Abstract
Genomic analyses have shown that adjacent genes are often coexpressed. However, it remains unclear whether the observed coexpression is a result of functional organization or a consequence of adjacent active chromatin or transcriptional read-through, which may be free of selective biases. Here, we compare temporal expression profiles of one-to-one orthologs in conserved or divergent genomic positions in two genetically distant nematode species—Caenorhabditis elegans and C. briggsae—that share a near-identical developmental program. We find, for all major patterns of temporal expression, a substantive amount of gene expression divergence. However, this divergence is not random: Genes that function in essential developmental processes show less divergence than genes whose functions are not required for viability. Coexpression of gene neighbors in either species is highly divergent in the other, in particular when the neighborhood is not conserved. Interestingly, essential genes appear to maintain their expression profiles despite changes in neighborhoods suggesting exposure to stronger selection. Our results suggest that a significant fraction of the coexpression observed among gene neighbors may be accounted for by neutral processes, and further that these may be distinguished by comparative gene expression analyses.
Gene regulation is central to embryonic developmental programs and their evolution (Carroll et al. 2001; Davidson 2006). In experimental developmental systems as diverse as sea urchins, fruit flies, and amphibians, precise temporal and spatial gene regulation is encoded in transcriptional gene networks (Davidson et al. 2002; Loose and Patient 2004; Stathopoulos and Levine 2005). Changes to these networks often lead to the evolution of the developmental processes, further emphasizing their importance (Davidson 2006; Prud'homme et al. 2007). For example, evidence suggests that regulatory evolution of Hox transcription factors are the key modifications leading to the relative difference in body plans of mouse, chicken, and python (Belting et al. 1998; Cohn and Tickle 1999; Carroll 2000). Evidence also suggests that the variation in the beak morphology of Darwin's finches results from changes in the relative expressions of BMP4 and calmodulin (Abzhanov et al. 2004, 2006; Wu et al. 2004). Such insights into gene expression regulation and its effect on development highlight the importance of studying the organization of the genome to unravel the principles by which regulation is encoded.
Analyses in yeast, plants, nematodes, fruit flies, and mammals show that chromosomal gene neighbors tend to be coexpressed (Cohen et al. 2000; Blumenthal et al. 2002; Reymond et al. 2002; Roy et al. 2002; Spellman and Rubin 2002; Hurst et al. 2002, 2004; Lercher et al. 2003; Khaitovich et al. 2004; Stolc et al. 2004; Williams and Bowles 2004; Denver et al. 2005). In Saccharomyces cerevisiae, for example, stress-induced genes tend to cluster together on chromosomes (Burhans et al. 2006), as well as transcription factors with their target genes (Janga et al. 2008). Notably, housekeeping genes tend to form gene clusters (Lercher et al. 2002). Gene clusters are conserved across genomes (Singer et al. 2005), with the distance being the best predictor (Poyatos and Hurst 2007). This may indicate a structural or functional organization to simplify regulation, as in bacterial operons. However, because a clear functional association between the gene neighbors is often not apparent, the functional nature of this coexpression has been questioned (Spellman and Rubin 2002; Yanai et al. 2006; Batada et al. 2007; Purmann et al. 2007).
Recent work has suggested that a significant degree of coexpression concomitant with physical proximity may be spurious (Batada et al. 2007). Thus, mechanisms such as chromatin remodeling and transcriptional read-through may better account for the coexpression of gene neighbors instead of coregulation by transcription factors (Yanai et al. 2006; Babu et al. 2008). Further support comes from a report identifying a “ripple effect” by which intensive transcription at one genetic locus spills over to the neighboring loci (Ebisuya et al. 2008). At another level, it has been shown that changes to a gene's expression profile tend to correlate with changes to its genomic neighborhood (Yanai et al. 2006; Xiao et al. 2008; De et al. 2009). Thus, the selective pressures impinging upon gene expression of coexpressed genes remain unclear, as well as the general mode of evolution of the transcriptome.
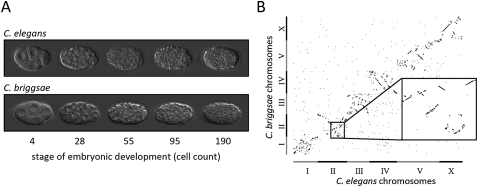
To study the evolution of gene expression and its relationship to genome evolution, we selected a pair of species with a vast discrepancy between their phenotypic and genomic similarities. The nematodes Caenorhabditis elegans and C. briggsae are morphologically nearly indistinguishable (Nigon and Dougherty 1949), sharing the same pattern of embryonic cell divisions (Zhao et al. 2008; Fig. 1A). However, these organisms are genetically very distant, suggesting >100 million years (Myr) of independent evolution (Stein et al. 2003). Analysis of the sequenced genomes of C. elegans and C. briggsae (Stein et al. 2003) showed that there are ∼800 genes present in C. briggsae that are absent in C. elegans and vice versa, with a myriad of chromosomal rearrangements (Fig. 1B). Such a pair of species enabled us to correlate gene expression changes with genomic rearrangements and other tractable genomic traits.
Figure 1.
C. elegans and C. briggsae development is indistinguishable despite a large genetic distance. (A) Micrographs showing C. elegans and C. briggsae embryos at the embryonic developmental stages examined in this study. The cell division patterns are remarkably similar in terms of cell deaths, relative division times, and cell locations within the embryo (Zhao et al. 2008). (B) Dot plot showing the genomic locations of one-to-one orthologs between the C. elegans and C. briggsae genomes, indicating numerous genomic rearrangements.
Results
To identify differences in gene expression profiles between C. elegans and C. briggsae, we designed whole-genome species-specific microarrays. We then used these to measure transcript abundances in precisely staged embryos that span the first quarter of embryonic development (Fig. 1A). The time course included a stage of almost exclusively maternal transcripts (four-cell), the transition to zygotic transcription (28-cell), and the presumptive commitment to the major cell fates (55-, 95-, and 190-cell stages). For 9619 C. elegans genes and 7104 C. briggsae genes, we found statistically significant variation of transcript abundance across the developmental stages (see Methods; Supplemental Fig. S1).
To estimate the reproducibility of the microarray data, we invoked the following controls. First, five probes on one of the designed microarrays were dedicated to each of 795 C. briggsae genes. Of the 390 genes, in which both the predefined best probe and at least one other associated probe passed our expression profile thresholds (Supplemental Fig. S1), only three (0.1%) had a probe that was not supported (Supplemental Fig. S2). Second, we compared our C. elegans data set with a previously reported microarray data set that used a commercially defined platform. Examining two comparable time points, we found that only 1.6% of the genes differ (Supplemental Fig. S3). To control against a general bias in our comparisons, we also examined the similarities among the transcriptomes (expression values of all genes) of the examined cell stages. We found that the correlations between cell stages in both the C. elegans and C. briggsae data sets are nearly identical, suggesting that observed changes are gene specific and do not disturb the overall pattern of the transcriptome (Supplemental Fig. S4). From these controls we concluded that our data set accurately captures temporal expression profiles across the two species.
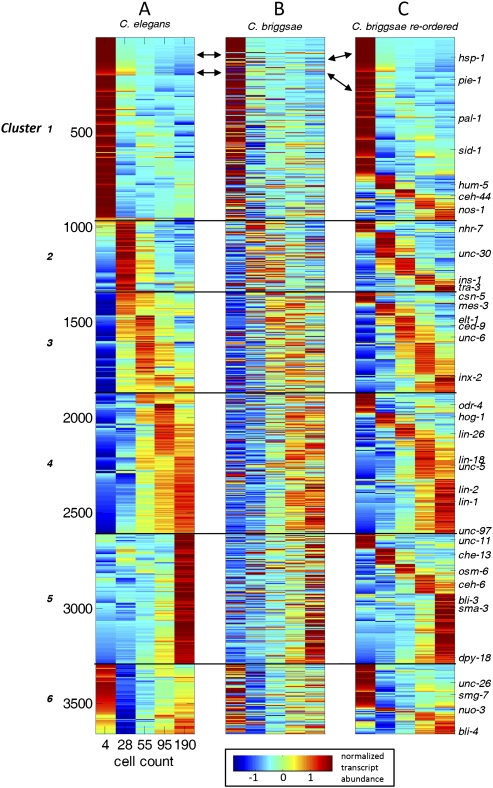
We limited our detailed analysis to 3658 one-to-one orthologous gene pairs with high-confidence expression profiles for both genes (see Methods). One-to-one orthologs are pairs of C. elegans and C. briggsae genes that do not have a recent (post-speciation) duplicate in their respective genome and consequently are among the most evolutionarily stable genes in the genomes (Lynch 2007). Figure 2A shows six C. elegans gene expression profile clusters enriched for expression in each developmental stage examined (clusters 1–5) as well as one cluster (cluster 6) with both maternal and embryonic expression. Figure 2B shows the expression profiles of the C. briggsae one-to-one orthologs. Although the overall signal is conserved, significant divergences are apparent. To refine this view we reordered the C. briggsae genes within each cluster (Fig. 2C). This shows that within each cluster, a sizeable fraction of the genes show expression divergence. Interestingly, significant gene-expression divergence is readily detected even among the earliest embryonically transcribed genes (cluster 2). For example, genes nhr-7, ins-1, and tra-3 each shows a different profile in C. briggsae (Fig. 2). Similar results were obtained when the analysis was initiated with C. briggsae generated clusters (Supplemental Fig. S5).
Figure 2.
Comparative transcriptomics of nematode embryonic development. (A) For 3658 C. elegans genes, the temporal gene expression profiles cluster by K-means (on correlation coefficients) into six general patterns. (B) Expression profiles of C. briggsae orthologs of the C. elegans genes shown in A. (C) C. briggsae orthologs are reordered within the C. elegans defined clusters. To the right, a few specific genes are indicated. The K-means cluster of each C. elegans and C. briggsae gene is provided in Supplemental Table S3.
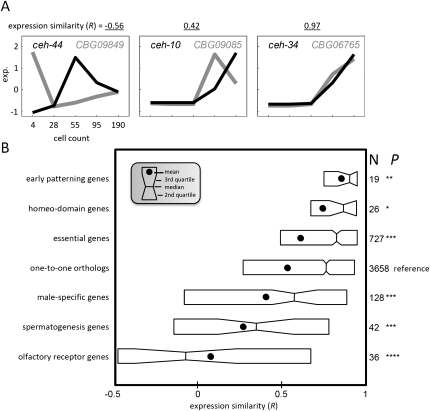
To quantify expression similarity, we computed Pearson's correlation coefficient (R) on pairs of expression profiles (Fig. 3A). We found that one-third of the one-to-one orthologs have diverged beyond a correlation of 0.5 (Fig. 3B). This sizeable fraction of change in gene regulation is likely an underestimate of the true change in gene regulation because we excluded ortholog pairs whose expression was not detected in both species and examined only expression during early embryonic development, a stage thought to be refractory to evolutionary change (Cutter and Ward 2005; Davis et al. 2005). Further, while we are cautious to extrapolate from the one-to-one expressed orthologs to the entire transcriptome, we note that since gene duplicates tend to exhibit significantly higher rates of gene expression evolution (Castillo-Davis and Hartl 2002), the actual degree of gene expression divergence between the C. elegans and C. briggsae transcriptomes is also likely to be higher than this analysis of one-to-one orthologs suggests.
Figure 3.
Conservation of expression similarity by gene function. (A) Normalized temporal gene expression (exp) profiles for three pairs of one-to-one orthologs. For each pair, the correlation coefficient (R) is indicated. (B) Distributions of expression similarities (R) between C. elegans and C. briggsae gene profiles for six gene categories and for all of the one-to-one orthologs in boxplot format. The middle vertical, notch, circle, and left and right boxes, respectively, indicate the median, 95% confidence interval, mean, and second and third quartiles. Shown to the right is the number of genes in each category and the associated P-value with respect to the set of all orthologs (*P < 0.05; **P < 0.005; ***P < 0.0005, Kolmogorov-Smirnov test).
We next asked how the distributions of expression correlations (R) compare among different functional gene categories (Fig. 3B). Genes whose functional perturbation by RNAi leads to lethality (henceforth “essential genes,” see Methods regarding identification of gene sets used in this analysis) and thus are enriched for developmental regulators are significantly enriched for high correlations (P < 10−3, Kolmogorov-Smirnov test, with respect to the one-to-one orthologs from which they are drawn). Further, genes that play known roles in the patterning of the embryo show remarkable expression similarity between orthologs (median R = 0.9, P < 0.005). In contrast, we find significantly lower levels of expression similarity among groups of genes not expected to function in early embryogenesis: olfactory receptors, spermiogenesis-enriched genes, and male-enriched genes (P < 10−5, P < 10−4, and P < 10−3, respectively). While the genes comprising these three gene sets are likely functional in other developmental stages, the divergence observed in their expression profiles between these near-identical nematodes may be the result of relaxed selective constraints.
To further explore the relationship between functional relevance and expression similarity, we compared the C. elegans maternal transcriptome (four-cell stage) of the Bristol N2 strain to the Hawaiian CB4856 strain. Again, among these geographic isolates that share ∼98.8% genomic identity (Wicks et al. 2001; Swan et al. 2002; Denver et al. 2003), we find that gene categories related more (less) to embryogenesis are depleted (enriched) in expression variation, respectively (Supplemental Fig. S6). For example, essential genes show a twofold reduction in the occurrence of variation in expression between the strains, while male-enriched genes show a twofold increase (P < 10−4 and P < 10−3, respectively). Thus, examining gene expression divergences between C. elegans and C. briggsae and between two C. elegans strains revealed that genes likely subject to increased purifying selection exhibit more evolutionarily stable gene expression profiles.
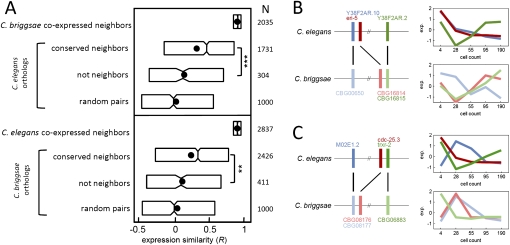
Our gene expression data set allows us to test whether coregulation between gene neighbors is dependent on conserved chromosomal proximity. To examine expression divergence of gene neighbors we identified pairs of coexpressed gene neighbors in C. briggsae and asked whether the C. elegans orthologs are also coexpressed. We found that the general level of expression similarity was dramatically lower, despite examining only C. elegans orthologs within conserved gene neighborhoods (Fig. 4A). We next examined those C. elegans orthologs that are not neighbors. Interestingly, these pairs show significant divergence beyond that shown by the conserved neighborhood pairs (P < 10−6, Kolmogorov-Smirnov test, Fig. 4A). In fact, the expression similarity among these is only marginally statistically different than that of randomly selected gene pairs (P = 0.04, Fig. 4A). These results indicate that the coexpression of gene neighbors is generally not observed for those orthologs where proximity is not conserved.
Figure 4.
The effect of gene-neighbor conservation on coexpression. (A) Boxplots indicating expression similarity among orthologs of coexpressed gene neighbors. The orthologs were parsed into two sets: Conserved neighbors are those orthologs that are also proximate (within four genes) and non-neighbors are orthologs beyond 100 genes apart. The last group is pairs of genes selected at random. Shown to the right are the number of genes in each category and the associated P-value of the difference among the indicated distributions (***P < 10−6; **P < 10−3, Kolmogorov-Smirnov test). For each set of coexpressed gene neighbors, thresholds for expression were applied (Supplemental Fig. S1) on the starting set, and correlation coefficients were examined for all orthologs. (B,C) The horizontal lines indicate C. elegans and C. briggsae chromosomes, and the vertical bars indicate genes; orthologs are linked by black lines. The expression profile for each gene is shown in the profiles to the right. Additional properties of coexpressed gene neighbors are given in Supplemental Tables S4 and S5.
Three lines of evidence further support divergence in expression between orthologs of coexpressed gene neighbors. First, we performed the inverse analysis starting with the C. elegans coexpressed gene neighbors and examining coexpression in the C. briggsae orthologs. Again, we found comparable results (Fig. 4A), even when C. elegans genes contained in operons are excluded (Supplemental Fig. S7). In this analysis we examined gene neighbors without setting a threshold for genomic distance. However, when restricting to only those gene neighbors within 1 kb, we found a greater divergence between orthologs of coexpressed neighbors relative to the divergence observed in more distant genes (Supplemental Fig. S8).
Second, we found evidence that non-neighboring C. briggsae gene pairs, whose C. elegans orthologs are coexpressed gene neighbors, are more similar in expression to their neighbor than expected at random (P < 10−4, see Supplemental Fig. S9). Figure 4B shows an example involving two coexpressed C. elegans gene neighbors (eri-5 and Y38F2AR.10) whose C. briggsae orthologs are neither neighbors nor coexpressed. Instead, the C. briggsae eri-5 ortholog exhibits a conserved expression profile while the Y38F2AR.10 ortholog is coexpressed with its specific neighbor as in C. elegans. Figure 4C shows another example of this scenario.
Third, analysis of a compendium of human and mouse tissue-specific gene expression data showed a similar correlation between changes in gene order and divergence in gene expression. In this data set we identified a common set of 26 tissues across a set of 15,025 one-to-one orthologs. We found that mouse orthologs of coexpressed human gene neighbors are significantly divergent in those instances where the neighborhood is not conserved (Supplemental Fig. S10). Collectively, these analyses support a significant correlation between chromosomal rearrangements and expression divergence.
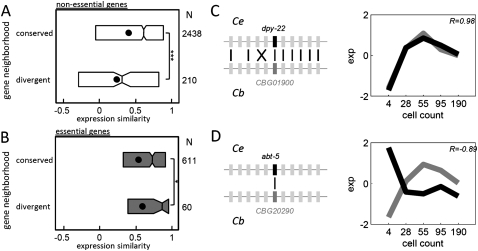
The analysis of divergence of expression in gene pairs does not indicate which partner of each pair is more likely to have diverged in expression. Thus, we asked whether gene expression conservation between orthologs correlates with changes to gene neighborhoods. For this we compared for each ortholog pair the gene neighborhoods (five genes both upstream and downstream) and looked for conservation. We defined two gene sets: (1) those with a high level of ortholog conservation (Fig. 5C); and (2) genes with no ortholog neighbors in common (Fig. 5D). For nonessential genes, orthologs with different neighborhoods show significantly more expression divergence than those with conserved order (P < 10−5, Fig. 5A). Interestingly, the trend is opposite for essential genes (P < 0.05, Fig. 5B), indicating that natural selection acts to constrain gene expression divergence among essential genes, independent of gene neighbor effects.
Figure 5.
Correlation between conservation of gene location and gene expression similarity. (A) Boxplots of expression divergences among nonessential orthologs with highly conserved and divergent genomic neighborhoods (***P < 10−5). (B) Same as A for essential genes. Interestingly, the trend is opposite for essential genes (*P < 0.05). (C) A gene with an overall conserved gene order; (D) a gene whose orthologs are situated in divergent neighborhoods. The horizontal lines indicate the C. elegans and C. briggsae chromosomes, and the vertical gray bars indicate neighboring genes. Black lines indicate a one-to-one orthologous relationship. The plots to the right show the standardized temporal expression profile for the one-to-one orthologs.
Finally, we asked whether differences in promoter sequences for orthologs in different neighborhoods may account for the observed enrichment of expression divergences. Indeed, for nonessential genes we found that promoters in nonconserved neighborhoods diverged significantly more than promoters in conserved neighborhoods (P < 10−9, Supplemental Fig. S11). Similar differences in promoter conservation were found for the essential orthologs (P < 10−3, Supplemental Fig. S11). This suggests that changes to the promoter sequence are concomitant with changes to gene neighborhoods, perhaps representing an adaptation to the local DNA composition. Since expression similarity for essential orthologs remained high irrespective of genomic neighborhood (Fig. 5), promoter divergence does not necessarily lead to gene expression evolution. Together, these results show that, irrespective of gene expression divergences, promoters evolve significantly when the genomic location of the gene changes and suggest that the gene expression changes correlated with genomic rearrangements may be generally independent of changes to the proximal promoter. We conclude that the substantial amount of observed expression changes between C. elegans and C. briggsae can be interpreted according to the principles of gene essentiality and genomic location.
Discussion
We have shown here that two species with near-identical development have a highly divergent set of gene expression profiles. Thus, the rapid rate of genomic evolution is reflected in the transcriptome. While the level of expression divergence we observe between C. elegans and C. briggsae is difficult to relate to other studied systems (Rifkin et al. 2003; Yanai et al. 2004; Khaitovich et al. 2005; Davidson and Erwin 2006), its apparent significance looms even larger given the absence of morphological change. It is highly plausible that natural selection, perhaps due to limited population sizes, cannot optimize gene expression regulation across time, space, and conditions (Yanai et al. 2004, 2006). Consequently, gene expression that is neither strongly deleterious nor advantageous—previously termed ”gratuitous expression” (Gerhart and Kirschner 1997)—may not be under selection and is free to evolve by drift.
While coexpression of gene neighbors may indeed reflect functional constraints under selection in many instances, our results suggest that a significant fraction may be attributed to leaky expression or profligate expression driven by proximity to active genes. The “ripple effect” of transcriptional read-through seems to be entirely consistent with this explanation (Ebisuya et al. 2008). Moreover, identical transgenes integrated in different chromosomal regions have been shown to acquire expression levels that strongly correlate with the expression levels of the gene neighborhood (Gierman et al. 2007). These transgenes often lack enhancers and detection of their expression may be largely dependent upon co-option of nearby regulatory elements. Thus, the features of enhancers that enable modular combinatorial evolution of gene expression appear to also enable unintended and nonoptimized expression for gene neighbors. Overall, the hierarchical manner by which the eukaryotic genomes are organized provides many levels of gene expression regulation (Felsenfeld and Groudine 2003; Wray et al. 2003), where nonoptimality at any of these may be propagated and amplified while remaining mostly invisible to natural selection.
Our results indicate that, even in the absence of morphological change, organisms undergo extensive changes in gene expression thereby providing genome-wide molecular evidence for the ”developmental systems drift” hypothesis (True and Haag 2001). This extensive and putatively unselected variation suggests that, similar to sequence analysis methods that identify unchanging and thereby functional components of coding regions, comparative transcriptomic methods may be able to distinguish functional from nonfunctional components of gene expression.
Methods
Sample preparation
C. elegans (N2 strain) and C. briggsae (AF16 strain) embryos were isolated and staged as described previously (Baugh et al. 2003). The five embryonic stages assayed—the four-, 28-, 55-, 95-, and 190-cell stages—were determined for each organism by DAPI (4′,6-diamidino-2-phenylindole) staining and differential interference contrast microscopy. For C. elegans and C. briggsae, the minutes following the four-cell stage for the four later stages are 80′, 115′, 155′, and 200′ and 55′, 80′, 120′, and 160′, respectively. Fifty embryos were isolated for each sample, and the total RNA was extracted as described previously (Baugh et al. 2003). To obtain sufficient RNA for microarray analysis, RNA was amplified by two rounds of in vitro transcription using the MessageAmpII kit (Ambion) with the following modifications designed to increase the average lengths of the aRNA populations (Baugh et al. 2001). First-round cDNA construction from total RNA was carried out at one-fifth of the recommended quantities and total volume, while in vitro transcription was carried out at one-half of the recommended volume. One hundred nanograms of aRNA from the first round was used as input for the second round of amplification following the manufacturer's protocol for amino-allyl modified nucleotides. Ten micrograms of aRNA was labeled with Cy3, and 1.65 μg of labeled aRNA was used to hybridize onto each microarray. Each embryonic stage was produced in biological triplicates, with the exception of C. elegans 55-cell and C. briggsae four-cell and 95-cell stages, which were hybridized in duplicates because of poor amplification of one of the triplicates. The same procedure was also used to identify the C. elegans Hawaiian (CB4856) four-cell stage transcriptome in triplicates.
Microarrays
We designed two organism-specific microarrays, which were then manufactured by Agilent as 44K arrays. Probes were designed to target the coding region, preferentially near the 3′ end, as well as in the presumptive 3′ UTR, up to 150 bp downstream from the stop codon. Probes 50-60 mer were determined using OligoWiz (Wernersson and Nielsen 2005), which selects oligos based upon their cross-hybridization to other coding sequences, Tm, position along the transcript, folding potential, and low complexity in the sequence. The probes were also restricted against spanning splice junctions to avoid missing transcripts due to errors in gene structure predictions. Each gene was assigned between one and five probes, where the probe with the best overall score was selected as the ”A-probe,” and the remaining ranked B through E. The hybridization and washing of the microarrays were performed following the Agilent protocol for single-channel arrays. The arrays were scanned at 10% laser power to avoid signal saturation. Agilent's Feature Extraction software was used to extract the data, which were then mean-normalized. The data are provided in Supplemental Tables S1 and S2.
Statistical analyses
Temporal gene expression profiles were standardized by subtracting the mean from each expression value and then dividing by the standard deviation, such that the final mean and standard deviation are 0 and 1. Each gene's expression profile was assigned a confidence based upon an error model (see Supplemental Fig. S1). Distributions of correlation coefficients were compared for significant differences using the Kolmogorov-Smirnov nonparametric two-sided test.
Bioinformatics
Predicted one-to-one orthologs were delineated using the Inparanoid algorithm (Remm et al. 2001), which uses sequence analysis comparisons among the two species. Synteny calculations are based upon the two genome sequences (Bieri et al. 2007). The gene sets used in the analyses were compiled as follows and are listed in Supplemental Table S3. Homeodomains were identified as genes annotated with Pfam domain PF00046 (Reece-Hoyes et al. 2005). Essential genes are those annotated as such by genome-wide RNAi screens (Kamath et al. 2003; Bieri et al. 2007). Early patterning genes are those genes reviewed in three publications (Schnabel and Priess 1997; Maduro and Rothman 2002; Baugh et al. 2005). Genes with transcripts enriched during spermatogenesis and in males were retrieved from a published microarray analysis (Kim et al. 2001). Olfactory receptor genes were identified using WormBase (Bieri et al. 2007). For each gene set only those that were one-to-one orthologs were examined. Coexpression among gene neighbors was defined as R ≥ 0.8, selected as the mean R (expression similarity) of the early patterning genes (Fig. 3). Gene neighbors were defined as separated in the genome by up to four genes. Identical trends are observed for adjacent genes and separation of up to four genes.
Acknowledgments
We thank David Yao, Michal Levin, and Ryan Baugh for technical assistance, and Daniel Schott, Antony Jose, Ran Kafri, Adnan Derti, Asher Cutter, and Martin Lercher for critical reading of the manuscript and constructive suggestions, and to three anonymous reviewers. We thank the Caenorhabditis Genetics Center for strains. This work was supported by NIH grants HD051137 to I.Y. and GM64429 to C.P.H.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE15551.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093815.109.
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Babu MM, Janga SC, de Santiago I, Pombo A. Eukaryotic gene regulation in three dimensions and its impact on genome evolution. Curr Opin Genet Dev. 2008;18:571–582. doi: 10.1016/j.gde.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2007;23:480–484. doi: 10.1016/j.tig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:e29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132:1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- Belting HG, Shashikant CS, Ruddle FH. Modification of expression and cis-regulation of Hoxc8 in the evolution of diverged axial morphology. Proc Natl Acad Sci. 1998;95:2355–2360. doi: 10.1073/pnas.95.5.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. WormBase: New content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Burhans DT, Ramachandran L, Wang J, Liang P, Patterton HG, Breitenbach M, Burhans WC. Non-random clustering of stress-related genes during evolution of the S. cerevisiae genome. BMC Evol Biol. 2006;6:58. doi: 10.1186/1471-2148-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Endless forms: The evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. doi: 10.1016/s0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: Molecular genetics and the evolution of animal design. Blackwell Science; Malden, MA: 2001. [Google Scholar]
- Castillo-Davis CI, Hartl DL. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol Biol Evol. 2002;19:728–735. doi: 10.1093/oxfordjournals.molbev.a004131. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Ward S. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol Biol Evol. 2005;22:178–188. doi: 10.1093/molbev/msh267. [DOI] [PubMed] [Google Scholar]
- Davidson EH. The regulatory genome: Gene regulatory networks in development and evolution. Academic Press; Burlington, VT: 2006. [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Davis JC, Brandman O, Petrov DA. Protein evolution in the context of Drosophila development. J Mol Evol. 2005;60:774–785. doi: 10.1007/s00239-004-0241-2. [DOI] [PubMed] [Google Scholar]
- De S, Teichmann SA, Babu MM. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 2009;19:785–794. doi: 10.1101/gr.086165.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Thomas WK. Phylogenetics in Caenorhabditis elegans: An analysis of divergence and outcrossing. Mol Biol Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, embryos, and evolution: Toward a cellular and developmental understanding of phenotypic variation and evolutionary adaptability. Blackwell Science; Malden, MA: 1997. [Google Scholar]
- Gierman HJ, Indemans MH, Koster J, Goetze S, Seppen J, Geerts D, van Driel R, Versteeg R. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17:1286–1295. doi: 10.1101/gr.6276007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Williams EJ, Pal C. Natural selection promotes the conservation of linkage of coexpressed genes. Trends Genet. 2002;18:604–606. doi: 10.1016/s0168-9525(02)02813-5. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Janga SC, Collado-Vides J, Babu MM. Transcriptional regulation constrains the organization of genes on eukaryotic chromosomes. Proc Natl Acad Sci. 2008;105:15761–15766. doi: 10.1073/pnas.0806317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Patient R. A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol. 2004;271:467–478. doi: 10.1016/j.ydbio.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: The gene regulatory network of the Caenorhabditis elegans endoderm. Dev Biol. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Nigon V, Dougherty EC. Reproductive patterns and attempts at reciprocal crossing of Rhabditis elegans Maupas, 1900, and Rhabditis briggsae Dougherty and Nigon, 1949 (Nematoda: Rhabditidae) J Exp Zool. 1949;112:485–503. doi: 10.1002/jez.1401120307. [DOI] [PubMed] [Google Scholar]
- Poyatos JF, Hurst LD. The determinants of gene order conservation in yeasts. Genome Biol. 2007;8:R233. doi: 10.1186/gb-2007-8-11-r233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci. 2007;104(Suppl. 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purmann A, Toedling J, Schueler M, Carninci P, Lehrach H, Hayashizaki Y, Huber W, Sperling S. Genomic organization of transcriptomes in mammals: Coregulation and cofunctionality. Genomics. 2007;89:580–587. doi: 10.1016/j.ygeno.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJ. A compendium of Caenorhabditis elegans regulatory transcription factors: A resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, Scamuffa N, Caccioppoli C, Dermitzakis ET, Lyle R, Banfi S, et al. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Priess JR. Specification of cell fates in the early embryo. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 361–382. [PubMed] [Google Scholar]
- Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of coexpressed genes in mammalian genomes are conserved by natural selection. Mol Biol Evol. 2005;22:767–775. doi: 10.1093/molbev/msi062. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Swan KA, Curtis DE, McKusick KB, Voinov AV, Mapa FA, Cancilla MR. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 2002;12:1100–1105. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Nielsen HB. OligoWiz 2.0—integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14:1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Wu P, Jiang TX, Suksaweang S, Widelitz RB, Chuong CM. Molecular shaping of the beak. Science. 2004;305:1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- Yanai I, Graur D, Ophir R. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. OMICS. 2004;8:15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- Yanai I, Korbel JO, Boue S, McWeeney SK, Bork P, Lercher MJ. Similar gene expression profiles do not imply similar tissue functions. Trends Genet. 2006;22:132–138. doi: 10.1016/j.tig.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]