Abstract
Candida dubliniensis is the closest known relative of Candida albicans, the most pathogenic yeast species in humans. However, despite both species sharing many phenotypic characteristics, including the ability to form true hyphae, C. dubliniensis is a significantly less virulent and less versatile pathogen. Therefore, to identify C. albicans-specific genes that may be responsible for an increased capacity to cause disease, we have sequenced the C. dubliniensis genome and compared it with the known C. albicans genome sequence. Although the two genome sequences are highly similar and synteny is conserved throughout, 168 species-specific genes are identified, including some encoding known hyphal-specific virulence factors, such as the aspartyl proteinases Sap4 and Sap5 and the proposed invasin Als3. Among the 115 pseudogenes confirmed in C. dubliniensis are orthologs of several filamentous growth regulator (FGR) genes that also have suspected roles in pathogenesis. However, the principal differences in genomic repertoire concern expansion of the TLO gene family of putative transcription factors and the IFA family of putative transmembrane proteins in C. albicans, which represent novel candidate virulence-associated factors. The results suggest that the recent evolutionary histories of C. albicans and C. dubliniensis are quite different. While gene families instrumental in pathogenesis have been elaborated in C. albicans, C. dubliniensis has lost genomic capacity and key pathogenic functions. This could explain why C. albicans is a more potent pathogen in humans than C. dubliniensis.
Candida species are pathogenic yeasts and are the most prevalent cause of opportunistic fungal infections in humans. The diseases caused by these yeasts include superficial infections of the oral cavity and vagina (commonly known as thrush) and deep-seated systemic infections that can affect a wide range of organs and are associated with high levels of morbidity and mortality. Candida albicans is the most frequent cause of superficial and systemic candidosis and is widely recognized as the most pathogenic yeast species. Candida dubliniensis is the most closely related species to C. albicans (Sullivan et al. 1995, 2004) and was routinely misidentified as C. albicans prior to its recognition, since it can produce germ tubes and chlamydospores, traits previously only associated with C. albicans. However, while C. albicans causes ∼50%–60% of cases of systemic candidosis, C. dubliniensis is not usually associated with haematogenous infection and accounts for only ∼2%–3% of Candida species recovered from blood samples (Kibbler et al. 2003; Odds et al. 2007).
Epidemiological and infection model data suggest that C. dubliniensis is less prevalent than C. albicans because it is substantially less pathogenic. In two separate studies using the murine systemic infection model, mice infected with C. dubliniensis have longer survival times than C. albicans (Gilfillan et al. 1998; Vilela et al. 2002). Similarly, in an infant mouse oral intragastric model of infection, C. dubliniensis was far less capable than C. albicans of colonizing the intestinal tract and disseminating to other organs (Stokes et al. 2007). The reasons for the apparently lower virulence of C. dubliniensis, in comparison with C. albicans, are not yet known. However, it is known that the species differ in their tolerance of various environmental stresses (Pinjon et al. 1998; Alves et al. 2002; Vilela et al. 2002; Enjalbert et al. 2009). Furthermore, although C. dubliniensis has the ability to produce hyphae, widely accepted as being an important virulence factor for C. albicans, it does so less efficiently than C. albicans under a wide range of hypha-induction conditions (Stokes et al. 2007) and exhibited low levels of filamentation in infection model experiments (Vilela et al. 2002; Moran et al. 2007; Stokes et al. 2007).
Recently, Butler et al. (2009) compared the genomes of seven Candida species with other parasitic and free-living yeasts, but not including C. dubliniensis. These data encompassed the global diversity of Candida spp. and identified genomic changes that have accompanied the evolution of parasitism, such as expansions of secreted and cell-wall proteins, including the secreted aspartyl proteinase (SAP) and agglutinin-like sequence (ALS) families. However, in addressing such ancient events associated with the origins of Candida and candidosis, the study could not identify those specific features of C. albicans that explain its particular virulence and medical relevance. Moran et al. (2004) analyzed the C. dubliniensis and C. albicans genomes using comparative genome hybridization (CDH). They identified numerous species-specific genes, indicating that a comparative analysis could elucidate the genetic factors responsible for reduced virulence in C. dubliniensis. Therefore, we have sequenced the genome of the type C. dubliniensis strain and compared it with the C. albicans genome sequence (Jones et al. 2004) to further characterize their differences and identify possible mechanisms of virulence in C. albicans. In doing so, we have exposed a much more recent episode in Candida evolution than was possible in the pan-Candida genome comparison, revealing disparities in copy number of gene families that, while present in both species, have evolved subtly different repertoires and may be involved in the increased pathogenicity of C. albicans.
Results
Genome order and content in C. dubliniensis and C. albicans are highly similar
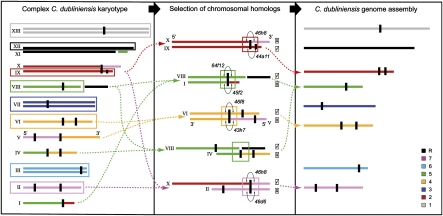
The C. dubliniensis genome (see Supplemental Table S1) was produced by whole genome shotgun sequencing to 11-fold average coverage, assembled de novo, and manually finished. Compared to C. albicans, the 14.6-megabase (Mb) diploid genome has a complex karyotype comprising 10 haploid and three diploid chromosomes (Magee et al. 2008). Various chromosomal translocations have occurred since the separation of C. dubliniensis and C. albicans, meaning that homologous chromosomes, or chromosomal blocks, may occupy two distinct genomic positions, as shown in Figure 1. For instance, the entire length of chromosome 5 in C. albicans corresponds to part of chromosome VIII in C. dubliniensis, along with a part of chromosome R. In addition, sequences corresponding to chromosome 5 in C. albicans are also found in chromosomes IV and I in C. dubliniensis. Furthermore, the sequences of chromosomal homologs are so similar that it was not possible to separate them into a true diploid representation of the sequence. Instead, for each chromosome a single haploid sequence was produced up to the point where two alternative junctions in the assembly were possible. Without duplicating the sequence, or arbitrarily splitting reads between the two assembly choices, a single junction was reconstructed by choosing the one that most closely resembled C. albicans. By ordering C. dubliniensis contigs onto the C. albicans genome sequence, we produced eight haploid chromosomal sequences that correspond to the eight-chromosome C. albicans karyotype. The resultant eight pseudochromosomes were submitted to EMBL (http://www.ebi.ac.uk/embl/) under accession numbers FM992688–FM992695, inclusive. Subsequent PCR walking demonstrated that these pseudochromosomes were real. The remaining chromosomes from the complex karyotype that are not represented were shown to exist through sequencing of selected BACs, as shown in Figure 1. Hence, the C. dubliniensis genome sequence consists of eight pseudochromosomes that reconstruct eight genuine chromosomes: Five haploid (II, VI, VIII, IX, and XII) and three diploid (III, VII, and XIII). While the remaining five haploid chromosomes are not shown, no genome sequence was discarded since each pseudochromosome is a haploid consensus of both chromosomal homologs. So for example, chromosome “5” in the C. dubliniensis genome sequence reconstructs chromosome VIII of the complex karyotype, but also incorporates sequence from chromosomes IV and I that are not represented structurally, essentially because they differ from chromosomal arrangements in C. albicans.
Figure 1.
The C. dubliniensis complex in vivo karyotype and genome sequence. The 14.6-Mb C. dubliniensis genome (left panel, redrawn with permission from Magee et al. 2008) has a complex karyotype of 10 haploid and three diploid chromosomes (Roman numerals), showing multiple chromosomal rearrangements relative to C. albicans. Vertical black marks denote MRS regions. In order to avoid redundancy in the assembly and to maximize comparability with C. albicans, the sequence was assembled into eight “pseudochromosomes” (right panel), which are numbered according to the karyotype of C. albicans strain SC5314 (Arabic numerals) and color-coded after C. albicans in order to illustrate the extent of chromosomal rearrangement in vivo. Note that each pseudochromosome in the right panel is a haploid consensus of the distinct chromosomal homologs apparent in the left panel. Components of the in vivo karyotype selected for the genome sequence are boxed. C. dubliniensis chromosomal homologs often adopted two alternative configurations, relative to the C. albicans karyotype. The middle panel illustrates the selection process by which one haploid chromosome was chosen over its homolog, to reproduce the C. albicans karyotype. Tick boxes indicate which chromosomal configuration was selected in the final genome representation (as submitted to the EMBL database). BAC clones were sequenced that bridged the two distinct chromosomal configurations to confirm that both existed (BAC clone identifiers are noted in italics where appropriate). Note that chromosome V is inverted.
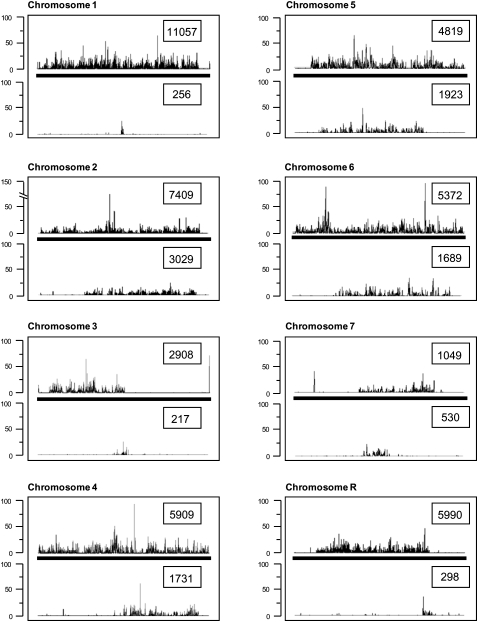
Since the C. dubliniensis genome sequence is a consensus of both chromosomal homologs, we can assess the scale and distribution of nucleotide heterogeneity. Polymorphism is unevenly distributed across all chromosomes of the C. albicans genome sequence and occurs at a frequency of one SNP per 330–390 bases (Butler et al. 2009). The distribution of sequence polymorphisms across the C. dubliniensis genome is presented in Figure 2. While the distribution pattern is not the same as in C. albicans, polymorphism is similarly distributed unevenly and does not correlate with major repeat sequence (MRS), centromeric, or telomeric-proximal regions in any distinctive manner. There are large chromosomal tracts, as in C. albicans, where polymorphism is entirely absent. In general, the C. dubliniensis genome is much less polymorphic than most Candida genome published to date, with SNP frequency ranging from every 635 bases (chromosome 6) to every 12,555 bases (chromosome 1); the latter rate is comparable with C. parapsilosis, which is virtually homozygous (one SNP per 15,553 base pair [bp]; Butler et al. 2009). Polymorphisms are available in Supplemental Table S2 of the Supplemental material.
Figure 2.
Comparison of sequence polymorphism (SNP) distribution between homologous chromosomes in C. albicans and C. dubliniensis. Each panel describes the distribution of SNPs along the haploid consensus for each chromosome in C. albicans (top) and C. dubliniensis (bottom). The absolute numbers of polymorphisms per chromosome are boxed in each panel.
Approximately 6% of the 5758 identified genes are spliced and putative functions were manually assigned to over 4000. Aside from karyotypic differences, the composition and gene order of the C. dubliniensis genome sequence is very similar to C. albicans. The conservation of genome structure extends to complex repeat regions, such as the subtelomeres and the MRSs (Joly et al. 2002). The level of finishing achieved here allows us to fully elaborate the structure of the chromosome end in C. dubliniensis for two instances: the left end of chromosome 6 and the right end of chromosome R (see Supplemental Fig. S1). The subtelomeres of C. dubliniensis possess an end-proximal mosaic of repetitive regions, including the terminal 22-mer repeat, which differ by only 1 base from that observed in C. albicans (Sadhu et al. 1991). The subtelomeres also contain various transposable elements (see below) and a subclass of RecQ-like helicase pseudogenes, which are most similar to the Saccharomyces cerevisiae Y′ helicase (as well as homologous subtelomeric RecQ helicases in C. albicans), rather than to internally located helicases. However, all but one RecQ helicase in C. dubliniensis were gene relics; the intact copy, on the right arm of chromosome 6, differs in its predicted transcriptional orientation, pointing away from the chromosome terminus, while all others point toward it.
The MRS is a feature unique to the genomes of C. albicans and C. dubliniensis, and may contribute to karyotypic variation in these species by acting as a “hot-spot” for chromosomal translocation (Lephart et al. 2005). The situation in C. dubliniensis is described in Supplemental Table S3 and noted in Figure 1. Each chromosome carried at least one MRS element, except for chromosome R, and the conserved HOK and RB2 units flanking the central RPS domains were also present. This is consistent with the situation previously described in C. albicans (Jones et al. 2004), except that C. albicans has no MRS on chromosome 3 and sequencing revealed an additional, smaller MRS on chromosome 2 in C. dubliniensis. The single MRS on chromosome 3 was very small, containing only a single RPS domain, and was flanked by a transposable element on either side; however, we found no evidence to associate the movement of MRS with transposable element insertion sites.
The absence of any substantial variation in these dynamic genomic features was matched by the generally high identity between protein-coding genes; of the 5569 orthologous gene pairs, 44.4% (2470) are >90% identical at the nucleotide level and 96.3% (5363) are >80% identical. Gene order is also largely collinear, with 98.1% (5647) of all C. dubliniensis putative genes positionally conserved in both species. This background of similarity means that any breaks in chromosomal colinearity are obvious and manual inspection of such putative species-specific loci identified 111 genes in C. dubliniensis and 191 in C. albicans that have either no heterospecific sequence match, or no reciprocal top BLASTX hit. This compares well with the 247 species-specific genes identified by CGH, which is sensitive only where a gene is absent from one species or where sequence divergence exceeds 60% (Moran et al. 2004). However, after removing transposable element genes, the number of “verified” species-specific genes (i.e., those with recognized homology) fell to 29 and 168, respectively (see Supplemental Table S4). Among these putative genes are factors with established roles in candidosis, which might contribute to increased virulence in C. albicans.
Exceptions to genomic colinearity concern genes with known associations to pathogenesis.
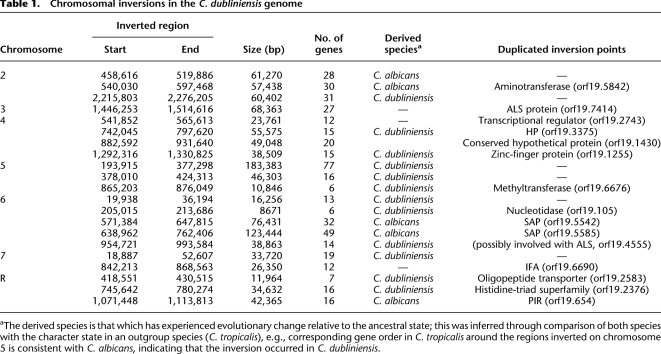
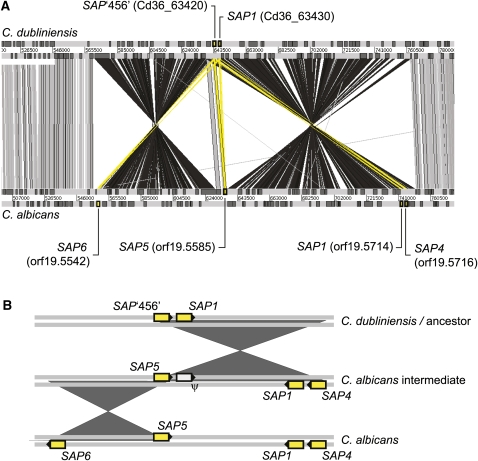
While the C. dubliniensis and C. albicans genomes largely correspond, there are various inversion, insertion–deletion, and transposition events that disrupt the otherwise collinear gene order. Importantly, some of these differences have affected genes known to have roles in Candida pathogenesis. Table 1 lists 21 major inversion events of between 8.5 and 185 kilobases (kb), (average ∼ 50 kb) that were observed across the genome. One of these events co-occurs with genes of the SAP family, which mediates hydrolytic responses to host tissues in various developmental stages and physical conditions, and is among the most important virulence factors in C. albicans (Naglik et al. 2003). Four SAP loci are found in close proximity on chromosome 6 in C. albicans and Figure 3 shows that, of these, C. dubliniensis lacks two loci, SAP4 and SAP5. One of the two existing SAP genes in C. dubliniensis is orthologous to SAP1 in C. albicans, while the second (labeled SAP‘456’ in Fig. 3A) has partial affinity with SAP4-6, so there is no strict ortholog in C. albicans. We suggest a hypothesis, depicted in Figure 3B, in which two segmental inversions have modified an ancestral tandem gene pair (retained in C. dubliniensis) to create the observed differences. This hypothesis predicts that SAP4-6, but not SAP1, in C. albicans should be monophyletic, which was confirmed by phylogenetic trees (data not shown).
Table 1.
Chromosomal inversions in the C. dubliniensis genome
aThe derived species is that which has experienced evolutionary change relative to the ancestral state; this was inferred through comparison of both species with the character state in an outgroup species (C. tropicalis), e.g., corresponding gene order in C. tropicalis around the regions inverted on chromosome 5 is consistent with C. albicans, indicating that the inversion occurred in C. dubliniensis.
Figure 3.
Segmental inversion and the evolution of secreted aspartyl proteinases (SAP). (A) Comparison of SAP genes on chromosome 6 (0.5–0.8 Mb) in C. albicans and C. dubliniensis generated using the Artemis Comparison Tool (ACT); DNA strands are represented by horizontal gray bars (scale in base pairs); genes are indicated by darker boxes. SAP genes are shaded yellow and linked to systematic IDs. Significant TBLASTX hits between genes are shown by vertical bars (gray, sense; black, anti-sense; yellow, SAPs). (B) A cartoon of the hypothesis that two segmental inversions created two additional SAP loci in C. albicans. The first inversion duplicated the original tandem gene pair (“C. dubliniensis/ancestor”) creating paralogs of SAP1 and SAP‘456’ in the opposite orientation (“C. albicans intermediate”); this is consistent with SAP1 and SAP4 in C. albicans and demands that the original SAP1 copy was subsequently lost (ψ). The second inversion duplicated the remaining SAP‘456’ copy (i.e., SAP5) to create a single gene in the opposite orientation (i.e., SAP6).
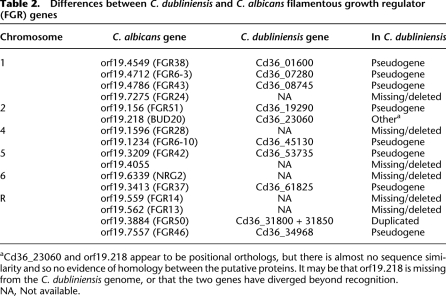
Breaks in chromosomal colinearity were more commonly caused by insertion–deletion events, and most of these were due to transposable elements. The C. dubliniensis genome contains numerous families of LTR and non-LTR retrotransposons and two types of DNA transposon; these are uniformly distributed, and have close affinity to those in C. albicans (see Supplemental Table S5). Although almost all elements are inactive, no elements were identified at the same locus in the two species, suggesting a complete loss of individual insertions from the last common ancestor, and extensive transposition subsequently. Beyond transposon activity, the remaining breaks in colinearity were due to insertions or deletions of individual genes, most of which are captured in the global gene family analysis below. For instance, C. dubliniensis lacks a member of the IFF gene family, the hyphal-associated HYR1 (orf19.4975). The IFF gene family encodes highly decorated, repetitive cell-wall proteins, which are induced during hyphal differentiation in C. albicans (Bailey et al. 1996). One member, IFF11 is secreted and contributes to virulence (Bates et al. 2007). Phylogenetic trees (data not shown) indicate that HYR1 is a relatively old lineage and not a recent duplication of an existing locus in C. albicans. A deletion in C. dubliniensis was confirmed by residual identity to HYR1 at the corresponding position in C. dubliniensis (e.g., ∼65% amino acid identity to last 30 residues). As later analysis confirms, gene deletion was a common phenomenon in C. dubliniensis, but rarely observed in C. albicans. Furthermore, genes continue to be lost from the C. dubliniensis genome, relative to its inferred ancestor; 115 pseudogenes were observed (see Supplemental Table S6), of which 78 have intact positional orthologs in C. albicans. These pseudogenes affect various gene functions but several were identified as orthologs of the filamentous growth regulator (FGR) genes that have suggested roles in C. albicans morphogenesis (Uhl et al. 2003). Table 2 describes 16 FGR genes that are functionally modified in C. dubliniensis, among them are eight pseudogenes and six deletions.
Table 2.
Differences between C. dubliniensis and C. albicans filamentous growth regulator (FGR) genes
aCd36_23060 and orf19.218 appear to be positional orthologs, but there is almost no sequence similarity and so no evidence of homology between the putative proteins. It may be that orf19.218 is missing from the C. dubliniensis genome, or that the two genes have diverged beyond recognition.
NA, Not available.
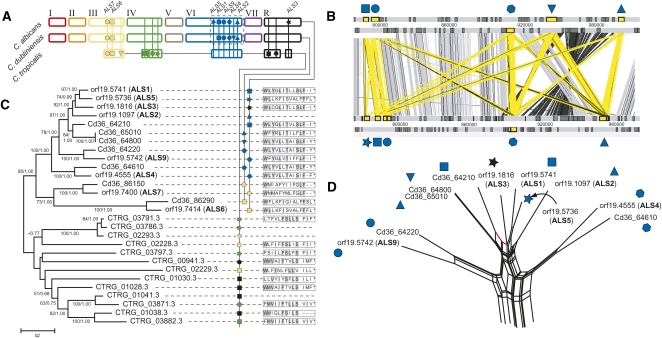
In contrast, C. albicans displayed various novel insertions, relative to C. dubliniensis and C. tropicalis; again, most of these were subsequently observed by the global gene family analysis described below. One particular example of note concerns the invasin-like ALS3 (orf19.1816; Hoyer et al. 1998; Phan et al. 2007; Almeida et al. 2008). ALS encodes a large family of repetitive cell-surface proteins that function primarily in host cell adhesion (Fu et al. 2002; Zhao et al. 2003, 2007; Sheppard et al. 2004); they are found throughout Candida (Hoyer 2001) and are instrumental in the initiation and maintenance of infection (Hoyer et al. 2008). ALS3 is found on chromosome R in C. albicans, but is entirely absent from the corresponding position in C. dubliniensis. A maximum likelihood phylogeny of all ALS genes in C. albicans, C. dubliniensis, and C. tropicalis was estimated from an alignment of the conserved 5′ domain and is shown in Figure 4B. It indicates that ALS3 clusters with ALS1 and 5, although it is most similar to ALS1, consistent with the origin of ALS3 in C. albicans through a unique transposition event from chromosome 6 to R.
Figure 4.
Comparative genomics and phylogeny of agglutinin-like sequences (ALS) in Candida spp. (A) A cartoon showing genomic distribution of ALS loci in C. albicans, C. dubliniensis, and C. tropicalis. The positions of loci are indicated by vertical lines and the presence of a gene(s) is represented by a unique symbol (which is also applied to positional orthologs in other species). A dashed line indicates the region of chromosome 6 expanded in panel C. Note that the chromosomes are drawn based on the C. albicans karyotype, with C. dubliniensis and C. tropicalis pseudochromosomes drawn for comparison, reflecting their conserved gene order, but not precise karyotypes. (B) Maximum likelihood phylogeny of ALS N-terminal nucleotide sequences in C. albicans (for which gene names ALS1-9 are given), C. dubliniensis (prefixed Cd36_) and C. tropicalis (prefixed CTRG_). Branch lengths are proportional to evolutionary change and measured in substitutions/site. This topology was concordant with an alternative Bayesian consensus tree; each node is attended by nonparametric bootstrap values and posterior probabilities from likelihood and Bayesian analyses respectively. Terminal nodes are labeled with gene IDs and locus symbols (as used in A), and then to the 14 amino acid sequence of the C terminus (where available). (C) ACT representation of ALS loci on chromosome 6 in C. dubliniensis (top) and C. albicans (bottom). ALS genes are shaded yellow and marked as in A. Significant TBLASTX hits between genes are represented by vertical bars (gray, sense; black, anti-sense; yellow, ALS). (D) Phylogenetic network showing a consensus of all possible relationships among chromosome 6 genes simultaneously. Splits supporting monophyly of Cd36_64210 and ALS1 are shown in red.
Subtle differences in C. dubliniensis and C. albicans ALS gene family repertoire
Transposition of ALS3 is not the only change to have affected this vital gene family. Superficially, C. dubliniensis appears to have a very similar ALS repertoire to C. albicans, in which eight ALS genes are distributed across three chromosomes (Jones et al. 2004; Sheppard et al. 2004). As shown in Figure 4, A and C, six C. albicans loci are shared with C. dubliniensis: ALS1, ALS2, ALS4, ALS6, ALS7, and ALS9. ALS3, as we have seen, has no corresponding locus, and neither do ALS5 or Cd36_64800. So, based on genomic position only, both species have evolved novel ALS. Turning to the apparently orthologous ALS genes, Figure 4B shows that: (1) C. albicans and C. dubliniensis sequences for ALS4, ALS6, ALS7, and ALS9 are siblings, supporting their orthology; (2) conversely, ALS1 and ALS2 cluster together rather than with their putative orthologs in C. dubliniensis; (3) Cd36_64800 is almost identical to the positional ortholog of ALS2 (Cd36_65010); and (4) ALS1 and ALS5 cluster together, suggesting a recent tandem duplication in C. albicans. Hence, phylogenetic analysis of C. albicans and C. dubliniensis ALS repertoires reveals a mixture of some positional orthologs displaying sequence orthology as expected (ALS4, ALS6, ALS7, and ALS9), and others showing none; either because they are species-specific acquisitions (ALS3, ALS5, Cd36_64800), or because structure has been altered post hoc (ALS1 and ALS2, Cd36_65010).
There is evidence that recombination between ALS genes at different loci is responsible for ALS genes at corresponding genomic positions lacking the expected sequence similarity. The phylogenetic network shown in Figure 4D presents a consensus of all competing relationship patterns within the sequence alignment for ALS genes on chromosome 6, and suggests that there are conflicting phylogenetic signals. For example, while ALS1 is most closely related to ALS2, there are some characters (highlighted in red) that still support orthology with its positional ortholog in C. dubliniensis (Cd36_64210). A Pairwise Homoplasy Index confirmed that the alignment contained significant phylogenetic incompatibility (P < 0.0001; Bruen et al. 2006). This apparent mosaicism among ALS 5′ regions was corroborated by the distribution of C terminus types, shown in Figure 4B; some closely related genes have dissimilar C termini (e.g., ALS9 and Cd36_64220), while some unrelated sequences have identical C termini (e.g., ALS5 and 6).
Principal differences in gene family repertoire concern hypothetical proteins with unspecified functions
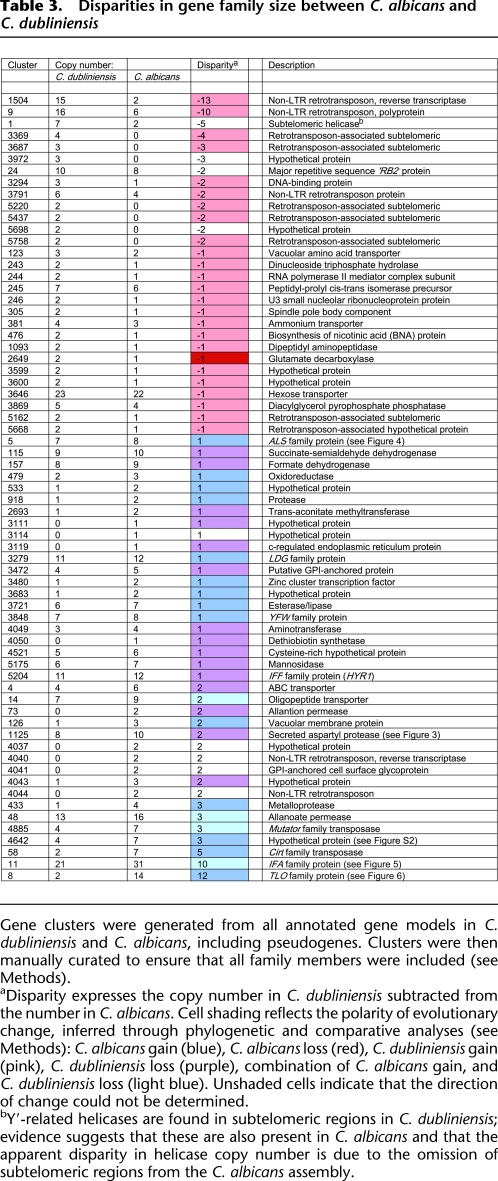
For a more comprehensive comparison of genomic content, we used OrthoMCL (Li et al. 2003) to cluster all homologous sequences by reciprocal BLAST searches. This reproduced the differences described above (ALS, cluster 5; IFF, cluster 5204; SAP, cluster 1125), but also identified the principal disparities, in terms of copy number, between the C. dubliniensis and C. albicans gene families. In Table 3, gene clusters for which there is a difference are ordered by disparity, with those in excess in C. dubliniensis at the top. Two general points can be made about these results. First, the principal interspecific differences do not concern the familiar cell-wall gene families with established roles at the host–pathogen interface; indeed, a survey of predicted GPI-anchored proteins produced only four differences (see Supplemental Table S7). Second, through comparison with C. tropicalis it was possible to determine the polarity of evolutionary change and show that excesses in C. dubliniensis were due to specific gains and not losses in C. albicans, while excesses in C. albicans were due to both C. dubliniensis losses and C. albicans gains.
Table 3.
Disparities in gene family size between C. albicans and C. dubliniensis
Gene clusters were generated from all annotated gene models in C. dubliniensis and C. albicans, including pseudogenes. Clusters were then manually curated to ensure that all family members were included (see Methods).
aDisparity expresses the copy number in C. dubliniensis subtracted from the number in C. albicans. Cell shading reflects the polarity of evolutionary change, inferred through phylogenetic and comparative analyses (see Methods): C. albicans gain (blue), C. albicans loss (red), C. dubliniensis gain (pink), C. dubliniensis loss (purple), combination of C. albicans gain, and C. dubliniensis loss (light blue). Unshaded cells indicate that the direction of change could not be determined.
bY′-related helicases are found in subtelomeric regions in C. dubliniensis; evidence suggests that these are also present in C. albicans and that the apparent disparity in helicase copy number is due to the omission of subtelomeric regions from the C. albicans assembly.
In terms of specific gene family differences, it is notable that while expansions in C. dubliniensis typically concern retrotransposons (clusters 1504 and 9) and associated genes (clusters 3369 and 3687), the largest excesses in C. albicans include various characterized genes, for example, metalloproteases (cluster 433), allanoate permeases (cluster 48), and oligopeptide transporters (cluster 14). Expansions of certain hypothetical genes are also suggested, for instance, cluster 4642 contains genes predicted to encode hypothetical proteins with four transmembrane domains and a signal peptide. These genes are arranged in a tandem array in both C. albicans and C. dubliniensis. The phylogeny shown in Supplemental Figure S2 describes four orthologous clades consistent with genomic position, indicating that the four paralogs are structurally distinct. The only departure from this scheme is the expansion of the fourth and last tandem duplicate in C. albicans through tandem duplication (to form orf19.3906), and through transposition within the array (to form orf19.3903) and to chromosome 4 (to form orf19.4961). Maintenance of orthology between tandem duplicates in the arrays, strongly suggests that the paralogs perform subtly different functions on the cell surface, and points to functional innovation of the last copy only in C. albicans.
The largest disparities in gene family copy number favoring C. albicans concern two uncharacterized gene families in particular: the leucine-rich repeat protein genes (IFA, cluster 11) and the telomere-associated genes (TLO, cluster 8), which are now described in detail.
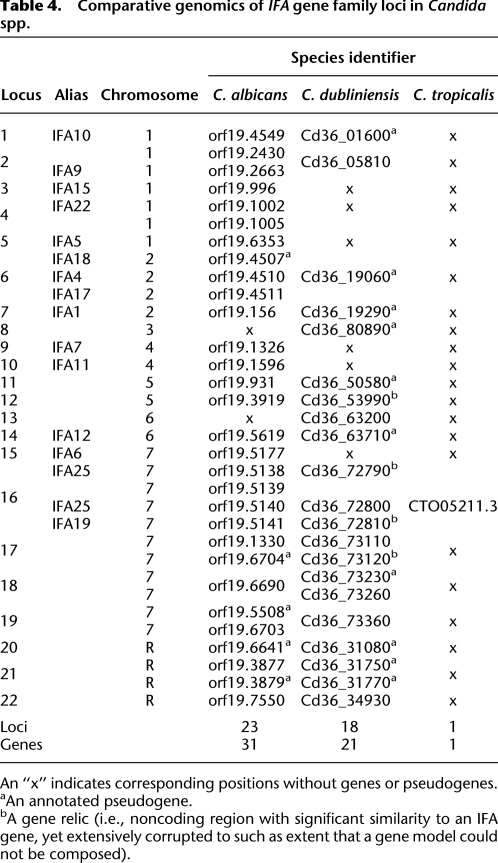
The IFA gene family has undergone widespread gene loss in C. dubliniensis
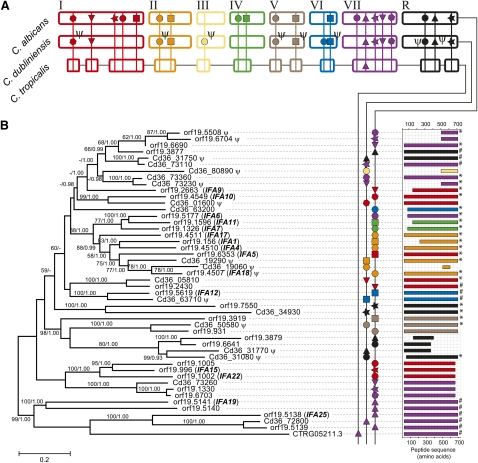
The IFA genes encode a large family of putative transmembrane proteins with weak affinity to “LPF” leucine repeat-rich proteins in viruses. Table 4 describes the 22 loci defined in C. albicans and C. dubliniensis. Locus 16 (IFA19/25) is conserved in both species on chromosome 7, as well as C. tropicalis. All other loci were either shared by C. albicans and C. dubliniensis (15), specific to C. albicans (6), or specific to C. dubliniensis (2). However, a substantial component of the C. dubliniensis repertoire was predicted to be fragmentary or nonfunctional (14/21 loci, compared with 6/31 in C. albicans). A maximum likelihood phylogeny estimated from the 648 amino acid multiple alignment, shown in Figure 5 indicates that (1) derived gene duplication in C. albicans has created several new loci; (2) gene relics at various stages of mutational decay demonstrate that gene loss is on-going in C. dubliniensis; and (3) while many positional orthologs cluster together as expected (e.g., locus 13, indicated by a blue square), other cases are poorly related suggesting that gene conversion may affect loci in close proximity (e.g., locus 17, purple stars). This is supported by the phylogenetic distribution of C-terminal types, which are described as “acidic” or “basic” (see Fig. 5B). As with the ALS C termini previously, the distribution is incompatible with the phylogenetic relationships; for instance, at locus 2, Cd36_05810 and orf19.2430 (red triangles) are positionally and structurally orthologous, but do not share the same C terminus type. In summary, the IFA gene family shows a marked dichotomy between evolutionary expansion in C. albicans and depletion in C. dubliniensis.
Table 4.
Comparative genomics of IFA gene family loci in Candida spp.
An “x” indicates corresponding positions without genes or pseudogenes.
aAn annotated pseudogene.
bA gene relic (i.e., noncoding region with significant similarity to an IFA gene, yet extensively corrupted to such as extent that a gene model could not be composed).
Figure 5.
Phylogeny and comparative genomics of the IFA gene family in Candida spp. (A) Cartoon of genomic distribution (note: not precise karyotypes). Pseudogenes are denoted by Ψ. (B) Maximum likelihood phylogeny of IFA protein sequences in C. albicans (prefixed orf19.), C. dubliniensis (prefixed Cd36_), and C. tropicalis (CTRG05211.3). Branch lengths are proportional to evolutionary change and measured in substitutions/site. This topology was concordant with an alternative Bayesian consensus tree. Dotted lines link gene IDs to a cartoon of a multiple sequence alignment. C termini are labeled according to amino acid composition: (#) long, acidic-type; (*) a shorter, nonacidic-type C-terminal. No label indicates that neither type is present or the C terminus is absent.
The TLO gene family has been uniquely elaborated in C. albicans
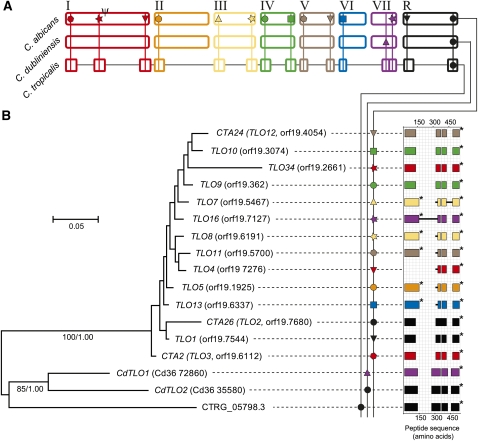
15 TLO genes were identified at the subtelomeres of the original C. albicans genome sequence (van het Hoog et al. 2007). As the OrthoMCL analysis suggests, C. dubliniensis lacks these genes and only two TLO homologs were identified: CdTLO1 (Cd36_72860; internally on chromosome 7) and CdTLO2 (Cd36_35580; subtelomerically on chromosome R). For the purposes of providing an outgroup, a single homolog was found in C. tropicalis (CTRG05798.3). Figure 6A shows that the only locus occupied by all three species is at the right arm of chromosome R. This is termed the “ancestral” locus, since a single gene at this position is the probable ancestral state. The phylogeny in Figure 6B shows that (1) C. albicans and C. dubliniensis copies are reciprocally monophyletic, supporting independent TLO expansions; (2) C. albicans copies are genetically homogeneous (average nucleotide identity = 96.5%), except for the length of repeat sequences and the apparent alternative splicing of a subset of sequences, whereas the two distinct C. dubliniensis paralogs are relatively well diverged (nucleotide identity = 74.9%); (3) in contradiction of the species tree, C. dubliniensis sequences are closer in average amino acid identity over 193 aligned positions to C. tropicalis (73.6%) than to C. albicans (67.2%); and (4) despite its shared position with the ancestral locus in C. dubliniensis and C. tropicalis, orf19.7680 does not branch basally, but instead is barely distinguishable from its paralogs. These observations indicate that beginning with a single gene at the ancestral locus, additional loci have evolved independently in C. dubliniensis, through a transposition event to create Cd36_72860 on chromosome 7, and in C. albicans, through the expansion to almost all telomeres.
Figure 6.
Phylogeny and comparative genomics of telomere-associated (TLO) genes in Candida spp. (A) Cartoon of genomic distribution (note: not precise karyotypes). One gene copy previously identified as TLO14 on the right arm of chromosome 6 in C. albicans (van het Hoog et al. 2007F), is not included in current assemblies or detected using BLAST. Pseudogenes are denoted by ψ. (B) Bayesian phylogeny of TLO nucleotide sequences in C. albicans, C. dubliniensis (prefixed Cd36_), and C. tropicalis (CTRG_05798.3). Branch lengths are proportional to evolutionary change. Monophyly of C. albicans and C. dubliniensis sequences is supported by nonparametric bootstrap values and posterior probabilities from maximum likelihood and Bayesian analyses, respectively. Dotted lines link individual genes to a cartoon of a multiple sequence alignment. Exons are represented by shaded rectangles, putative introns with heavy black lines; spaces between exons represent gaps introduced by multiple alignment. An asterisk (*) denotes a stop codon.
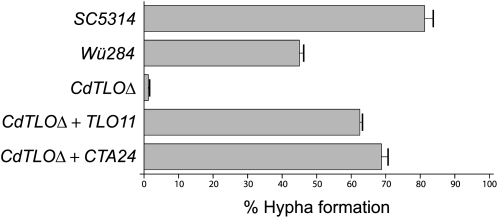
As the principal disparity in gene content between C. dubliniensis and C. albicans, we sought to investigate TLO function by creating null mutants in C. dubliniensis. Null mutants for CdTLO1 showed a major reduction in hyphal formation in response to serum, as shown in Figure 7. Complementation of the CdTLO1Δ with either of two C. albicans TLO genes (i.e., TLO11 and CTA24 [also known as TLO12]) restored the defect in hyphal formation, indicating that these TLO genes have a common role in morphogenesis in both species. Further experiments are on-going to determine if all TLO gene copies have the same effects when ablated and restored, and to define the precise function of the TLO families in C. albicans and C. dubliniensis and their role in pathogenesis.
Figure 7.
Formation of hyphae by wild-type C. albicans (SC5314), wild-type C. dubliniensis (Wü284), Cdtlo1Δ (Cd36_35580 deleted), Cdtlo1Δ expressing CaTLO11 (orf19.5700), and Cdtlo1Δ expressing caCTA24 (orf19.4054). The percentage of yeast cells producing germ tubes was calculated following incubation at 37°C for 1 h.
Absence of adaptive evolution among orthologous gene sequences
Orthologous gene sequences in C. albicans and C. dubliniensis are generally well conserved, as described previously. While the rates of both synonymous and nonsynonymous (i.e., amino acid replacing) substitutions were low, the ratio of these two rates could still reveal the action of adaptive evolution. However, the dN/dS rates shown in Supplemental Table S8 demonstrate that there is little evidence for positive selection among 5569 orthologous gene pairs. For most genes (5177) dN/dS < 0.3, indicating strong purifying selection and conservation of protein structure. Only 1 gene (orf19.6601) had a dN/dS value significantly greater than 1 (3.077), indicative of positive selection. Therefore, the expansion of particular gene families described above is not accompanied by a concomitant innovation in protein structures.
Discussion
This study is the first to compare the genome sequences of C. dubliniensis and C. albicans, a comparison that is particularly valuable because these closely related organisms are very similar, but differ markedly in pathogenicity. In our analysis of their differences in genetic repertoire, one might have expected gene families with known roles in virulence to be specific to C. albicans or nonfunctional in C. dubliniensis. However, we find that the two gene complements are highly similar, with only subtle differences in repertoire between such characterized gene families. Perhaps surprisingly, it is the unfamiliar TLO and IFA gene families that have expanded most in C. albicans, and these now provide compelling candidates for pathogenicity-associated factors. This picture of the relative complement of specific gene families was produced by an earlier genomic analysis using CGH (Moran et al. 2004), indicating that this technique can provide an accurate impression of genomic differences.
C. albicans infection is associated with the developmental transition from the yeast to hyphal growth stage, and genes regulating, or specifically expressed during hyphal differentiation, are therefore implicated in pathogenesis. Both C. dubliniensis and C. albicans possess various hydrolytic enzymes, such as secretory lipases and phospholipase B, which are expressed at the onset of hyphal differentiation and when infection is established (Naglik et al. 2003). Both genomes also include diverse SAP gene families, although additional SAP genes have evolved in C. albicans, which we propose is a result of sequential segmental inversions. Duplication of genes at the breakpoints of chromosomal inversions has been observed previously, for instance, in Drosophila melanogaster (Matzkin et al. 2005) and Anopheles gambiae (Sharakhov et al. 2006), and in yeast, recently duplicated genes co-occur with chromosomal breakpoints more frequently than expected by chance (Gordon et al. 2009). This is thought to occur during the repair of chromosomal breaks, either single- or double-stranded, during which nonreciprocal recombination can occur with the template used in DNA repair, which is proposed to contain the duplicated gene (Sharakhov et al. 2006; Ranz et al. 2007; Meisel 2009). In this case, the genes concerned, SAP5 and SAP6, encode proteins known to have hypha-specific expression and may have increased the ability of C. albicans to cause systemic infection. In almost all respects C. dubliniensis and C. albicans correspond in their repertoires of GPI-anchored proteins, which mediate interactions at the host–pathogen interface. Thus, their common ancestor was probably a competent opportunist, with the molecular tools to cause disease, among them at least six ALS genes that are instrumental in host cell adhesion and are unambiguously associated with pathogenesis (Hoyer 2001; Hoyer et al. 2008).
Given the prominent role of ALS in disease (Fu et al. 2002; Zhao et al. 2007), their known functional differentiation (Sheppard et al. 2004; Zhao et al. 2004; Cheng et al. 2005; Zhao et al. 2005) and genetic variability, perhaps even hypervariability (Zhang et al. 2003; Zhao et al. 2003; Oh et al. 2005; Zhao et al. 2007,), it is important to clearly and correctly interpret genomic variation in ALS repertoire. While ALS loci are positioned in a roughly similar manner in C. dubliniensis and C. albicans, the phylogeny in Figure 4B often showed that there is no simple orthology between ALS genes on chromosome 6. ALS2 and 4 from C. albicans did not cluster with their positional orthologs in C. dubliniensis because, as Figure 4D demonstrates, recombination between ALS genes has altered some sequences since speciation. This is supported by the distribution of distinct C-terminal types, which clearly shows that ALS genes can be chimaeric. While this “loss of orthology” has only affected certain genes in C. albicans and C. dubliniensis, it is worth noting that all C. tropicalis sequences are monophyletic in Figure 4B, despite ALS6 and 7 having positional orthologs in this species (CTRG_02229.3 and CTRG_02228.3, respectively). This indicates that, ultimately, recombination is likely to affect all ALS genes and result in concerted evolution. Hence, although both C. albicans and C. dubliniensis have five genes within the region of chromosome 6 in Figure 4C, positional orthology is no guarantee of structural, or perhaps functional, correspondence. Where sequence and positional identities both suggest orthology, as with ALS1, ALS6, and ALS7, we might reliably transfer functional information between genes; but where sequences are very different, function may also have changed.
Since it shared an ancestor with C. albicans, C. dubliniensis has experienced reductive evolution that has probably diminished the genetic repertoire it inherited. With the exception of conspicuous transposon activity, there has been little expansion of existing gene families, or derivation of novel genes. Instead, C. dubliniensis has lost genes with important functions, such as HYR1, and is in the process of losing other genes, some of which have suggested roles in filamentous growth and perhaps virulence, through pseudogenization. The most dramatic loss of genetic capacity has occurred, and continues to occur, in the IFA gene family, which clearly originated in Candida, and could be a specific adaptation to parasitism (Butler et al. 2009). C. dubliniensis and C. albicans are known to differ in their IFA gene repertoire (Moran et al. 2004); the exhaustive analysis presented here shows that 14 out of 21 loci are predicted to be nonfunctional. While some genes are corrupted by only one or a few frame-shifts, others are genuine relics with heavily corrupted coding sequences that betray a recent history of mutational decay. However, this process of unmistakable degradation in C. dubliniensis is only half the story: it cannot explain the greatest differences between the species.
For its part, C. albicans has lost relatively little and has expanded its genomic repertoire of selected gene families. In Table 3, only one case of excess copy number in C. dubliniensis is due to a C. albicans loss (glutamate decarboxylase; cluster 2649). Relatively minor evolutionary acquisitions, such as SAP4 and 5 through inversion, or ALS3 through duplicative transposition, may yet turn out to be of great importance. But it is the TLO gene family that stands out as the largest copy number disparity that has evolved specifically in C. albicans, rather than through deletion in C. dubliniensis. This disparity was suggested in previously reported CGH experiments (Moran et al. 2007), but was not observed in pan-Candida genomic comparisons (Butler et al. 2009), despite only one TLO gene being present in C. tropicalis. TLO are thought to encode transcription factors (Kaiser et al. 1999; van het Hoog et al. 2007), since they contain a putative Med2 domain (Hallberg et al. 2004; Pfam: PF11214), which is associated with the RNA polymerase II mediator complex in yeast (Björklund and Gustafsson 2005). In preliminary assays, we have shown that loss of TLO1 in C. dubliniensis significantly reduces hyphal formation in response to serum, and that this can be restored by complementation with certain C. albicans TLO genes. Therefore, TLO genes may have a regulatory function during morphogenesis in both species, perhaps affecting their relative abilities to cause disease.
As Figure 6 suggests, C. albicans has derived novel TLO gene copies through duplication at its subtelomeres, perhaps through telomeric exchange. Since their sequences are virtually identical, this suggests that subtelomeric TLO genes diverge as a unit under concerted evolution, and evolved for dosage rather than functional diversity. Contrary to expectation, the C. albicans TLO gene sequence is less related to the C. dubliniensis homolog than the C. tropicalis gene. Substitution patterns within the TLO multiple alignments (see Supplemental Fig. S3) show that this unexpected similarity is due to amino acid motifs shared by C. dubliniensis and C. tropicalis (shown in red), which have been modified in C. albicans. Hence, the “closeness” of C. dubliniensis and C. tropicalis TLO sequences, which contradicts the overall species relationships, probably owes more to the derived state of C. albicans: in short, the TLO family has been uniquely modified in number and structure in C. albicans, while C. dubliniensis retains a TLO repertoire not unlike the ancestral state.
The differences between the genomic repertoires of C. albicans and C. dubliniensis demonstrate that they have taken divergent evolutionary trajectories since speciation. While C. albicans has augmented its parasitic capacity with expansions of the SAP, ALS, and IFF gene families (among many others), C. dubliniensis has experienced widespread deletions, many of which have affected its capacity to cause disease, at least in humans. Since there is no comparable innovation in the C. dubliniensis genome, this suggests a process of reductive evolution and the deletion of redundant loci after C. dubliniensis. We exploited the diminished pathogenicity of C. dubliniensis to identify the candidate genes in C. albicans that could account for the phenotypic disparity between these otherwise very similar organisms. We have shown that the principal disparities favoring C. albicans concern two relatively unknown gene families, the TLO and IFA proteins, providing substantial new leads in the molecular characterization of candidal virulence.
Methods
Genome sequencing and assembly
C. dubliniensis has a complex diploid karyotype consisting of 13 haploid chromosomes, some of which correspond in their entirety to a C. albicans chromosome, and some of which are chimaeras of C. albicans chromosomal blocks (Magee et al. 2008). This karyotype could not be recovered during genome sequencing, or reproduced in the final assembly, because chromosomal homologs are virtually indistinguishable. Therefore, the C. dubliniensis genome sequence is a haploid consensus of both chromosomal homologs, which was assembled de novo without reference to the C. albicans genome sequence. Genome assembly produced 1110 contigs with an N50 value of 86 kb, meaning that at least half of all bases belonged to contigs of this size or greater. Given that the actual karyotype could not be resolved, and the purpose of assembly was for comparison with C. albicans, at this point the order and orientation of C. dubliniensis contigs was established by mapping them onto the C. albicans genome sequence, to produce pseudochromosomes. This inevitably meant that those haploid components of the C. dubliniensis karyotype that departed from the eight chromosome karyotype of C. albicans were not represented in our C. dubliniensis assembly. However, no genome data were discarded since these “missing” chromosomes contributed their nucleotide sequences to the haploid consensus. PCR walking was used to validate the contig scaffolds produced in reference to C. albicans. Specific BACs were sequenced to demonstrate that those haploid components contained in the in vivo karyotype, but omitted from our genome assembly, did exist.
Sequence polymorphism
To identify heterozygous sites in the C. dubliniensis assembly, we realigned all of the shotgun reads using SSAHA2 (Ning et al. 2001) onto the final genome sequence. We then identified all reads that uniquely align and are paired in the correct orientation within the expected insert size. Sites were identified where a base differed from the reference base and where the cumulative phred score for the alternative base was greater than 42, as described previously (Jeffares et al. 2007). This was also done for C. albicans reads for comparison.
Gene prediction and annotation
Open reading frames (ORFs) over 300 bp were marked up in Artemis (Rutherford et al. 2000). ORFs were manually checked using BLASTX to select the most plausible coding sequence where multiple overlapping ORFs existed, and to identify likely start and stop codons. All splice donor and acceptor sequences and lariat (TACTAAC) sites were manually marked up, enabling a refinement of intron–exon structures of appropriate gene models. Additional coding sequences were annotated, and existing gene models were refined based on a base-wise comparison with C. albicans using the Artemis Comparison Tool (ACT; Carver et al. 2005). Functions were ascribed by searching UNIPROT and PFam databases for homologous proteins using FASTA and BLASTP analyses. All protein alignments were manually reviewed. Pseudogenes were only confirmed after exhaustive manual base checking to prove that both alleles were predicted to be nonfunctional. Where only one allele was nonfunctional, the haploid consensus, (which contains sequences from both chromosomal homologs), was resolved into two haplotypes and the full-length allele was retained in the consensus.
Annotation of the major repeat sequence (MRS)
A gene-poor region of chromosome 2 was found to contain a cluster of five SfiI restriction sites, known to be located in the C. albicans MRS (Chibana et al. 1994; Jones et al. 2004), and to contain a coding sequence which, in C. albicans, was described as being located in the MRS RB2 region. Dot-plot analysis (Dotter) of this region showed that it contained four tandem repeat units of 2106 bp each, plus one partial repeat of 1535 bp. Each of these repeat units contained a single SfiI site. This represents the RPS “core” of the MRS region. A single RPS unit from this region was compared to each C. dubliniensis chromosome in turn using BLASTN, in order to locate the other MRS regions. The conserved flanking HOK and RB2 regions, previously noted in C. albicans MRS (Chindamporn et al. 1998), were located by aligning 10 kb of sequence upstream and downstream of the core RPS unit from each MRS using ClustalW (Larkin et al. 2007).
Comparative analysis of genome content
The OrthoMCL algorithm (Li et al. 2003; Chen et al. 2006) was used to obtain a global view of the differences between C. dubliniensis and C. albicans genomic complement. OrthoMCL groups homologous sequences into clusters that range in size from single species-specific genes to large gene families conserved across species. We used OrthoMCL to obtain a preliminary list of gene clusters that was then manually curated to identify gene families with interspecific disparities in copy number. After inspecting all such clusters using BLASTX to ensure that gene families were not subdivided into distinct clusters, and that all family members were included, we then inspected the genomic positions of each cluster member in ACT to confirm presence or absence in C. dubliniensis and C. albicans. The preliminary list of disparities produced by OrthoMCL did not include many species-specific singleton genes that were evident from chromosomal comparisons. Therefore, we manually inspected each chromosome for disruptions to the colinear gene order. C. dubliniensis coding sequences (including transposable element genes, but not pseudogenes) at these breakpoints were then compared with the C. albicans genome using BLASTX to obtain the top sequence match. Genes without matches in C. albicans, or where the top hit in C. albicans was not reciprocal, were confirmed as C. dubliniensis specific. The process was repeated for putative C. albicans-specific genes. Where possible, the polarity of evolutionary change (i.e., the species in which it occurred) was assessed through comparison with the character state in an outgroup, in this case, the draft C. tropicalis genome sequence (Butler et al. 2009).
Comparative analysis of predicted GPI-anchored proteins
Genes that encode putative GPI proteins were selected by identifying coding sequences that were predicted to have an N-terminal signal peptide and a GPI-anchor attachment site, but were negative for transmembrane spanning regions. C-terminal GPI signal peptides were predicted using Big-PI (Eisenhaber et al. 2004) and a complementary pattern search method that searched for the GPI-specific sequence (gasndc)(gasvietkdlf)(gasv)-x(4,19)-(filmvagpstcywn) (Prosite format) using the fuzzpro (de Groot et al. 2003) program from EMBOSS. Absence of internal transmembrane domains was analyzed using TMHMM (Krogh et al. 2001), and presence of an N-terminal signal peptide predicted using SignalP (Emanuelsson et al. 2007). Proteins fulfilling the criteria described above were analyzed further by BLAST to identify C. albicans orthologs. The C. dubliniensis genome was surveyed for orthologs to all known GPI-anchored proteins and protein families in C. albicans.
Phylogenetic analysis
Comparative genomics and phylogenetic estimation were used to characterize the evolutionary changes affecting several gene families (TLO, IFA, ALS, IFF, SAP, and cluster 4629). Sequences were retrieved through BLASTX analysis and subsequent inspection of each locus in ACT. Where possible, outgroup sequences for C. albicans and C. dubliniensis were provided by homologs from C. tropicalis (Butler et al. 2009). All gene sequences were translated and aligned using ClustalW (Larkin et al. 2007) and then edited manually. It was necessary to remove highly divergent repeat regions from the TLO (789 bp) and ALS (1326 bp) alignments. The SAP (1857 bp), IFA (1941 bp), IFF (1311 bp), and cluster 4642 (577 bp) alignments comprised the entire coding sequences after minor adjustments to ensure equal length. Phylogenies for the TLO, ALS, and cluster 4642 nucleotide alignments were estimated using maximum likelihood (ML) and Bayesian inference (BI). The ML phylogeny was estimated with PHYML v2.4.4 (Guindon and Gascuel 2003; Guindon et al. 2005), using a general-time reversible (GTR) model (Yang 1994) with six rate categories. Empirical base frequencies and a gamma distribution parameter (α) were optimized from the data. The BI phylogeny was estimated with MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). Four parallel MCMC chains were run for 1,000,000 generations, with a sample frequency of 100 generations; a burn-in of 1000 generations was found to be sufficient to achieve stationary model parameters using Tracer v1.4.1 (http://tree.bio.ed.ac.uk/software/tracer/). Due to greater sequence divergence and available characters, phylogenies for the SAP, IFA, and IFF families were estimated from protein sequences. ML and BI phylogenies employed a WAG model (Whelan and Goldman 2001) with an additional rate heterogeneity parameter. Otherwise, operating conditions were as described previously. In all cases, topological robustness was assessed through 100 nonparametric bootstrap replicates (Felsenstein 1985).
Experimental disruption of TLO genes
Growth and transformation of C. dubliniensis strain Wü284 was performed by electroporation as described previously (Moran et al. 2007). Disruption of the CdTLO1 gene was achieved using the SAT1-flipper cassette system (Reuß et al. 2004). A deletion construct was created by amplifying the flanking regions of CdTLO1 with the primer pairs CTA21KF/CTA1X and CTA1S/CTA21SIR (see Supplemental Table S9), followed by ligation of the products into plasmid pSFS2A to yield plasmid pTY101. This construct was then used to transform C. dubliniensis Wü284, and deletion of the CdTLO1 gene was confirmed by Southern analysis. Reintroduction of wild-type TLO genes was achieved by amplification of the entire ORF plus their upstream and downstream regulatory sequences using primer pairs CdTLO1FP/CdTLO1RP (for CdTLO1), CdTLO2FP/CdTLO2RP (for CdTLO2), CaTLO11FP/CaTLO11RP (for CaTLO11), and CaTLO12FP/CaTLO12RP (for caCTA24) (see Supplemental Table S9) and ligation of these into the C. dubliniensis integrating vector pCDRI to yield plasmids pCdTLO1, pCdTLO2, pCaTLO11, and pCaTLO12, respectively. These plasmids were transformed into the CdTLO1Δ strain as described above.
Calculation of dN/dS ratios
Five thousand five hundred sixty-nine (5569) pairs of orthologous gene sequences were extracted from the OrthoMCL analysis described above. Each of these pairs was aligned using ClustalX. The ratio of the nonsynonymous substitutions per site to synonymous substitutions per site (dN/dS) was estimated for each orthologous pair, averaged over the entire alignment, using Ka/Ks Calculator v1.2 (Zhang et al. 2006). This program implements several candidate models of codon substitution in a maximum likelihood framework; we used the NG method to estimate dN/dS values.
Acknowledgments
We thank Lee Murphy and members of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute (WTSI). This work was supported by the Wellcome Trust (WT085775/Z/08/Z) through their support of the Pathogen Genomics Group at the WTSI, and grants from Science Foundation Ireland (O4/IN3/B463), the European Union (MRTN-CT-2004-512481), and the Irish Health Research Board (RP/2004/235). A.P.J. is supported by a Postdoctoral Fellowship from the Wellcome Trust Sanger Institute. C.A.M. was supported by a Medical Research Council New Investigator Award. We thank two anonymous referees for their helpful comments.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to EMBL (http://www.ebi.ac.uk/embl/) under accession nos. FM992688–FM992695.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.097501.109.
References
- Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Jr, Filler SG, Hube B. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves SH, Milan EP, de Laet Sant'Ana P, Oliveira LO, Santurio JM, Colombo AL. Hypertonic sabouraud broth as a simple and powerful test for Candida dubliniensis screening. Diagn Microbiol Infect Dis. 2002;43:85–86. doi: 10.1016/s0732-8893(02)00368-1. [DOI] [PubMed] [Google Scholar]
- Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, de la Rosa JM, MacCallum DM, Brown AJ, Gow NA, Odds FC. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect Immun. 2007;75:2922–2928. doi: 10.1128/IAI.00102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr, Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun. 2005;73:1656–1663. doi: 10.1128/IAI.73.3.1656-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana H, Iwaguchi S, Homma M, Chindamporn A, Nakagawa Y, Tanaka K. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. J Bacteriol. 1994;176:3851–3858. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chindamporn A, Nakagawa Y, Mizuguchi I, Chibana H, Doi M, Tanaka K. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144:849–857. doi: 10.1099/00221287-144-4-849. [DOI] [PubMed] [Google Scholar]
- de Groot PW, Hellingwerf KJ, Klis FM. Genome-wide identification of fungal GPI proteins. Yeast. 2003;20:781–796. doi: 10.1002/yea.1007. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Mol Biol. 2004;337:243–253. doi: 10.1016/j.jmb.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Enjalbert B, Moran GP, Vaughan C, Yeomans T, Maccallum DM, Quinn J, Coleman DC, Brown AJ, Sullivan DJ. Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Mol Microbiol. 2009;72:216–228. doi: 10.1111/j.1365-2958.2009.06640.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr Candida albicans Als1p: An adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol. 2002;44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- Gilfillan GD, Sullivan DJ, Haynes K, Parkinson T, Coleman DC, Gow NA. Candida dubliniensis: Phylogeny and putative virulence factors. Microbiology. 1998;144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online: A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg M, Polozkov GV, Hu GZ, Beve J, Gustafsson CM, Ronne H, Björklund S. Site-specific Srb10-dependent phosphorylation of the yeast Mediator subunit Med2 regulates gene expression from the 2-microm plasmid. Proc Natl Acad Sci. 2004;101:3370–3375. doi: 10.1073/pnas.0400221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly S, Pujol C, Soll DR. Microevolutionary changes and chromosomal translocations are more frequent at RPS loci in Candida dubliniensis than in Candida albicans. Infect Genet Evol. 2002;2:19–37. doi: 10.1016/s1567-1348(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser B, Munder T, Saluz HP, Kunkel W, Eck R. Identification of a gene encoding the pyruvate decarboxylase gene regulator CaPdc2p from Candida albicans. Yeast. 1999;15:585–591. doi: 10.1002/(SICI)1097-0061(199905)15:7<585::AID-YEA401>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Kibbler CC, Seaton S, Barnes RA, Gransden WR, Holliman RE, Johnson EM, Perry JD, Sullivan DJ, Wilson JA. Management and outcome of bloodstream infections due to Candida species in England and Wales. J Hosp Infect. 2003;54:18–24. doi: 10.1016/s0195-6701(03)00085-9. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lephart PR, Chibana H, Magee PT. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot Cell. 2005;4:733–741. doi: 10.1128/EC.4.4.733-741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Sanchez MD, Saunders D, Harris D, Berriman M, Magee PT. Extensive chromosome rearrangements distinguish the karyotype of the hypovirulent species Candida dubliniensis from the virulent Candida albicans. Fungal Genet Biol. 2008;45:338–350. doi: 10.1016/j.fgb.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJ, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Evolutionary dynamics of recently duplicated genes: Selective constraints on diverging paralogs in the Drosophila pseudoobscura genome. J Mol Evol. 2009;69:81–93. doi: 10.1007/s00239-009-9254-1. [DOI] [PubMed] [Google Scholar]
- Moran G, Stokes C, Thewes S, Hube B, Coleman DC, Sullivan D. Comparative genomics using Candida albicans DNA microarrays reveals absence and divergence of virulence-associated genes in Candida dubliniensis. Microbiology. 2004;150:3363–3382. doi: 10.1099/mic.0.27221-0. [DOI] [PubMed] [Google Scholar]
- Moran GP, MacCallum DM, Spiering MJ, Coleman DC, Sullivan DJ. Differential regulation of the transcriptional repressor NRG1 accounts for altered host-cell interactions in Candida albicans and Candida dubliniensis. Mol Microbiol. 2007;66:915–929. doi: 10.1111/j.1365-2958.2007.05965.x. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC, Hanson MF, Davidson AD, Jacobsen MD, Wright P, Whyte JA, Gow NA, Jones BL. One year prospective survey of Candida bloodstream infections in Scotland. J Med Microbiol. 2007;56:1066–1075. doi: 10.1099/jmm.0.47239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SH, Cheng G, Nuessen JA, Jajko R, Yeater KM, Zhao X, Pujol C, Soll DR, Hoyer LL. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology. 2005;151:673–681. doi: 10.1099/mic.0.27680-0. [DOI] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, Ashburner M, Bergman CM. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sadhu C, McEachern MJ, Rustchenko-Bulgac EP, Schmid J, Soll DR, Hicks JB. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo NF, Santolamazza F, Della Torre A, Simard F, Collins FH, Besansky NJ. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci. 2006;103:6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE., Jr Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–30489. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- Stokes C, Moran GP, Spiering MJ, Cole GT, Coleman DC, Sullivan DJ. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet Biol. 2007;44:920–931. doi: 10.1016/j.fgb.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: Phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- Sullivan DJ, Moran GP, Pinjon E, Al-Mosaid A, Stokes C, Vaughan C, Coleman DC. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 2004;4:369–376. doi: 10.1016/S1567-1356(03)00240-X. [DOI] [PubMed] [Google Scholar]
- Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, Cuomo C, Berriman M, Scherer S, Magee BB, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela MM, Kamei K, Sano A, Tanaka R, Uno J, Takahashi I, Ito J, Yarita K, Miyaji M. Pathogenicity and virulence of Candida dubliniensis: Comparison with C. albicans. Med Mycol. 2002;40:249–257. doi: 10.1080/mmy.40.3.249.257. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Zhang N, Harrex AL, Holland BR, Fenton LE, Cannon RD, Schmid J. Sixty alleles of the ALS7 open reading frame in Candida albicans: ALS7 is a hypermutable contingency locus. Genome Res. 2003;13:2005–2017. doi: 10.1101/gr.1024903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomic Proteomic Bioinform. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Pujol C, Soll DR, Hoyer LL. Allelic variation in the contiguous loci encoding Candida albicans ALS5, ALS1 and ALS9. Microbiology. 2003;149:2947–2960. doi: 10.1099/mic.0.26495-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Oh SH, Yeater KM, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology. 2005;151:1619–1630. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oh SH, Jajko R, Diekema DJ, Pfaller MA, Pujol C, Soll DR, Hoyer LL. Analysis of ALS5 and ALS6 allelic variability in a geographically diverse collection of Candida albicans isolates. Fungal Genet Biol. 2007;44:1298–1309. doi: 10.1016/j.fgb.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]