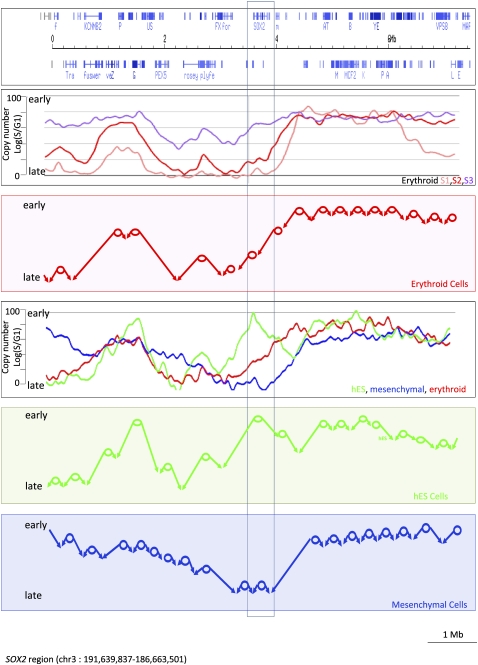
We propose here a comprehensive model for the regulation of the timing of DNA replication in mammalian cells (Fig. 6). This model is compatible with most of the data reported here. It also recapitulates and extends conclusions that have been accumulated over the years by many different groups. The main features of the model are that origins or zones of replication initiation are highly regulated, are unevenly distributed throughout the genome, and fire in defined narrow temporal windows. Origins of replication firing early are preferentially located near highly expressed genes; origins firing in mid S or in late S are preferentially located near genes expressed at a lower level, while late origins are located far from active genes. Once an origin has fired, elongation proceeds at a relatively constant rate until another fork is met. We did not detect any evidence that timing of replication is regulated by pause sites that would delay fork progression for a significant portion of the cell cycle.
Figure 6.
Model of DNA replication in mammalian cells. (Top panel) An 8-Mb region centered on the SOX2 gene region; the second panel is the result of a TimEX analysis of sorted early, middle, and late S fractions from basophilic erythroblasts. The red circles in the third panel represent the major zones containing origins of replication that can be deduced from an analysis of the second panel; the arrows show the predicted fork direction in erythroid cells starting from these origins. The main features of the model are that (1) active origins of replication are unevenly distributed in the genome and relatively rare in parts of the genome, creating large initiation free regions, (2) origins are programmed to fire within narrow time windows during S phase, (3) lack of pause sites creates long transition regions in which the forks progress unidirectionally. The fourth panel represents the TimEX profiles of whole S fractions from basophilic erythroblasts (red), mesenchymal cells (blue), and hESC (green) in the same region. The two bottom panels illustrate the predicted organization of the replication of this genomic segment in hESC and mesenchymal cells. One segment with major differences in timing between the three cell lines is boxed in the immediate neighborhood of the SOX2 gene, which is expressed at a high level in hESC and silent in erythroid and mesenchymal cells (data not shown). Because of the paucity of active origins in the region, the tissue-specific activation in hESC of an early origin or zone of initiation, which appears to coincide with the SOX2 gene, has dramatic repercussions in both the timing of replication and the fork directions in a megabase-wide region.
A consequence of this organization is that many genomic segments are replicated by forks progressing unidirectionally in TTR similar to the one first described in the mouse Igh locus.
Interestingly, in transition regions, DNA strands always replicate either as leading or as lagging strands. Whether this has any effect on gene expression is not yet known in mammalian cells, but reports in fission yeast have shown that the direction in which fork progression occurs can be a regulatory developmental mechanism (Dalgaard and Klar 1999).
Another consequence of this organization is that tissue-specific activation or silencing of a single early origin can have long-distance effects on the timing of replication and on fork direction, because timing of replication in temporal transition regions is regulated passively by the absence of firing of any origin. Again, whether this can affect gene expression is unknown, but reports based either on microinjection of DNA at a different time point in S phase of the cell cycle (Zhang et al. 2002) or on the study of transgene silencing (Fu et al. 2006) have suggested a mechanistic connection between timing of replication and gene expression. Timing of replication might also affect other nuclear processes such as DNA repair, recombination, and transposition.
We propose that the tissue-specific replication “programs” that we can accurately measure using TimEX, and predict using a straightforward algorithm have evolved in concert with developmental programs to allow replication of the genome in multicellular organisms without disruption of increasingly complex transcriptional and other nuclear processes. We also propose that these replication programs function as regulatory mechanisms to facilitate or limit transcriptional activity and other nuclear processes in different segments of the genome, and therefore help orchestrate the complex implementation of the overall genetic program that allows a fertilized egg to develop into an adult. DNA methylation patterns can also be predicted from DNA sequences (Zhao and Han 2009). Of course, because these patterns change during differentiation, the coefficients of correlation cannot reach 1 and are generally in the range of from 0.6 to 0.8. As the quality of the high-throughput data improve, combination of predictions based on different epigenetic marks might increase these coefficients of correlation and allow the definition of an expected epigenetic status for any genomic regions. Such a tool would allow the detection of anomalies and could be useful to assess the efficiency of reprogramming in induced pluripotent stem cells or to predict tumor progression.
Methods
Cell culture
H1 hESC, mesenchymal stem cell, and basophilic erythroblasts were cultured as described by Qiu et al. (2008) and Olivier et al. (2006)
Cell cycle analysis
Exponentially growing cells were fixed in ethanol, RNase treated, and stained for DNA content by addition of propidium iodide (50 μg/mL).
Microarray hybridization
Labeling of genomic DNA and hybridization to tiling arrays were performed using standard Roche NimbleGen protocols. The HG18 release of the human genome was used to design the arrays (see Supplemental Table S1)
Massively parallel sequencing
Library preparation and sequencing was performed using standard manufacturer recommended procedures.
SMARD analysis
SMARD analysis was performed as previously described (Norio et al. 2005).
Data processing
Roche NimbleGen microarrays
The S and G1 signals were normalized using a log2 transformation and two adjustable parameters per track: α and δ.
δ, a damping factor, was added to the signal to minimize large fluctuations of the log ratio when the signal is very weak.
Slope calculation
In order to measure the slopes of the TTR regions, we searched for genomic regions that were at least 250 kb in length and that had constant slopes defined as slopes that did not differ by more than 0.1 kb/min over the entire length of the TTR. Outliers, defined as group of at most two consecutive windows that did not respect above criteria, were eliminated. The algorithm is available on request.
Visualization of the data
A tiling array visualizer that includes several filters to eliminate repetitive or atypical probes, allows for various smoothing methods, and produces fully scalable displays was developed and used to generate most of the figures.
Smoothing algorithms
Small differences in copy number were detected using a method classical in image analysis: Gaussian convolution. The smoothed signal Z(x) at any position x on the genome is evaluated as the weighted average of the signal S(y) at all the neighboring positions y, with the weights decreasing as the exponential of the square of the distance between x and y, (x − y)2:
Massively parallel sequencing
Reads were mapped to the HG18 genome assembly using an algorithm developed by Thierry-Mieg and Thierry-Mieg (2006) or using the ELAND alignment program (Illumina). Unique reads were defined as 35-nt long reads that matched the genome only once with no, one, or two mismatches.
Raw data and wiggles files, complete methods, and 10 supplemental figures are provided in the online Supplemental materials.
Acknowledgments
R.D., E.E.B., and N.L. are supported in part by grants GM075037 and HL088467. E.E.B., N.L., and J.L. are supported in parts by grants NYSTEM N08S-001 and N08T-006. None of the investigators declared any conflict of interest. D.T.M. and J.T.M. are supported by the Intramural Research Program of the NIH, National Library of Medicine. We thank Dr. David Lipman for discussions.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data and the microarray data from this study have been submitted to NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE18679.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094060.109.
References
- Aladjem MI. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature. 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML. The search for origins of DNA replication. Methods. 1997;13:211–219. doi: 10.1006/meth.1997.0521. [DOI] [PubMed] [Google Scholar]
- Dutta D, Ensminger AW, Zucker JP, Chess A. Asynchronous replication and autosome-pair non-equivalence in human embryonic stem cells. PLoS One. 2009;4:e4970. doi: 10.1371/journal.pone.0004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova OV, Nguyen LH, Little RD, Chevillard C, Riblet R, Ashouian N, Birshtein BK, Schildkraut CL. Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I. Global organization of replication time zones of the mouse genome. Genome Res. 2008;18:1562–1570. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Wang L, Lin CM, Singhania S, Bouhassira EE, Aladjem MI. Preventing gene silencing with human replicators. Nat Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Cedar H. Replicating by the clock. Nat Rev Mol Cell Biol. 2003;4:25–32. doi: 10.1038/nrm1008. [DOI] [PubMed] [Google Scholar]
- Hamlin JL, Mesner LD, Lar O, Torres R, Chodaparambil SV, Wang L. A revisionist replicon model for higher eukaryotic genomes. J Cell Biochem. 2008;105:321–329. doi: 10.1002/jcb.21828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Bekiranov S, Karnani N, Kapranov P, Ghosh S, MacAlpine D, Lee C, Hwang DS, Gingeras TR, Dutta A. Temporal profile of replication of human chromosomes. Proc Natl Acad Sci. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H, Satoh H, Obuse C, Okazaki T. Autonomous replication of human chromosomal DNA fragments in human cells. Mol Biol Cell. 1993;4:1121–1132. doi: 10.1091/mbc.4.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P. DNA replication: The unbearable lightness of origins. EMBO Rep. 2006;7:779–781. doi: 10.1038/sj.embor.7400766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24:1914–1922. doi: 10.1634/stemcells.2005-0648. [DOI] [PubMed] [Google Scholar]
- Qiu C, Olivier EN, Velho M, Bouhassira EE. Globin switches in yolk sac-like primitive and fetal-like definitive red blood cells produced from human embryonic stem cells. Blood. 2008;111:2400–2408. doi: 10.1182/blood-2007-07-102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Schubeler D. A question of timing: Emerging links between transcription and replication. Curr Opin Genet Dev. 2006;16:177–183. doi: 10.1016/j.gde.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Sugimura K, Saito T, Sato C, Fukushima Y, Taguchi H, Okumura K. Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp Cell Res. 2005;304:162–174. doi: 10.1016/j.yexcr.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR. Cis-acting sequences regulating expression of the human alpha-globin cluster lies within constitutively open chromatin. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Han L. CpG islands: Algorithms and applications in methylation studies. Biochem Biophys Res Commun. 2009;382:643–645. doi: 10.1016/j.bbrc.2009.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]