Abstract
The Y centromere sequence of house mouse, Mus musculus, remains unknown despite our otherwise significant knowledge of the genome sequence of this important mammalian model organism. Here, we report the complete molecular characterization of the C57BL/6J chromosome Y centromere, which comprises a highly diverged minor satellite-like sequence (designated Ymin) with higher-order repeat (HOR) sequence organization previously undescribed at mouse centromeres. The Ymin array is ∼90 kb in length and resides within a single BAC clone that provides sequence information spanning an endogenous animal centromere for the first time. By exploiting direct patrilineal inheritance of the Y chromosome, we demonstrate stability of the Y centromere DNA structure spanning at least 175 inbred generations to beyond the time of domestication of the East Asian M.m. molossinus “fancy” mouse through which the Y chromosome was first introduced into the classical inbred laboratory mouse strains. Despite this stability, at least three unequal genetic exchange events have altered Ymin HOR unit length and sequence structure since divergence of the ancestral Mus musculus subspecies around 900,000 yr ago, with major turnover of the HOR arrays driving rapid divergence of sequence and higher-order structure at the mouse Y centromere. A comparative sequence analysis between the human and chimpanzee centromeres indicates a similar rapid divergence of the primate Y centromere. Our data point to a unique DNA sequence and organizational architecture for the mouse Y centromere that has evolved independently of all other mouse centromeres.
The centromere is a specialized structure that plays a fundamental role in the faithful segregation and transmission of eukaryotic chromosomes during cell division (Choo 1997). In house mouse, Mus musculus, the centromere consists of two highly conserved, tandemly repeated sequences known as minor and major satellite DNA. Minor satellite DNA comprises an AT-rich, 120-bp monomer that occupies 300–600 kb of the terminal region of all mouse telocentric (Kipling et al. 1991; Kalitsis et al. 2006) (single-armed) chromosomes and is the site of kinetochore formation and spindle microtubule attachment (Wong and Rattner 1988; Broccoli et al. 1990). Major satellite DNA is a more abundant, 234-bp tandem repeat (Horz and Altenburger 1981) that resides adjacent to a minor satellite and has a role in heterochromatin formation and sister chromatid cohesion (Guenatri et al. 2004). Neither of these satellite sequences has been identified at the centromere of the morphologically distinct acrocentric Y chromosome (Broccoli et al. 1990), which has a very small short arm that distinguishes it from the telocentric autosomes and X chromosome. The high degree of minor and major satellite sequence conservation that exists across the telocentric domain of all mouse telocentric chromosomes argues strongly for frequent recombinational exchanges between nonhomologous chromosomes driving sequence homogenization at mouse centromeres (Vissel and Choo 1989; Kalitsis et al. 2006). Lack of homology at the Y centromere suggests it has been excluded from such recombinational events, but an absence of sequence information precludes a comparative analysis and therefore a more complete understanding of centromere biology and chromosome evolution in mouse. Here, we identify a fully sequenced BAC clone from the male-specific RPCI-24 mouse (C57BL/6J) genomic DNA library that enables the complete molecular characterization of the mouse Y centromere, and comparative analyses with the mouse telocentric centromeres and the primate Y centromeres.
Results and Discussion
Identification of the putative mouse Y centromere DNA (Ymin)
Although no genomic positional information exists for the mouse Y centromere, combined immunofluorescence and fluorescence in situ hybridization (immuno-FISH) localizes it to a region at least 3–5 Mb from the short arm telomere. This region lies beyond the multicopy Rbmy1a1 gene, which is currently the most proximally annotated gene to the unmapped Y centromere (Fig. 1A–C). Using a bioinformatics approach, we identified a 90-kb Mus musculus sequence in a fully sequenced BAC clone, RP24-110P17, as the putative Y centromere DNA as described in Methods. This DNA contains an AT-rich (68.9%) tandem repeat with distant homology (76.8%) to mouse minor satellite (Table 1). Like minor satellite, the AT residues are asymmetrically distributed between the two DNA strands, with adenine comprising 46.6% of all bases in the A-rich strand in a ratio of 2.1 adenines to 1.0 thymine. We designated this novel satellite repeat Ymin (Y minor satellite), confirmed its specificity for the Y centromere, and obtained evidence that BAC 110P17 harbors the entire C57BL/6J Ymin array, thereby enabling complete molecular characterization of the mouse Y centromere.
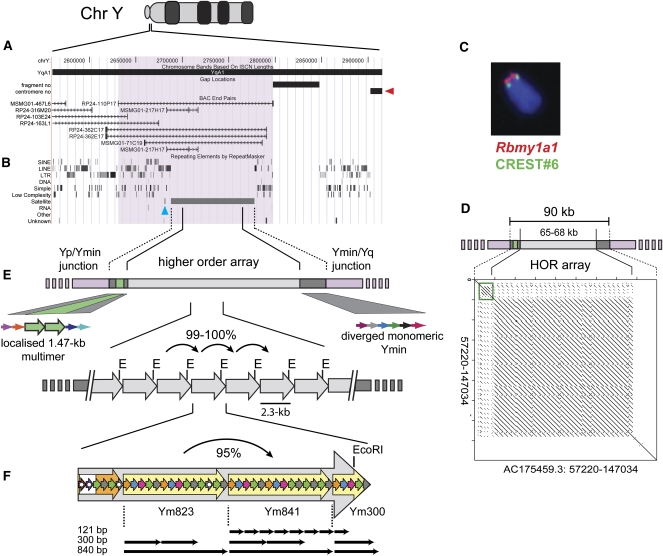
Figure 1.
Structural and sequence map spanning the C57BL/6J mouse Y centromere. (A) UCSC mouse Genome Browser view (July 2007, Build 37 assembly) at the unmapped Y centromere. BAC 110P17 (coverage highlighted mauve) is the most proximally annotated BAC to the unanchored Y centromere (arbitrarily indicated at right, red arrowhead). (B) Repeat elements identified by RepeatMasker show Ymin as a 90-kb tract of uninterrupted satellite DNA except for a single, mouse-specific LINE1 element (L1Md_F) that isolates ∼620 bp of Ymin (blue arrowhead) at the 5′ edge of the array. The most closely annotated gene, a multicopy RNA-binding motif protein (Rbmy1a1), lies ∼290 kb on the short arm-side of the Y centromere, as shown in (C) by immuno-FISH using Rbmy1a1 DNA FISH probe (red) and CREST#6 anti-centromere antibody (green). (D) Self-alignment dot-plot of Ymin reveals a 65–68-kb tandem array of 2.3-kb HOR units flanked by 10–12 kb of diverged, localized multimeric (green box) and monomeric Ymin. (E) HOR periodicity of 2.3 kb is also indicated by regularly-spaced restriction sites (e.g., E, EcoRI; see Supplemental Fig. S1) within the homogeneous Ymin array, with individual units sharing 99%–100% sequence identity. (F) Each canonical HOR unit comprises 2.4 copies of an ∼840-bp periodicity subunit repeat (large yellow arrows designated Ym823, Ym841, and Ym300; see Supplemental Fig. S2) and a less uniform region lacking repeat periodicity or with 60-bp periodicity (large composite white/orange arrow, respectively). Highly diverged ∼60–61-bp monomers (small colored arrows representing monomers with similar homology) or 121-bp monomers are arranged in HOR subunits with periodicities of ∼300–840 bp (examples shown by black arrows). No subunit repeat extends the full length of the HOR unit domain (Supplemental Table S3A). Monomers harboring deletions or other rearrangement are highlighted by solid white circles.
Table 1.
Summary analysis of BAC RP24-110P17 (GenBank accession no. AC175459)
aC57BL/6J carries a M.m. musculus Y chromosome of Asian (M.m. molossinus) origin on a predominantly M.m. domesticus genetic background.
bIndelA position calculated from EcoRI consensus start site.
cMonomers within 5% range of 60 bp (57–63 bp) used in calculation. Includes all monomers in HOR.
dMonomers within 5% range of 120 bp (114–126 bp) used in calculation. Includes all monomers in HOR.
eCENPB box motif is found in a subset of mouse minor satellite monomers and a subset of human HOR α-satellite monomers at all centromeres except the Y.
fCpG methylation sensitive in genomic DNA.
Ymin has higher-order repeat sequence organization
Bioinformatics analysis found Ymin to be both structurally and organizationally distinct from minor satellite. Rather than housing a simple reiterated monomeric repeat, self-alignment dot-plot (Rice et al. 2000; Brodie et al. 2004) and in silico restriction analyses (Vincze et al. 2003) determined the 90-kb array to consist of multiple copies of a 2.3-kb higher-order repeat (HOR) unit (Fig. 1; Supplemental Fig. S1). The HOR units are homogeneous (>99%–100% pairwise identities) (Supplemental Table S1), tandemly repeated across the core 65–68 kb of the array and flanked by 10–12 kb of diverged multimeric and/or monomeric Ymin that lacks higher-order periodicity. The same sequence directionality is maintained across the length of the array. A small region of localized sequence homogenization at the 5′ edge of the HOR array comprises 2.8 copies of a truncated 1.47-kb multimer (Fig. 1D,E). Complete copies of this multimer are 100% identical, which indicates a recent homogenization event, but are diverged (97.5% identity) from colinear segments of canonical Ymin at the core of the array. Such regions of localized sequence homogenization are postulated to represent the very first stages in the possible evolution of new HOR arrays (Rudd and Willard 2004).
The Ymin HOR array is devoid of transposable elements as determined by RepeatMasker analysis. Only a single, mouse-specific LINE1 element (L1Md_F) punctuates the 5′ edge of the flanking monomeric repeats, and isolates ∼620 bp of monomeric Ymin from the otherwise uninterrupted array (Fig. 1B). An absence of interspersed repetitive elements is a characteristic feature of homogeneous HOR arrays, and reflects both efficient mechanisms of sequence homogenization that operate within the core of the arrays (Smith 1976), as well as their recent evolution (Alexandrov et al. 2001; Schueler et al. 2005). Beyond the Ymin boundary, transposable elements and other repetitive sequences are abundant (Fig. 1B).
Ymin monomer structure is unusually complex
The multimeric HOR α-satellite arrays found at human centromeres are characterized by substantial intermonomer sequence divergence. The monomers within the HOR units typically share about 65%–90% sequence identity, although little sequence divergence exists between the HOR units of a particular array (Waye and Willard 1985; Willard 1991). This contrasts with minor satellite where monomers share high (>95%) pairwise identity (Kalitsis et al. 2006), but lack HOR unit structure (see below). To assess the sequence substructure at the mouse Y centromere we examined the canonical Ymin HOR unit with sequence-alignment (Larkin et al. 2007) and tandem-repeat-finding tools (Gelfand et al. 2007) and found its structure to be remarkably complex (Table 1; Fig. 1; Supplemental Table S2A). The HOR unit consists of an amalgam of highly diverged (<70% mean pairwise identity) monomers with a periodicity of 60–61 or 121 bp. The majority of the monomers form progressively larger and less diverged repeating HOR subunits, the largest comprising 2.4 copies of an 840-bp periodicity repeat (with 95% pairwise identity) that spans the greater part of the HOR unit domain (Fig. 1F; Supplemental Fig. S2). The remaining HOR sequence (∼330 bp) is less uniform, and contains several monomers harboring deletions and/or other rearrangement. This highly diverged and irregular structure is dramatically depicted by using a self-alignment dot plot, and is seen to lack the uniformity of the homogeneous minor satellite, and exhibit greater sequence divergence and organizational complexity than the monomers that comprise the human Y centromere HOR α-satellite (Supplemental Fig. S3; Tyler-Smith and Brown 1987). Despite this complexity, the mouse Ymin data are typical of HOR unit structure, with high intermonomer sequence divergence within the HOR unit, but low sequence divergence between HOR units.
In summary, our bioinformatics analysis has shown that the Y centromere DNA of mouse is organized as a chromosome specific, multimeric HOR array. This sequence organization is best documented at the human centromeres (e.g., Willard 1991; Schueler et al. 2001; Rudd et al. 2003), but has not previously been observed in mouse. The mouse telocentric minor satellite DNA exhibits no evidence for HOR unit sequence organization, either by restriction enzyme digest (Pietras et al. 1983; Wong and Rattner 1988; Kipling et al. 1991), or by sequence or computational analysis (Kalitsis et al. 2006; MD Pertile, AN Graham, KHA Choo, and P Kalitsis, unpubl.). Furthermore, its sequence is highly conserved across the telocentric domain of the mouse telocentric chromosomes, including the X chromosome. This very high level of sequence conservation supports a mechanism of frequent recombinational exchanges between nonhomologous chromosomes causing extensive sequence homogenization at mouse centromeres (Kalitsis et al. 2006). In contrast, the unique multimeric HOR unit organization and chromosome-specific sequence present at the mouse Y centromere argues for an intrachromosomal mode of sequence homogenization (Smith 1976) driving Y centromere evolution in isolation of the frequent interchromosomal recombinational exchanges that have characterized minor satellite evolution at other mouse centromeres. In this respect the mouse Y centromere sequence architecture has more in common with the human centromeres than it does other mouse centromeres.
PCR amplification of Ymin differentiates the Y centromeres of M.m. musculus and M.m. domesticus
PCR amplification of genomic DNA from male and female inbred mice confirmed Ymin as a male-specific satellite repeat (Fig. 2A). Moreover, this assay differentiated the Y centromeres of M.m. musculus (0.82-kb Ymin PCR product) and its closely related subspecies M.m. domesticus (1.6 kb product), providing evidence for sequence divergence at the mouse Y centromere. Wild-derived mice of M.m. musculus, M.m. castaneus, and M.m. molossinus origin all exhibit a “musculus-type” Y centromere by PCR amplification (Supplemental Fig. S4; Supplemental Table S3), consistent with phylogenetic and SNP resequencing data that support M.m. musculus and M.m. castaneus being sister subspecies (Prager et al. 1998; Yang et al. 2007) with more recent extensive hybridization between the two leading to the M.m. molossinus hybrid in Japan (Yonekawa et al. 1986, 1988). A majority of classical inbred laboratory mouse strains, including C57BL/6J, carry the M.m. molossinus Y chromosome, introduced through East Asian “fancy” mice early last century (Nagamine et al. 1992; Abe et al. 2004), while others carry a M.m. domesticus Y chromosome of Western European (or North American) origin (Tucker et al. 1989; Atchley and Fitch 1993). The ability of the Ymin PCR assay to accurately distinguish the Y centromeres of these mice reflects a fundamental difference in their Ymin HOR unit sequence structure (Supplemental Fig. S4; see below).
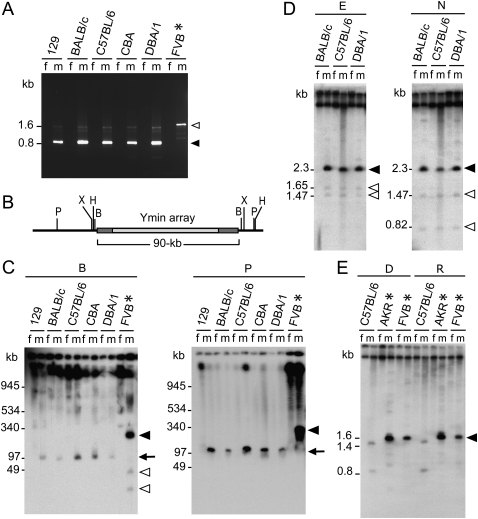
Figure 2.
Molecular characterization of Ymin in inbred mice. (A) Ymin PCR assay amplifies male (m) but not female (f) paired genomic DNA samples and differentiates the Y centromeres of M.m. musculus (molossinus) (0.82 kb) and M.m. domesticus (1.6 kb) (also see Supplemental Fig. S4). (B) Restriction map of Ymin domain showing enzymes BamHI (B), HindIII (H), PacI (P), and XbaI (X) predicted to release the 90-kb Ymin array. (C) PFGE Southern hybridization of genomic DNAs probed with Ymin exhibits male-specific fragment lengths of predicted size (arrow) in five strains carrying a M.m. molossinus Y chromosome after digestion with enzymes BamHI (90.7 kb expected) and PacI (124 kb expected) (HindIII and XbaI data not shown; also see Supplemental Fig. S6). Strain FVB (asterisk), carries a M.m. domesticus Y chromosome with an array length of 325–350 kb (sum total of fragments). Additional, smaller fragments (open arrowheads) produced by enzymes BamHI (and HindIII data not shown) suggests increased sequence complexity at the flanking edges of the larger M.m. domesticus Ymin array (solid arrowhead). (D) Short-range Southern analysis of Ymin in the same five M.m. molossinus strains (129 and CBA data not shown) following EcoRI (E) and NcoI (N) digestion confirms a HOR unit length of 2.3 kb (solid arrowhead). Predicted rare variant fragment lengths of 1.47 and 1.65 kb (EcoRI) and 0.82 and 1.47 kb (NcoI) are seen as faint hybridization bands (open arrowheads). (E) Mice carrying a M.m. domesticus Y chromosome (AKR and FVB, *) reveal a Ymin HOR unit length of 1.6 kb following digestion of genomic DNA with DraI (D), RsaI (R), and TaqI (data not shown). The C57BL/6 (molossinus) Ymin HOR cleaves twice, as predicted by BAC 110P17 sequence data.
Genomic restriction mapping supports the Y centromere sequence structure as predicted by BAC 110P17
We verified the gross sequence integrity of BAC RP24-110P17 by restriction mapping using pulsed-field gel electrophoresis (PFGE). Annotation of this clone on the UCSC July 2007 (Build 37) assembly of the mouse genome browser enabled us to identify an independent, partly overlapping, end-sequenced BAC clone (RP24-362E17) that was also predicted to contain the C57BL/6J Ymin array (see Fig. 1A). Southern hybridization analysis of PFGE fractionated DNA from both clones, run in parallel, and probed with Ymin (pYmin2.3a), verified the predicted 90-kb Ymin array length (Supplemental Fig. S5A,C). Furthermore, enzymes cutting within the Ymin array (EcoRI and NcoI) produced multiple copies of the 2.3-kb HOR unit, together with flanking fragments, at a size predicted by the 110P17 BAC sequence data (Supplemental Fig. S5C). At the resolution of PFGE, the apparently identical Ymin restriction fragment lengths produced by these independent clones provide evidence that the BAC insert DNA is not chimeric or otherwise rearranged relative to the genomic state.
We then used the BAC sequence data for long-range mapping of Ymin in mouse genomic DNA. PFGE fractionation of genomic DNA, Southern blotted and probed with Ymin, confirmed an array length of ∼90 kb in C57BL/6J (Fig. 2C; Supplemental Fig. S5B,D) and in four other inbred strains (129, BALB/c, CBA, DBA/1) known to carry a M.m. molossinus Y chromosome (Fig. 2C; Tucker et al. 1992; Atchley and Fitch 1993; Frazer et al. 2007; Yang et al. 2007). Short-range mapping confirmed a HOR unit length of 2.3 kb in the same strains using enzymes predicted to release the HOR, as well as confirming variant restriction fragment lengths as predicted by the BAC sequence data (Fig. 2D; data not shown). Taken together, these data indicate the overall sequence integrity of BAC 110P17 and provide compelling evidence the entire C57BL/6J Ymin array is present within this BAC clone. They also provide evidence that the unusually small 90-kb Ymin array is not peculiar to the C57BL/6J strain.
Kinetochore specific protein CENPA colocalizes with Ymin and is enriched within multimeric Ymin DNA
Minor satellite DNA is found at all mouse centromeres except the Y (Fig. 3A). To investigate an association between Ymin DNA and the functional Y centromere we performed immuno-FISH and chromatin immunoprecipitation (ChIP) array experiments. Immuno-FISH analysis of male 129/1 mouse ES cell metaphase chromosomes demonstrated the precise colocalization of anti-centromere autoserum (CREST#6) and anti-kinetochore (anti-mouse CENPA) antibody signals with Ymin DNA FISH probe (Fig. 3B,C). Furthermore, this precise colocalization persisted in interphase cells (data not shown), providing firm evidence that Ymin is the Y centromere sequence of mouse. Immuno-FISH analysis was also performed on stretched interphase chromatin associated with the telocentric minor satellite DNA and the Ymin DNA. The kinetochore-specific histone H3 variant CENPA was found to consistently occupy a subdomain of stretched telocentric minor satellite chromatin (Fig. 3D), confirming the kinetochore forms within a subset of this DNA (Kalitsis et al. 2006) and agreeing with a similar CENPA occupancy of a subset of α-satellite DNA at the human centromeres (Warburton et al. 1997; Blower et al. 2002). At the Y centromere, stretched Ymin chromatin continued to tightly colocalize with CENPA, further demonstrating its close association with the Y chromosome kinetochore (Fig. 3E,F). At the resolution and sensitivity of our stretched chromatin assay, CENPA occupied most, if not all of the Ymin domain, although the degree of chromatin stretching was consistently less than that obtained for the stretched minor satellite chromatin (Fig. 3F).
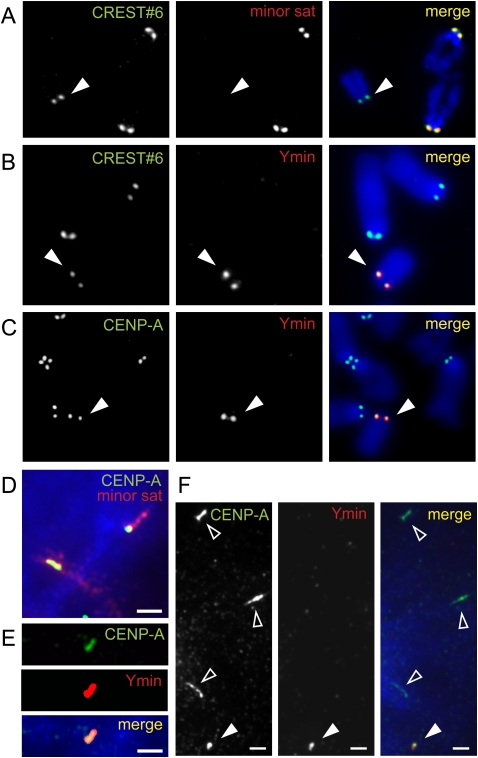
Figure 3.
Ymin colocalizes with kinetochore protein CENPA in 129/1 mouse embryonic stem (ES) cells using combined immuno-FISH. (A) Anti-centromere autoserum CREST#6 colocalizes with minor satellite DNA at all mouse telocentric centromeres, but not at the centromere of the acrocentric Y (solid arrowhead). (B,C) Ymin DNA FISH probe is highly specific for the Y centromere, showing precise colocalization with anti-centromere autoserum CREST#6 and anti-kinetochore anti-mouse CENPA antibody, respectively. (D) CENPA (green/yellow) consistently occupies a subdomain of the telocentric minor satellite DNA (red) on stretched interphase chromatin. (E) Stretched Ymin chromatin shows direct overlap of CENPA immunofluorescence signal and Ymin FISH probe. (F) Stretched Ymin chromatin (solid arrowhead) typically displays less stretching compared to other stretched telocentric centromeres (open arrowheads) by indirect immunofluorescence using antibodies to CENPA. D–F, scale bars, 2 μm.
In order to more precisely investigate the extent of CENPA distribution across the Y centromere region we performed anti-mouse CENPA ChIP array experiments on male 129/1 ES cells, containing a M.m. molossinus Y chromosome (Fig. 4). Flanking probes, Yp1, Yp2, and Yq1 showed no CENPA enrichment between input and CENPA bound chromatin. In contrast, the Ymin HOR unit probe showed a threefold increase in CENPA binding, which was comparable to the centromeric minor satellite control. The non-HOR or monomeric form of Ymin (Ymin-mon) exhibited a relatively small enrichment. These data indicate that CENPA predominantly occupies the multimeric form of the Ymin DNA.
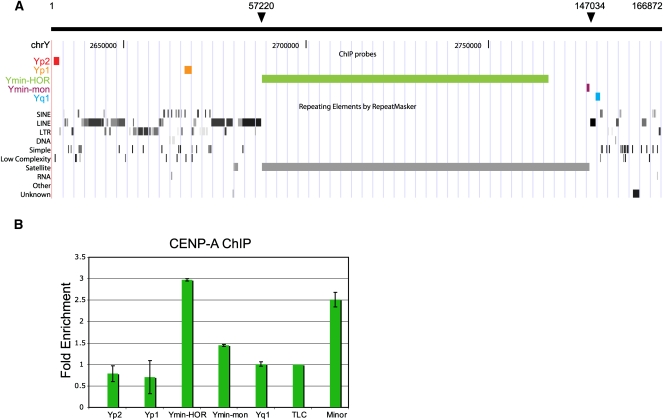
Figure 4.
Distribution of CENPA across the Y pericentromeric domain. (A) The positions of dot blot array probes are shown against the UCSC mouse Genome Browser view (Build 37, July 2007): Yp2 and Yp1, unique DNA located on the proximal p-side of the centromere; Ymin-HOR, recognizing ∼65–79 kb of Y centromere HOR/multimeric Ymin DNA; Ymin-mon, monomeric form of Ymin at the Ymin/euchromatin boundary on the q-side of the centromere; Yq1, homologous to the Yq-arm repeat, pY353/B (Bradbury et al. 1990), and the telocentric satellite repeat, TLC. Nucleotide coordinates from BAC RP24-110P17 are annotated at the top; solid arrows mark boundaries of the Ymin array. (B) CENPA enrichment is displayed as a ratio of CENPA bound/input signal averaged over three experiments, error bars are shown as standard error. Four contiguous monomers of minor satellite DNA were used as a positive centromere DNA control (Kalitsis et al. 2003). Data are normalized against the TLC satellite repeat.
Rapid divergence of sequence and HOR unit structure at the mouse Y centromere
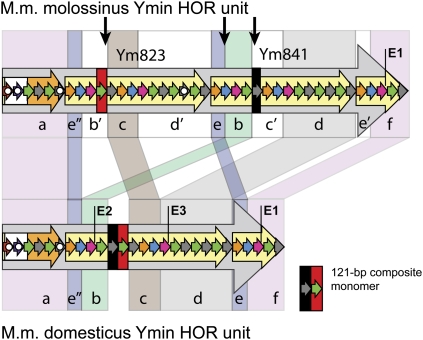
To further investigate the conservation of Ymin among closely related subspecies of mice, we performed short-range restriction mapping in three inbred strains (AKR, FVB, and SJL) known to carry a M.m. domesticus Y chromosome (Tucker et al. 1992; Atchley and Fitch 1993; Frazer et al. 2007; Yang et al. 2007) and found each to have a shortened Ymin HOR unit length of 1.6 kb, with no evidence for a 2.3-kb unit. We sequenced the HOR units from strains AKR and FVB and determined an actual unit length of 1597 bp, some 700 bp shorter than the M.m. molossinus Ymin HOR, and consistent with all short-range mapping data (Fig. 2E; Supplemental Fig. S6). Long-range mapping indicated strains FVB and SJL have an array length of at least 325–350 kb, substantially larger than the molossinus Ymin array (Fig. 2C; data not shown). We then performed comparative sequence analyses to investigate the relationship between the M.m. molossinus and M.m. domesticus HOR units. We found the domesticus unit lacks the equivalent of a large (823 bp) internal subunit repeat present in the molossinus HOR, replaced by a 121-bp composite monomer (Fig. 5; Supplemental Fig. S7). The two HOR units maintain an essentially colinear arrangement of DNA, albeit in a rearranged (shuffled) form. We estimate that at least three unequal genetic exchange events (presumably unequal sister chromatid exchanges) are required to reconcile key differences between the two HOR units (Fig. 5; Supplemental Fig. S8). Given the Mus musculus subspecies shared a common ancestor no more than 900,000 yr ago (Boursot et al. 1996; Geraldes et al. 2008), these unequal exchange events have been accompanied by an extremely rapid divergence of sequence and HOR unit structure at the mouse Y centromere, with orthologous M.m. molossinus and M.m. domesticus HOR monomers sharing just 95.6 ± 3.5% sequence identity. Of particular interest is the apparent lack of evidence for a common-length ancestral HOR unit in any of the mouse strains tested, which one might expect to have persisted given their recent common ancestry. Furthermore, the availability of sequence data from the M.m. molossinus Y centromere provides even greater confidence that a small number of the domesticus-type HOR units have not persisted within the C57BL/6J Y centromere array. This suggests an extremely rapid turnover of the ancestral HOR arrays in one or both subspecies.
Figure 5.
Comparative sequence analysis of M.m. molossinus (2.3 kb) and M.m. domesticus (1.6 kb) Ymin HOR units. Regions of highest sequence homology are grouped (a–f) and shaded accordingly. Landmark EcoRI restriction sites are labeled E1–E3; monomers harboring deletions or other rearrangements are highlighted by solid white circles (see Fig. 1F for full description). The M.m. domesticus HOR unit contains a 121-bp composite monomer that is absent from the equivalent regions of the M.m. molossinus Ym823 and Ym841 subunit repeats. Monomers with highest sequence identity to this composite monomer are highlighted in the molossinus HOR using red and black bars, and border regions of putative unequal exchange (vertical black arrows). Differences between the two HOR unit structures can be explained by a series of unequal sister chromatid exchange events occurring in the ancestral HOR arrays, followed by amplification and fixation of the present day canonical HOR units in each subspecies. At least three unequal exchange events are required to reconcile key differences between the two HOR units (for proposed model of unequal exchange, see Supplemental Fig. S8).
Comparative analysis indicates a similar rapid divergence of the primate Y centromere
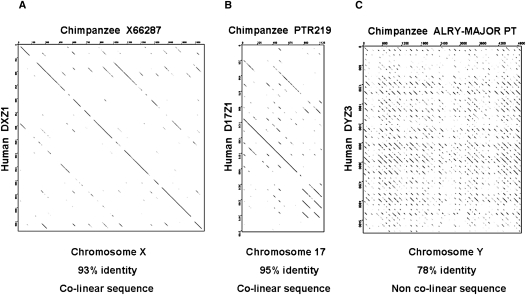
The above data exhibit an extremely rapid divergence of the mouse Y-centromere HOR. A comparison of human (DYZ3) and chimpanzee (ALRY-MAJOR_PT) orthologous sequences demonstrates a similar rapid divergence of the primate Y centromere (Fig. 6). We could find no evidence of a conserved colinear relationship of higher-order α-satellite DNA, accompanied instead by a best local contiguous alignment of 78.1% across the entire chimp Y centromere HOR. In contrast, orthologous human and chimpanzee X centromere higher-order α-satellite units maintain their structural (colinear) integrity and are 93% identical (Laursen et al. 1992) following at least five million years of evolutionary divergence. Similarly, the human chromosome 17 centromere (D17Z1) and its chimp ortholog (PTR219) maintain 95 ± 0.8% sequence identity (Rudd et al. 2006), and a conserved colinear relationship of α-satellite DNA for the four chimp PTR219 HOR monomers available for analysis. The greater sequence divergence and lack of sequence colinearity between the human and chimpanzee Y centromeres provides evidence for their accelerated divergence relative to other primate centromeres. This finding mirrors the greater sequence divergence that exists between the human and chimpanzee Y chromosomes when compared to the remainder of their genomes (Kuroki et al. 2006), and suggests that the accelerated evolutionary rate of the Y chromosome is also impacting the rate of Y centromere evolution.
Figure 6.
Comparative analysis of human and chimpanzee centromere sequences. (A) JDotter alignment of human chromosome X HOR unit (DXZ1; accession X02418) plotted against the chimpanzee orthologous sequence (accession no. X66287). HOR units exhibit a high degree of sequence homology and maintain sequence colinearity across their 2.0-kb lengths. (B) Human chromosome 17 HOR unit (D17Z1; accession no. M13882) plotted against an available segment of chimpanzee orthologous sequence (PTR219; accession no. F L08574) exhibits substantial sequence homology and conservation of sequence colinearity. (C) Increased sequence divergence and an absence of sequence colinearity is exhibited when the human Y-centromere HOR (DYZ3; accession no. AF522078Y) is plotted against the chimpanzee orthologous sequence (Repbase ALRY-MAJOR_PT). JDotter plots were created using a 50-bp sliding window with grayscale set at identical parameters.
The Y chromosome and Y centromere evolution
Centromeric satellite repeat families are subject to a number of genomic mechanisms that govern the evolution of tandem repeat arrays including mutation, deletion, transposition, unequal chromatid exchange, and sequence conversion (e.g., Smith 1976; Dover 1982; Charlesworth et al. 1994) (see below). However, the Y centromere must also come under the influence of evolutionary forces that act on the Y chromosome as a whole. In particular, the smaller effective population size of the Y chromosome (one quarter that of the autosomes) makes it more susceptible to genetic drift, increasing the probability that neutral sequence variants will be fixed (or lost) by chance (Kimura 1968). Furthermore, an absence of recombination makes the Y susceptible to selection as a single unit. Positive selection on a Y haplotype (selective sweep) will increase its spread in a population, causing the loss of other variant haplotypes, particularly if selection is strong. Likewise, negative or purifying selection also reduces variation. Other factors including migration (founder effects), population bottlenecks, and mating behavior (e.g., dominant males) will also reduce the effective population size of the Y chromosome, increasing its susceptibility to drift, and accelerating its divergence (Jobling and Tyler-Smith 1995; Hurles and Jobling 2001). On an evolutionary scale, these forces are likely to impact the rate of Y centromere evolution, as any newly evolved or existing variant Y centromere haplotype (e.g., a variant length HOR unit with the potential for further homogenization) will be linked to the evolutionary fate of that particular Y chromosome. Other factors, including high mutational rates, might also play a role in Y centromere divergence, with extensive structural changes documented for the Y chromosome in humans (Repping et al. 2006) and chimpanzee (Kuroki et al. 2006) pointing to a relocation of the human Y centromere, possibly in association with neocentromere formation (Tyler-Smith et al. 2006). It is perhaps not surprising then that the Y chromosome of many mammals, including those of mouse and humans, exhibit greater divergence of their Y centromere sequence in comparison to their other centromeres.
Evolution of the mouse Y centromere HOR array
The turnover of sequence variants that drive the rapid evolution of higher-order arrays can result from both stochastic and directional changes (e.g., unequal crossing-over, sequence conversion, duplicative transposition) in a process known as molecular drive (Dover 1982). Such genomic turnover mechanisms efficiently introduce and homogenize new sequence variants into tandem repeat arrays. We found evidence for the putative involvement of sequence conversion within the assembled C57BL/6J Ymin array, with at least 200 bp of Ymin DNA undergoing conversion to produce a “variant” HOR unit (Supplemental Fig. S9). The unit contains an additional NcoI restriction site, as well as several other sequence polymorphisms (Fig. 2D; Supplemental Fig. S9). A small number of these variant units have been dispersed throughout the HOR array by unequal sister-chromatid exchange or some other duplicating mechanism (Supplemental Fig. S9C), and might be considered to be in a “transient” state of fixation.
While sequence conversion may act locally to homogenize sequence variants, unequal chromatid exchange is the mechanism most frequently invoked to explain the evolution and ongoing sequence homogenization that occurs within tandem repeat arrays (Smith 1976; Willard 1991; Schueler et al. 2001; Henikoff 2002). Evidence for this mechanism is pervasive at the C57BL/6J mouse Y centromere, with the Ymin HOR unit structure exhibiting considerable evidence for unequal exchange between locally misaligned sister chromatids. Such exchange events are postulated to homogenize small tandem repeats into larger units, which then expand more efficiently into even larger tandem arrays. The internal sequence organization of Ymin is consistent with this process, with progressively larger and less diverged (evolutionarily more recent) repeating HOR subunits culminating in the fixation of the canonical Ymin HOR, which itself has become an efficient unit for sequence homogenization.
Unequal sister chromatid exchange between out-of-register HOR units is also the likely mechanism that has precipitated the divergence of the molossinus and domesticus Ymin HOR arrays. An out-of-register exchange will partly duplicate or delete the HOR unit, creating a newly sized HOR with the potential to be eliminated or further homogenized within the HOR array (Tyler-Smith and Brown 1987; Warburton et al. 1993; Schueler et al. 2005). In mouse, these genetic exchange events have occurred at the Y centromere in the absence of recombinational exchange with a paired homolog, and without apparent recent transposition of satellite DNA from elsewhere in the genome. This clearly demonstrates that HOR sequence organization can evolve from a haploid state (Smith 1976), having been initiated and maintained at a single locus in isolation of comparable sequence architecture at other mouse centromeres. But how frequently do such exchange events occur during the evolution of HOR arrays?
Evidence for recent sequence stability at the mouse Y centromere
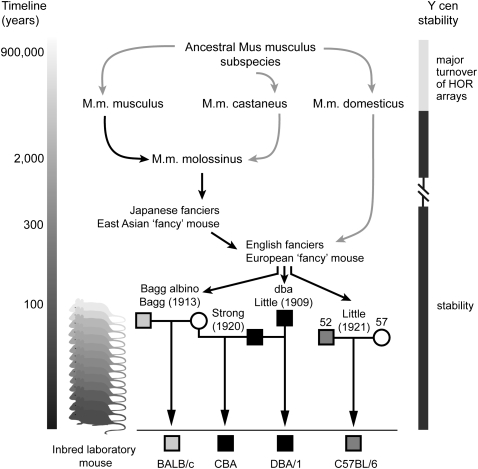
While HOR array-length polymorphisms are common at human centromeres, no evidence of array-length change has been observed in three-generation families, suggesting relative meiotic and mitotic stability at those centromeres studied, including the Y (Wevrick and Willard 1989). In mouse, direct patrilineal inheritance of the Y chromosome through inbred males provides a unique opportunity to study Y centromere array stability over many more successive generations. The DBA/1 and CBA mice used in this study carry an identical M.m. molossinus Y chromosome inherited from a common ancestral male earlier last century (Atchley and Fitch 1993) (Fig. 7). The apparently identical Ymin mapping data obtained for these mice provides direct evidence for the gross sequence stability of their Ymin DNA structure spanning almost 90 yr and at least 167 (DBA/1) and 175 (CBA) inbred generations. Furthermore, C57BL/6J, inbred for over 200 generations and carrying a phylogenetically distinct M.m. molossinus Y chromosome from those above (Tucker et al. 1992; Frazer et al. 2007), exhibits the same restriction fragment-length data, as do BALB/c (and 129 strains). These data reflect a total of 734 recorded inbred generations without detectable change to gross Ymin DNA sequence structure. Moreover, the apparently identical mapping results for three phylogenetically distinct M.m. molossinus Y chromosomes suggests stability of the Ymin array structure over a much longer time period, at least since prior to the domestication of the East Asian “fancy” mice that contributed their Y chromosomes to the first classical inbred strains; a period of several centuries to possibly more than 1000 years. This sequence stability belies the rapid turnover of Ymin higher-order arrays on an evolutionary scale that is apparent at the M.m. molossinus and M.m. domesticus Y centromeres, and argues that unequal intrachromosomal exchange leading to detectable changes in Ymin array structure is a rare event. Alternatively, these exchange events may occur more regularly, but in a small, stepwise fashion below the resolution of PFGE.
Figure 7.
Origins of inbred laboratory mice and evidence for sequence stability at the mouse Y centromere. Present day Mus musculus subspecies diverged from a common ancestor around 900,000 yr ago. Recent extensive hybridization in Japan between M.m. musculus and M.m. castaneus subspecies has given rise to the M.m. molossinus hybrid, which carries the M.m. musculus Y chromosome and M.m. castaneus mitochondrial DNA (M.m. musculus Y transmission depicted by black arrows). Many classical inbred laboratory mouse strains have a M.m. musculus (molossinus) Y chromosome following inbreeding of pet “fancy mice” of East Asian and European ancestry by mouse geneticists (e.g., Bagg, Strong, Little) early last century. The origin of four inbred laboratory strains carrying three phylogenetically distinct molossinus Y chromosomes is shown: light gray box, male BALB/c; black box; male DBA/1 and CBA; dark gray box, male C57BL/6J. Strains DBA/1 and CBA carry the same ancestral Y chromosome inherited from a dba male (the first inbred strain established by Clarence C. Little in 1909; cascading mice at left representing clonal inheritance). At the resolution of PFGE, these strains carry an identical 90-kb Ymin array length, suggesting stability of the same Y centromere array over at least 90 yr and 167 (DBA/1) and 175 (CBA) inbred generations. Strains C57BL/6J, BALB/c, and 129 (data not shown) also show 90-kb array lengths, increasing total recorded inbred generations to 734 without a detectable change to the Y centromere array length. These data suggest changes to the array occur below the resolution of PFGE (possibly in a small, stepwise fashion) or they occur very rarely. Despite this apparent stability, rapid divergence of Ymin DNA sequence and HOR unit structure is evident at the Y centromeres of M.m. molossinus and M.m. domesticus mice, driven by a major turnover of the Ymin HOR arrays (see main text for details).
Numerous phylogenetic studies have identified two main Y chromosome lineages in house mouse, one involving M.m. musculus (and the closely related M.m. castaneus) and the other M.m. domesticus (e.g., Tucker et al. 1989; Boissinot and Boursot 1997; Prager et al. 1998). Although these subspecies have evolved very recently from a common ancestor, their Y chromosomes have rapidly accumulated significant differences in several satellite repeat families (Bishop et al. 1985; Nishioka and Lamothe 1986; Boursot et al. 1989; Tucker et al. 1989). Our data indicate the Y centromere satellite DNA is undergoing a similar rapid evolution, and is likely at an intermediate stage toward a more diverged state, having not yet reached the same degree of sequence divergence and lack of sequence colinearity that exists between closely related primate Y centromeres.
In conclusion, the identification and molecular characterization of the mouse Y centromere fills a significant and long-standing void in mouse centromere biology and chromosome evolution (Pardue and Gall 1970), and provides an important physical landmark for ongoing efforts aimed at completing the repeat-rich sequence of the mouse Y chromosome. Importantly, these data also provide a basis for the ongoing research of Y centromere sequence organization and satellite repeat evolution in other species of mice. The unusually compact nature of the M.m. molossinus Y centromere array and the accelerated divergence of the Y centromere DNA are testament to the ongoing challenges for evolutionary and chromosome stability faced by the recombinationally isolated Y chromosome.
Methods
Bioinformatics analysis
Fully sequenced bacterial artificial chromosome (BAC) clone RP24-110P17 (GenBank accession no. AC175459) was identified by BLAST searching (http://www.ncbi.nlm.nih.gov/) publicly available mouse genomic DNA libraries using the 3′ end sequence of BAC RP24-103E24, which at that time was the most closely annotated BAC to the unmapped Y-centromere according to Build 36, February 2006 assembly of the UCSC Mouse Genome Browser (http://genome.ucsc.edu). Repeat element characterization of BAC 110P17 was carried out by using RepeatMasker (http://repeatmasker.org). Characterization of Ymin sequence structure was determined by using Dottup (Rice et al. 2000) or JDotter (Brodie et al. 2004) for dot-plot analyses and ClustalW2 (Larkin et al. 2007) set at default parameters for computation of pairwise sequence alignments of Ymin HOR units and their component monomers. Monomers comprising the canonical C57BL/6J Ymin HOR unit were manually binned into groups of ∼60 bp and 120 bp, and all monomer lengths within a 5% range of these sizes used for pairwise alignment calculations (Table 1). Tandem Repeats Finder Database (Gelfand et al. 2007) was used to analyze the monomer structure of C57BL/6J canonical Ymin HOR (Supplemental Table S2A), mouse minor satellite (Supplemental Table S2B) and human Y HOR α-satellite (data not shown). Comparative sequence analyses of M.m. molossinus and M.m. domesticus Ymin HOR units were undertaken using BLAST (Tatusova and Madden 1999), ClustalW2 (Larkin et al. 2007), and dot-plot (Rice et al. 2000; Brodie et al. 2004) alignment tools. The consensus sequence of the chimpanzee Y centromere HOR unit, ALRY-MAJOR_PT, was obtained from Repbase (http://www.girinst.org/).
Mouse DNA and BAC clones
Lung and spleen samples were dissected from male and female 6–8-wk-old mice (Laboratory strains; Supplemental Table S3) obtained from the Walter and Eliza Hall Institute, Melbourne, Australia. Genomic DNA was extracted from lung using standard molecular techniques. High molecular weight DNA for PFGE was obtained by embedding spleen cells in agarose plugs using standard methods. Genomic DNAs from wild-derived inbred strains (Supplemental Table S3) were obtained from The Jackson Laboratory (http://www.jax.org/). BAC clones from the male specific RPCI-24 genomic DNA library were obtained from BACPAC Resource Center, Children's Hospital Oakland Research Institute (http://bacpac.chori.org/) and are derived from the tissue of a single C57BL/6J mouse.
PCR amplification of Ymin
Ymin primers were designed “back-to-back” around a consensus NcoI restriction site within canonical Ymin HOR (Supplemental Fig. S4). Forward primer, YminF 5′-CCATGGAAAGGAGTAATGCAC-3′; reverse primer, YminR 5′-TTTTTGTTGTAACTCCTTTCCATGC-3′. PCR amplification of M.m. domesticus Ymin HOR was also undertaken using forward primer, decoF1 5′-CACAGTGTAGAACACCGTAC-3′; reverse primer, decoR1 5′-CGTTTCTCATTATATATGTTTTTCTTC-3′. HotStar polymerase (QIAGEN) or Expand Long Template PCR System (Roche) were used for all PCR reactions.
Isolation of C57BL/6J (M.m.molossinus) Ymin HOR unit
BAC 110P17 DNA was purified using a QIAGEN Maxi Prep kit, 2 μg digested with 20 U of EcoRI (Roche), and the fragments separated by electrophoresis on a 0.8% agarose gel. The 2.3-kb Ymin HOR fragment was excised, purified (QIAquick Gel Extraction Kit, QIAGEN) and cloned into plasmid vector pALTER-1 (Promega). A representative positive HOR clone pYmin2.3a was used for all Southern blot hybridization and FISH experiments.
Isolation of M.m. domesticus Ymin HOR unit
Genomic DNA from strains FVB and AKR was PCR amplified using Ymin and deco1 primers and products run on a 0.9% agarose gel. The 1.6-kb HOR fragment was excised, purified (QIAquick Gel Extraction kit, QIAGEN) and cloned into pCRII-TOPO vector (Invitrogen). The M.m. domesticus Ymin HOR consensus sequence used for comparative analysis was derived from pooled data (n = 15 forward and n = 17 reverse sequence reads) as minimal sequence variation existed between the two strains. Sequencing employed ABI BigDye v3.1 dye terminator chemistry (Applied Biosystems) and was undertaken at the Australian Genome Research Facility, Melbourne, Australia.
PFGE and Southern blot hybridization
Digested BAC 110P17 DNA (200 ng) was separated by PFGE on a 0.9% agarose gel run in 0.5× TBE at 6 V/cm, 14°C, included angle of 120°, pulse time 1–30 sec for 17 h using a Bio-Rad Chef Mapper. Long-range mapping of Ymin from inbred strains involved overnight digestion of agarose plugs with 40 U of restriction enzyme. Digested plugs were subjected to PFGE as described above except a pulse time of 2–150 sec was used for 18–21 h. Gels were transferred to Hybond-N+ nylon membranes (GE Healthcare) and Southern hybridization performed using standard methodology. Ymin sequences were detected by using a 32P-labeled Ymin probe made from 20 ng of PCR-amplified pYmin2.3a. Hybridization was performed overnight at 65°C in Church buffer and filters washed the next morning in 1× SSC/0.1% SDS at 68–72°C. Short-range mapping of Ymin HOR involved overnight digestion of 2 μg of genomic DNA with 20 U of appropriate enzyme. Digested DNA was run on a 0.9% agarose gel, transferred to nylon membranes and hybridized with labeled Ymin probe as above. Additional high stringency washes, using 0.1× SSC/0.1% SDS at 70°C–72°C, were also undertaken.
FISH and immuno-FISH
129/1 Mouse embryonic stem (ES) cells were exposed to 0.1 μg/mL colcemid (GIBCO/BRL) for 1–2 h, cells trypsinized, washed in PBS and transferred to 75 mM KCl hypotonic solution for 12 min at a concentration of 1 × 105 cells/mL. Cells (200 μL) were cytospun onto glass slides and immunofluorescence carried out as described (Kalitsis et al. 2003) using human CREST#6 antiserum (du Sart et al. 1997) and anti-mouse CENPA primary antibody (Lo et al. 2001b) (also see “ChIP dot blot array” section, below). Slides for immuno-FISH were then soaked in PBT (PBS + 0.1% Tween 20) to remove coverslip, air dried, fixed in 3:1 methanol/acetic acid for 10 min, air dried again and baked at 50°C for 45 min. Two microliters of directly-labeled FISH probe was applied to the cytospun cells, the slide covered with a 10-mm circular coverslip, sealed with rubber cement, codenatured at 75°C for 3 min and hybridized overnight at 37°C in a sealed, moist chamber. The coverslip was then removed and slides washed in 0.4× SSC/0.3% NP40 at 73°C for 2 min followed by rinsing in 2× SSC at RT for 2 min. Slides were mounted in Vectorshield (Vector Laboratories) containing DAPI prior to viewing. FISH probes were directly labeled with Alexa Fluor dyes using a FISH Tag DNA labeling kit (Invitrogen) in accordance with the manufacturer's instructions. Ymin FISH probe (2 ng/μL) was prepared from pYmin2.3a, minor satellite FISH probe (1 ng/μL) from a plasmid derived from a previous study (Kalitsis et al. 2003) and Rbmy1a1 FISH probe (2 ng/μL) from BAC RP24-318N19. Images were captured using an Axio Imager microscope (Zeiss) fitted with an AxioCam MRm CCD camera and processed using AxioVision (AxioVs40 V4.6.1.0) software (Zeiss).
Stretched chromatin immuno-FISH
Stretched chromatin was prepared as described (Blower et al. 2002) with minor modifications. Briefly, 129/1 mouse ES cells without colcemid exposure were transferred to 75 mM KCl hypotonic solution at 1 × 104 cells/mL for 12 min and 200 μL was cytospun onto glass slides at 1000 rpm for 5 min. Semidried slides were lowered into lysis buffer (25 mM Tris, 0.5 M NaCl, 1% Triton X-100, 0.5 M urea) for 15 min, slowly removed, placed vertically for 1 min and then carefully lowered into fixative (4% PFA, 0.5% Tween 20 in PBS) for 10 min. Following fixation, slides were washed 3 × 15 min in PBS prior to immuno-FISH as described.
ChIP dot blot array
ChIP dot blot array was performed as described in Lo et al. (2001b) with some minor modifications. Briefly, nuclear extracts were purified from 2 × 107 male mouse 129/1 ES cells. Chromatin was digested to mainly mono- and dinucleosomes with micrococcal nuclease. An aliquot was saved for the input fraction. The remaining chromatin was immunoprecipitated with a polyclonal rabbit anti-mouse CENPA antibody raised to a 25 amino acid peptide located at the amino end (positions 6–30) (Lo et al. 2001a). The DNA was extracted from the chromatin with an equal volume of phenol:chloroform (1:1) and then ethanol precipitated and resuspended in TE buffer. DNA probes spanning the chromosome Y centromere were generated by PCR and cloned into pGEM-T Easy (Promega) and verified by sequencing. Purified insert DNA (10 ng) was dot blotted onto Hybond-N+ membrane in duplicate. Separate blots were then hybridized in Church buffer at 67°C overnight with labeled DNA from unbound and CENPA associated chromatin from three independent experiments. Filters were then washed under high stringency conditions in 1× SSC, 0.1% SDS at 67°C, and exposed onto a phospho screen. Signals were measured with a Typhoon 9400 Variable Mode Imager (GE Healthcare Life Sciences) and quantified using ImageQuant. To calculate CENPA enrichment, the CENPA bound signal was divided by the input signal and average values were calculated for the three experiments. The average bound/input ratios were normalized against the mouse telocentric satellite repeat, TLC (Kalitsis et al. 2006). This low-copy repeat (∼200 kb per diploid genome) was chosen to ensure that the dot blots were hybridized and washed at high enough stringency since the TLC repeat shares distant homology (77%) to minor satellite DNA.
Acknowledgments
We thank Emma Northrop for technical advice with ChIP, and John Mitchell and Charles Robin for helpful discussions. The sequence and positional information of BAC clone RP24-110P17 was established by the Mouse Chromosome Y Mapping Project (Jessica E. Alfoldi, Helen Skaletsky, Steve Rozen, and David C. Page) at the Whitehead Institute for Biomedical Research, Cambridge, MA in collaboration with the Washington University Genome Sequencing Center, St. Louis, MO. Research in our laboratory has been supported by the National Health and Medical Research Council of Australia (NHMRC). P.K. is an RD Wright Fellow and K.H.A.C. is a Senior Principal Research Fellow of the NHMRC. M.D.P. is the recipient of NHMRC Scholarship ID 437024.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession no. FJ465504.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092080.109.
References
- Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, Ezawa K, Saitou N, Hattori M, Sakaki Y, et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. α-Satellite DNA of primates: Old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch W. Genetic affinities of inbred mouse strains of uncertain origin. Mol Biol Evol. 1993;10:1150–1169. doi: 10.1093/oxfordjournals.molbev.a040070. [DOI] [PubMed] [Google Scholar]
- Bishop CE, Boursot P, Baron B, Bonhomme F, Hatat D. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature. 1985;315:70–72. doi: 10.1038/315070a0. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Boursot P. Discordant phylogeographic patterns between the Y chromosome and mitochondrial DNA in the house mouse: Selection on the Y chromosome? Genetics. 1997;146:1019–1034. doi: 10.1093/genetics/146.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P, Bonhomme F, Catalan J, Moriwaki K. Variations of a Y chromosome repeated sequence across subspecies of Mus musculus. Heredity. 1989;63:289–297. doi: 10.1038/hdy.1989.101. [DOI] [PubMed] [Google Scholar]
- Boursot P, Din W, Anand R, Darviche D, Dod B, Von Deimling F, Talwar GP, Bonhomme F. Origin and radiation of the house mouse: Mitochondrial DNA phylogeny. J Evol Biol. 1996;9:391–415. [Google Scholar]
- Bradbury MW, Isola LM, Gordon JW. Enzymatic amplification of a Y chromosome repeat in a single blastomere allows identification of the sex of preimplantation mouse embryos. Proc Natl Acad Sci. 1990;87:4053–4057. doi: 10.1073/pnas.87.11.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Miller OJ, Miller DA. Relationship of mouse minor satellite DNA to centromere activity. Cytogenet Cell Genet. 1990;54:182–186. doi: 10.1159/000132989. [DOI] [PubMed] [Google Scholar]
- Brodie R, Roper RL, Upton C. JDotter: A Java interface to multiple dotplots generated by dotter. Bioinformatics. 2004;20:279–281. doi: 10.1093/bioinformatics/btg406. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Choo KHA. The centromere. Oxford University Press; Oxford, New York: 1997. [Google Scholar]
- Dover G. Molecular drive: A cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KHA. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Gelfand Y, Rodriguez A, Benson G. TRDB—the Tandem Repeats Database. Nucleic Acids Res. 2007;35:D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Near the edge of a chromosome's “black hole”. Trends Genet. 2002;18:165–167. doi: 10.1016/s0168-9525(01)02622-1. [DOI] [PubMed] [Google Scholar]
- Horz W, Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981;9:683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Jobling MA. Haploid chromosomes in molecular ecology: Lessons from the human Y. Mol Ecol. 2001;10:1599–1613. doi: 10.1046/j.0962-1083.2001.01314.x. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. Fathers and sons: The Y chromosome and human evolution. Trends Genet. 1995;11:449–456. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Griffiths B, Howman E, Newson AJ, Choo KHA. Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res. 2003;11:345–357. doi: 10.1023/a:1024044008009. [DOI] [PubMed] [Google Scholar]
- Kalitsis P, Griffiths B, Choo KHA. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci. 2006;103:8786–8791. doi: 10.1073/pnas.0600250103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kipling D, Ackford HE, Taylor BA, Cooke HJ. Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics. 1991;11:235–241. doi: 10.1016/0888-7543(91)90128-2. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Toyoda A, Noguchi H, Taylor TD, Itoh T, Kim DS, Kim DW, Choi SH, Kim IC, Choi HH, et al. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat Genet. 2006;38:158–167. doi: 10.1038/ng1729. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laursen HB, Jorgensen AL, Jones C, Bak AL. Higher rate of evolution of X chromosome α-repeat DNA in human than in the great apes. EMBO J. 1992;11:2367–2372. doi: 10.1002/j.1460-2075.1992.tb05300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KHA. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001a;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KHA. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001b;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine CM, Nishioka Y, Moriwaki K, Boursot P, Bonhomme F, Lau YF. The musculus-type Y chromosome of the laboratory mouse is of Asian origin. Mamm Genome. 1992;3:84–91. doi: 10.1007/BF00431251. [DOI] [PubMed] [Google Scholar]
- Nishioka Y, Lamothe E. Isolation and characterization of a mouse Y chromosomal repetitive sequence. Genetics. 1986;113:417–432. doi: 10.1093/genetics/113.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, Gall JG. Chromosomal localization of mouse satellite DNA. Science. 1970;168:1356–1358. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Pietras DF, Bennett KL, Siracusa LD, Woodworth-Gutai M, Chapman VM, Gross KW, Kane-Haas C, Hastie ND. Construction of a small Mus musculus repetitive DNA library: Identification of a new satellite sequence in Mus musculus. Nucleic Acids Res. 1983;11:6965–6983. doi: 10.1093/nar/11.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager EM, Orrego C, Sage RD. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics. 1998;150:835–861. doi: 10.1093/genetics/150.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol. 2003;68:141–149. doi: 10.1101/sqb.2003.68.141. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF. The evolutionary dynamics of α-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L, Rocchi M, Willard HF, Green ED. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tucker PK, Lee BK, Eicher EM. Y chromosome evolution in the subgenus Mus (genus Mus) Genetics. 1989;122:169–179. doi: 10.1093/genetics/122.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Lee BK, Lundrigan BL, Eicher EM. Geographic origin of the Y chromosomes in “old” inbred strains of mice. Mamm Genome. 1992;3:254–261. doi: 10.1007/BF00292153. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Brown WR. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J Mol Biol. 1987;195:457–470. doi: 10.1016/0022-2836(87)90175-6. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Howe K, Santos FR. The rise and fall of the ape Y chromosome? Nat Genet. 2006;38:141–143. doi: 10.1038/ng0206-141. [DOI] [PubMed] [Google Scholar]
- Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KHA. Mouse major (gamma) satellite DNA is highly conserved and organized into extremely long tandem arrays: Implications for recombination between nonhomologous chromosomes. Genomics. 1989;5:407–414. doi: 10.1016/0888-7543(89)90003-7. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Waye JS, Willard HF. Nonrandom localization of recombination events in human αsatellite repeat unit variants: Implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol. 1993;13:6520–6529. doi: 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Chromosome-specific αsatellite DNA: Nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. Long-range organization of tandem arrays of α satellite DNA at the centromeres of human chromosomes: High-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF. Evolution of alpha satellite. Curr Opin Genet Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- Wong AK, Rattner JB. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yonekawa H, Gotoh O, Tagashira Y, Matsushima Y, Shi LI, Cho WS, Miyashita N, Moriwaki K. A hybrid origin of Japanese mice “Mus musculus molossinus”. Curr Top Microbiol Immunol. 1986;127:62–67. [PubMed] [Google Scholar]
- Yonekawa H, Moriwaki K, Gotoh O, Miyashita N, Matsushima Y, Shi LM, Cho WS, Zhen XL, Tagashira Y. Hybrid origin of Japanese mice “Mus musculus molossinus”: Evidence from restriction analysis of mitochondrial DNA. Mol Biol Evol. 1988;5:63–78. doi: 10.1093/oxfordjournals.molbev.a040476. [DOI] [PubMed] [Google Scholar]