Abstract
The transcription factor GATA1 regulates an extensive program of gene activation and repression during erythroid development. However, the associated mechanisms, including the contributions of distal versus proximal cis-regulatory modules, co-occupancy with other transcription factors, and the effects of histone modifications, are poorly understood. We studied these problems genome-wide in a Gata1 knockout erythroblast cell line that undergoes GATA1-dependent terminal maturation, identifying 2616 GATA1-responsive genes and 15,360 GATA1-occupied DNA segments after restoration of GATA1. Virtually all occupied DNA segments have high levels of H3K4 monomethylation and low levels of H3K27me3 around the canonical GATA binding motif, regardless of whether the nearby gene is induced or repressed. Induced genes tend to be bound by GATA1 close to the transcription start site (most frequently in the first intron), have multiple GATA1-occupied segments that are also bound by TAL1, and show evolutionary constraint on the GATA1-binding site motif. In contrast, repressed genes are further away from GATA1-occupied segments, and a subset shows reduced TAL1 occupancy and increased H3K27me3 at the transcription start site. Our data expand the repertoire of GATA1 action in erythropoiesis by defining a new cohort of target genes and determining the spatial distribution of cis-regulatory modules throughout the genome. In addition, we begin to establish functional criteria and mechanisms that distinguish GATA1 activation from repression at specific target genes. More broadly, these studies illustrate how a “master regulator” transcription factor coordinates tissue differentiation through a panoply of DNA and protein interactions.
Control of gene expression occurs initially through regulatory proteins binding to specific DNA sequences and modulating the ability of RNA polymerase to transcribe a target gene (Pardee et al. 1959). Diverse mechanisms are employed for this regulation, ranging from direct interactions with RNA polymerase to recruitment of large enzymatic complexes that lead to activation or repression by altering histone modifications or chromatin structure (Maston et al. 2006; Jiang and Pugh 2009). Some cis-regulatory modules (CRMs), containing clusters of binding sites for transcription factors (Maniatis et al. 1987), are close to the transcription start sites (TSSs) of genes. A survey of 1% of the human genome (The ENCODE Project Consortium 2007) showed that the major biochemical signatures of gene regulatory regions are strongest within 1 to 2 kb of TSSs. Other CRMs are distal to the TSS. Genes encoding developmental regulatory proteins frequently have multiple enhancers (Jiang et al. 1991; Gottgens et al. 2002; Nobrega et al. 2003), which can be as much as 1000 kb from the TSS (Lettice et al. 2003).
Detailed investigations of individual genes have revealed many of the proteins and basic mechanisms that regulate gene expression. For example, hematopoietic transcription factors such as GATA1 and TAL1 bind both proximal and distal CRMs to regulate erythroid genes such as those encoding hemoglobin (Cantor and Orkin 2002). Among the distal CRMs are locus control regions that are needed for activation of all the genes within a locus, such as the beta-globin gene (HBB) cluster (Grosveld et al. 1987) and the alpha-globin gene (HBA) cluster (Vyas et al. 1992). While these range from 15 to 70 kb away from their target promoters, the effects of regulatory proteins binding to distal CRMs are exerted by close interaction with the core promoters in the interphase nucleus (Carter et al. 2002; Tolhuis et al. 2002; Vakoc et al. 2005; Dostie et al. 2006).
However, these studies do not provide a comprehensive view of how CRMs and transcription factors interact to orchestrate tissue development. This can only be approached through large-scale, genome-wide studies to define the spatiotemporal relationships among transcription factor occupancy to DNA, chromatin modifications, gene expression, and cellular responses. Recent studies of steroid hormone receptors (Carroll et al. 2006; So et al. 2007) and STAT proteins (Hartman et al. 2005; Robertson et al. 2007) revealed correlations between positions of DNA segments occupied by a transcription factor and the response in gene expression, but the results appear to be distinctive to each transcription factor. Additional comprehensive studies of transcription factor occupancy and clearly defined biological responses are needed to determine common and distinctive features of regulation in mammals.
In this paper, we studied the genome-wide effects of GATA1, a transcription factor that regulates many erythroid genes and is essential for the differentiation and survival of this lineage. We used a mouse erythroid cell line (G1E) derived from Gata1 knockout embryonic stem cells. G1E cells are frozen at the proerythroblast stage of differentiation and undergo GATA1-dependent terminal maturation (Weiss et al. 1997). Restoration of GATA1 function in G1E cells, by gene transfer and activation of an estrogen-inducible fusion protein (GATA1-ER), triggers an extensive program of gene activation and repression, in many cases due to direct transcriptional effects. Simultaneously, the cells undergo G1 arrest and acquire a late-stage erythroid phenotype (Welch et al. 2004). In this paper, we measured the levels of stable RNA from 19,000 genes in a time course after activation of GATA1 and concurrently ascertained genome-wide DNA occupancy of GATA1. In addition, a large segment of chromosome 7 was interrogated for occupancy by TAL1 (SCL), a transcription factor that participates in erythroid differentiation, at least in part by complexing with GATA1 (Shivdasani et al. 1995); this latter feature is associated with positive regulation of target genes (Tripic et al. 2009). Furthermore, given the important role of histone modifications in gene regulation and the established functions for GATA1 in this process, we examined the levels of histone H3K4 monomethylation, which is associated with gene activation through enhancers (The ENCODE Project Consortium 2007; Heintzman et al. 2007), and trimethylation of histone H3K27, a modification catalyzed by the Polycomb repressor complex 2 (Muller et al. 2002) and associated with down-regulation.
Our study is among the first in mammals that correlates spatial patterns in occupancy genome-wide with the effects of a transcription factor on gene expression. The results reveal consistent features for positive regulation but indicate multiple pathways for negative regulation.
Results
Gene expression profile after activation of GATA1 in G1E-ER4 cells
Earlier transcriptome studies after restoration of GATA1 activity in G1E-ER4 cells interrogated only about half the genes that are represented on current microarrays (Welch et al. 2004). Thus, we determined the kinetics of GATA1-regulated gene expression using 45,000 probe sets on the GeneChip Mouse Genome 430 2.0 Array from Affymetrix, representing 19,000 mouse genes. The mRNA of G1E-ER4 cells was extracted at progressive time points after estradiol-induced activation of the GATA1-ER hybrid protein. The expression responses were determined by hybridization of triplicate samples of cDNA from each time point to the microarray.
The new data confirm previous expression results and expand the number of interrogated genes considerably. For the probe sets that could be mapped across the two platforms, the expression signals of the earlier and the current data sets are substantially correlated (Pearson's correlation coefficient R = 0.6; Supplemental Fig. 1). In the previous microarray study (Welch et al. 2004), the change in expression level of six genes was validated by a Northern blot analysis; these six genes show the same response in the new data set. Also, the categories of genes responding to GATA1 are similar to those observed before (Yu et al. 2009). Thus, we conclude that the new expression profiles accurately reflect the trends in gene expression, and they are much more comprehensive than the previous data sets (45,000 probe sets compared with 12,500; 19,000 genes compared with 9266).
Genes whose expression level changed significantly in response to restoration and activation of GATA1-ER were identified in a multistep process (for details, see Methods and Supplemental Tables 1, 2). First, probe sets with a change in expression level at any time after activation of GATA1-ER of at least twofold (for most analyses) or exceeding a false discovery rate (FDR) threshold of 0.001 (compared with the level at zero time) were identified. The responsive probe sets were then divided into those with either a continuous upward or a continuous downward trend, using the program Oriogen (Peddada et al. 2005). These comprised 84% of the responsive probe sets; only a small minority had a biphasic response, and these were not considered further. The responsive probe sets were then filtered to remove those with low signals throughout the time course. Finally, the filtered probe sets were matched back to the RefSeq (Pruitt and Maglott 2001) and Ensembl (Curwen et al. 2004) gene models for mouse genome assembly mm8 to find annotated genes corresponding to the probe sets. This mapping produced 1048 up-regulated genes, 1568 down-regulated genes, and 5903 genes with no response (less than a 1.1-fold change at any point in the time course). Over half the genes (10,418) had expression changes between the thresholds for no response (1.1-fold) and change in expression (twofold); these are not assigned to any of the three categories.
Confirming the previous studies of Welch et al. (2004), the number of repressed genes is notably higher than the number of activated genes after restoration of GATA1. This is consistent with a terminal differentiation process in which the repertoire of gene expression becomes streamlined for erythroid cell functions. Within the up- or down-regulated cohorts, the groups revealed by k-means clustering (MacQeen 1967) differ primarily in the kinetics of the response, with some groups responding earlier and others later (Fig. 1A).
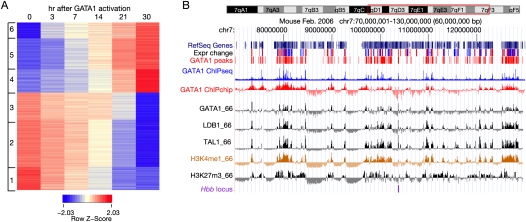
Figure 1.
Gene expression response and chromosomal DNA occupancy after restoring GATA1 in erythroid cells. (A) The expression patterns of GATA1 responsive genes are portrayed as a heat map, with red indicating higher levels and blue indicating lower levels of expression for each gene. Each row represents the expression level of one gene at the time points after induction indicated for each column. The hybridization signals from three replicates at each time point were averaged, and the log (base 2) of the average signals were normalized in each row to generate a Z-score. The data matrix was clustered using the k-means method with k = 6; the results show three clusters of up-regulated genes and three clusters of down-regulated genes (indicated on the left). (B) Large-scale view of expression response, occupancy by transcription factors, and repressive histone modification in erythroid cells. For a 60-Mb region of mouse chromosome 7 centered on the Hbb gene complex (outlined in red on the ideogram at the top), the tracks of data show (in order) RefSeq genes, indicators of the change in expression level (red for up- and blue for down-regulation) in response to restoration of GATA1, the genome-wide GATA1 peak calls, the ChIP-seq data for GATA1 after peak calling by MACS (blue), the raw ChIP-chip hybridization signals for GATA1 (tracks labeled GATA1_HD2 for the genome-wide data and GATA1_66 for chromosome 7 data), TAL1 and LDB1 occupancy, the raw ChIP-chip hybridization data for monomethylation of H3K4 and trimethylation of H3K27, and the location of the Hbb locus (purple). (Expr change) Change in expression level; (GATA1 peaks) those deduced from the genome-wide ChIP-chip data. The image was generated on a customized installation of the UCSC Genome Browser (Kent et al. 2002).
Occupancy of DNA segments by GATA1 in G1E-ER4 cells
To investigate how well the expression patterns could be explained by binding of GATA1 to specific sites, we mapped the locations of DNA segments occupied by this transcription factor by chromatin immunoprecipitation (ChIP) (Fig. 1B). The GATA1 ChIP experiments were performed on G1E-ER4 cells, 24 h after activation of the GATA1-ER hybrid protein. This time was chosen because the largest changes in expression occur by 21 and 30 h (Fig. 1A) and it is expected that GATA1 occupancy will coincide with or precede the expression response. We produced two genome-wide GATA1 ChIP data sets. First, we employed the sequence census methodology of ChIP with massively parallel sequencing (ChIP-seq) (Wold and Myers 2008), using Illumina GAII technology to produce 23 million reads (36 nucleotides long) uniquely mapped to the mouse genome (mm8 assembly) for the GATA1 ChIP DNA and 15 million mapped reads for the input DNA. Second, we hybridized an independent GATA1 ChIP sample to the NimbleGen HD2 tiling array for the mouse genome (mm8 assembly). These data, along with the expression results, can be viewed and downloaded from a custom genome browser at http://main.genome-browser.bx.psu.edu/, on the mouse mm8 (Feb 2006) genome assembly. A summary view from a 66-Mb region of mouse chromosome 7 studied previously (Cheng et al. 2008; Zhang et al. 2009) shows substantial ChIP signal above the background and a striking congruence of the signals for GATA1 occupancy between the two genome-wide data sets (Fig. 1B).
The peak-calling program MACS (Zhang et al. 2008) was applied to the GATA1 ChIP-seq and input sequence data to identify 14,351 potential GATA1 occupied segments (GATA1 OSs) (Fig. 2A). This set includes 56 out of the 63 validated GATA1 OSs determined previously (Cheng et al. 2008), indicating a sensitivity of 90%, while none of the 70 false-positive regions (i.e., ChIP with microarray hybridization [ChIP-chip] peaks not validated by quantitative PCR) are included, indicating high specificity.
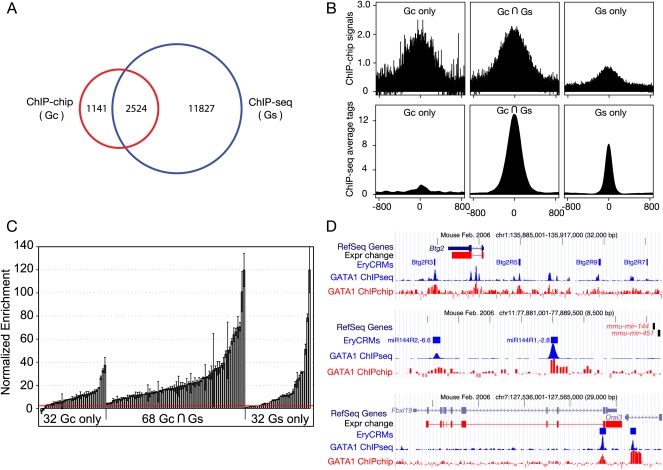
Figure 2.
Accuracy of GATA1 peaks from high-throughput analysis of ChIPs. (A) The Venn diagram shows the relationships among peaks called from ChIP-seq and ChIP-chip data on GATA1 in G1E-ER4 cells. (Gc) G1E-ER4 ChIP-chip; (Gs) G1E-ER4 ChIP-seq. (B) Support of raw ChIP-chip and ChIP-seq data for peaks called from different technologies. The graphs show the mean ChIP-chip and ChIP-seq signals for occupancy for common and unique peaks, centered on the middle of the called peak and extending 800 bp on each side. (C) Validation of GATA1 peaks by quantitative PCR. From the peaks called for the genome-wide GATA1 ChIP data, 68 from the set common to ChIP-seq and ChIP-chip peaks, 32 from the ChIP-chip only peaks, and 32 from the ChIP-seq only peaks were chosen randomly for validation of occupancy by GATA1 using a qPCR assay, along with 20 negative control regions (not called as peaks). The bar-plot shows the mean of two determinations of the enrichment for each tested DNA segment in the GATA1 ChIP material (error bars cover the range), expressed as the number of standard deviations above the normalized mean of the negative controls (see Supplemental material). The red line indicates the threshold for validation (two standard deviations above the mean of the negative controls). (D) Data for previously studied genes, showing strong correspondence between validated erythroid CRMs and the new ChIP-seq and ChIP-chip data for GATA1. Data tracks show genes, expression response, positions of experimentally validated cis-regulatory modules, and ChIP-seq and ChIP-chip data for GATA1.
The results of analyzing the ChIP-chip data by three different peak-calling programs, Mpeak (Zheng et al. 2007), TAMALPAIS (Bieda et al. 2006), and PASS (Zhang 2008), were combined to generate a set of 3558 potential GATA1 OSs across the erythroid mouse genome (see Supplemental material). Over two-thirds of the ChIP-chip peak data set overlaps with the ChIP-seq peak data set (2524 out of 3558, or 71%, Fig. 2A). As expected, the raw ChIP-chip and ChIP-seq signals are both high in the peaks in the intersection of the two data sets (Fig. 2B). In addition, GATA1 OSs identified by only one technology are also supported by the signal from the other; e.g., peaks identified only by ChIP-seq also have notable signal in the ChIP-chip data (Fig. 2B). Given this strong support for the GATA1 OSs identified by either technology, we then tested DNA intervals in all three categories for validation by independent quantitative PCR assays. The peaks were validated at a high rate. Occupancy was confirmed for all the 68 tested peaks identified by both technologies, for 29 of the 32 tested peaks (91%) found only by ChIP-chip, and for 28 of the 32 tested peaks (88%) identified only by ChIP-seq (Fig. 2C).
The high validation rates and supportive evidence from both methods indicate that peaks present in only one of the two genome-wide data sets are not false positives, but many are low-occupancy sites that are hard to detect consistently, using different peak calling programs set for high stringency (Johnson et al. 2008). Indeed, the aggregated raw signals (Fig. 2B) and the quantitative PCR results (Fig. 2C) suggest lower occupancy for most of the GATA1 OSs in the nonintersection sets. Therefore, we made a union of the ChIP-seq and ChIP-chip peaks, followed by merging of overlaps to generate a set of 15,360 GATA1 OSs. This set of GATA1 OSs is highly accurate based on comparisons with previously characterized DNA segments occupied by GATA1, including the well-known CRMs for globin genes (data not shown). Three examples (Fig. 2D) show that the genome-wide GATA1 occupancy data match well with CRMs predicted and experimentally validated as enhancers (e.g., Btg2; Wang et al. 2006), the GATA1-bound CRMs regulating the gene for miR-144 and miR-451 (Dore et al. 2008), and validated GATA1-occupied segments active as enhancers (e.g., Fbxl19; Cheng et al. 2008).
The sequences of GATA1 OSs contain several motifs that distinguish bound from unbound sites. Using both Discriminatory Motif Enumerator (Smith et al. 2005) and a hexamer enumeration method, good discrimination was found for a variant of the canonical binding site motif (AGATAA), multiple canonical binding site motifs, and matches to binding sites for transcription factors of the Krüppel-like zinc finger class and CP2 (for details, see Supplemental material). These data confirm the results of a separate study of GATA1 OSs in the 66-Mb region of mouse chromosome 7 (Zhang et al. 2009).
Proximity of target genes to DNA segments occupied by GATA1
The GATA1-responsive genes tend to occur in clusters separated by long genomic regions with no responsive genes (Fig. 1B, track “Expr change”). This level of clustering of responsive genes is greater than what would be expected from the gene density (Supplemental material; Supplemental Fig. 2). A localized region with responsive genes could represent a regulatory domain. In support of this, the peaks for occupancy by GATA1 correspond well to the locations of responsive genes (Fig. 1B, track “GATA1 peaks”).
Ideally, the correspondence between occupancy of DNA segments by GATA1 and the response in gene expression would be studied in a situation in which the target gene for each occupied DNA segment is known without ambiguity. Ultimately, that can only be determined rigorously by genetic knockouts or other manipulations of all the occupied DNA segments. For this study, we analyzed the distance between each responsive gene and the nearest DNA segment occupied by GATA1 to ascertain how the behavior of a (presumptive) target gene correlates with factor occupancy at varying distances. The distributions of distances between GATA1 OSs and the TSS of the nearest genes were compared between responsive and nonresponsive genes.
Across the mouse genome, over 88% of the responsive genes have a GATA1 OS within 100 kb of the TSS (Fig. 3A, top). The distance between the TSS and GATA1 OS required to capture 60% of the genes in a response category is less than 5 kb for induced genes, but it is over 33 kb for repressed genes (Fig. 3A, middle). Half of the induced genes have a GATA1 OS very close to the TSS, shown by the portion of the cumulative distribution curve with almost vertical slope. In contrast, the portion of down-regulated genes with a GATA1 OS is dramatically lower than that for induced genes at all distances. Both categories of target genes are closer to a GATA1 OS than are nonresponsive genes.
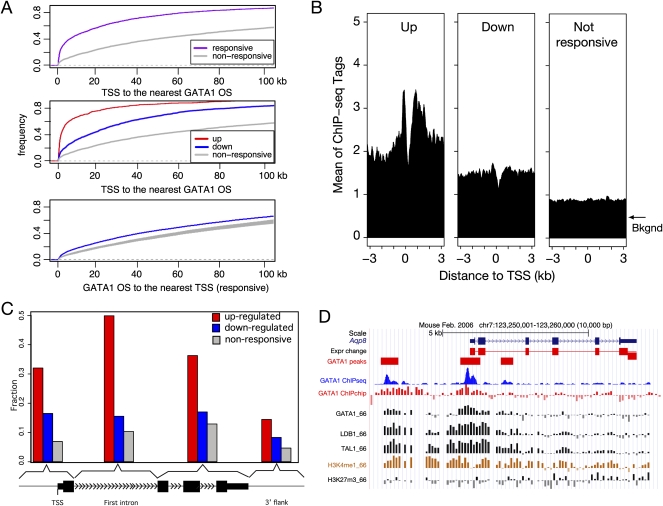
Figure 3.
Proximity of induced genes to GATA1-occupied DNA segments. (A) The cumulative distribution of the distance from the TSS of each gene in a response category to the nearest GATA1-occupied DNA segment (GATA1 OS) is shown in each panel, with the y-axis showing the fraction of genes whose nearest GATA1 OS is within the designated distance. The color of each distribution line is distinctive for each response category (purple for all responsive genes, red for up-regulated, blue for down-regulated, and gray for nonresponsive genes). The distributions of distances from the GATA1 OSs to the TSS of responsive genes are shown in the bottom panel, with the gray lines for 1000 iterations of random selection of TSSs from 2616 nonresponsive genes. (B) GATA1 occupancy signals near the TSSs. The distribution of raw GATA1 ChIP-seq signals (mean number of ChIP-seq tags in 100-bp windows) is graphed as a function of distance on either side of the TSS of genes in three response categories. DNA upstream of the TSS is given a negative value for the distance from the TSS. All the genes in a designated category were first centered by the TSS and then windows were extended along each side of TSS up to 3 kb. (C) Preferred locations of GATA1-occupied segments with respect to genes. The bar graph presents the fraction of genes in each response category that has at least one GATA1 OS in the indicated subregion of a gene. These subregions are segments around the TSS (−5 kb from the TSS to the end of the first exon), the first intron, the remaining exons and introns, and 5 kb past the poly(A) addition site. (D) An example of newly discovered GATA1 OSs close to the TSS of the GATA1-incuded gene Aqp8. Tracks are as in Fig. 1B.
The converse relationship also holds, i.e., GATA1 OSs are closer to responsive genes than they are to nonresponsive genes (Fig. 3A, bottom). About 42% of GATA1 OSs are within 100 kb of a responsive gene, leaving about 58% implicated in a function (if any) quite distal to the regulated genes.
Genes activated by GATA1 tend to be bound by this factor near the TSS
We then examined more thoroughly the pattern of GATA1 occupancy near the TSS of genes in each response category. First, we computed the mean of the GATA1-ChIP-seq tag counts in small bins within 3 kb on each side of the TSS for all the genes in each response category. Note that this analysis is not restricted to the peak calls; by including all the mapped reads, it is not limited by the sensitivity of peak calls. The results reveal a striking accumulation of GATA1 ChIP hybridization signal around the TSSs of induced genes, much greater than the signal for genes with no expression change (Fig. 3B). Interestingly, the signal for GATA1 binding dramatically dips close to the TSS and then rises substantially on both sides of the TSS. Thus, while GATA1 tends to bind in the vicinity of the TSS, it tends to be excluded from a small DNA segment just 3′ to the TSS, perhaps by a protein complex such as a paused RNA polymerase. The signal for GATA1 occupancy is also higher for repressed genes than for nonresponsive genes, but the levels of ChIP-seq tags are substantially lower than for induced genes (Fig. 3B). In addition, the fraction of genes that are occupied by GATA1 proximally, i.e., within a region from 10 kb upstream of the TSS to 10 kb downstream of the polyA addition signal, is larger for induced genes (866 out of 1048 genes, 83%) than for repressed genes (802 out of 1568 genes, 51%).
The enrichment for GATA1 occupancy in induced genes is also seen in distinctive subregions of genes (Fig. 3C). A substantial fraction of up-regulated genes are occupied by GATA1 in the first intron (50%), and about 32%–36% are occupied in the region around the TSSs or in other internal regions. In contrast, a much smaller proportion of down-regulated or nonresponsive genes are occupied by GATA1 in any of the subregions of the gene. The Aqp8 gene, encoding an aquaporin, is a newly discovered example of an induced gene occupied by GATA1 both in the proximal 5′ flanking region and in the second intron (Fig. 3D). A similar pattern is found for the induced gene Btg2, and the Mir144 and Mir451 genes (also known as mmu-mir-144 and mmu-mir-451) have GATA1-bound CRMs in their proximal 5′ flanking regions (Fig. 2D).
Responsive genes tend to have multiple GATA1-occupied DNA segments
For a notable fraction of GATA1-responsive genes (38%), more than one GATA1 OS is found in the proximal neighborhood extending to 10 kb before the TSS and 10 kb after the polyA addition site. This feature is more common in the set of up-regulated genes (58% of them) than in the set of down-regulated genes (only 24%). For example, the ChIP-seq data show two GATA1 OSs in the 5′ flanking region of the Aqp8 gene, each of which has two adjacent peaks, suggesting two pairs of GATA1 OSs in addition to the one in the second intron (Fig. 3D). Similarly, the Btg2, miR-144/451, and Fbxl19 genes all have multiple GATA1 OSs (Fig. 2D). The number of GATA1 OSs in the proximal neighborhood of genes is significantly higher for up-regulated genes than for down-regulated genes (Supplemental Fig. 3; the P-value for these two distributions being indistinguishable is 0.002 using a two-tailed Student's t-test).
Co-occupancy by TAL1 and LDB1 is strongly associated with positive regulation by GATA1
GATA1 can form a large multiprotein complex with TAL1, E47, LDB1, and LMO2 (Wadman et al. 1997) that occupies several erythroid CRMs (e.g., Anguita et al. 2004; Wozniak et al. 2008). In contrast, other GATA1-occupied DNA segments do not have the other components of the large complex, and the absence of TAL1 from GATA1-occupied DNA segments is strongly associated with down-regulation (Tripic et al. 2009). We examined the co-occurrence of these proteins in more detail using the new genome-wide ChIP data sets for GATA1 and the ChIP-chip data for TAL1 in the 66-Mb region of mouse chromosome 7 (Tripic et al. 2009) .
We find a strong positive correlation between occupancy of DNA by GATA1 and TAL1 (Fig. 4A). The overall similarity in occupancy between GATA1 and TAL1 is apparent in the 60-Mb overview of Figure 1B, and it is seen at higher resolution in the specific example of Aqp8 (Fig. 3D). We partitioned the GATA1 OSs in this 66-Mb region of chromosome 7 into those in the proximal neighborhoods of either up-regulated genes or down-regulated genes. The correlation between level of occupancy by TAL1 and GATA1 is equally strong for both classes of GATA1 OSs (Fig. 4A).
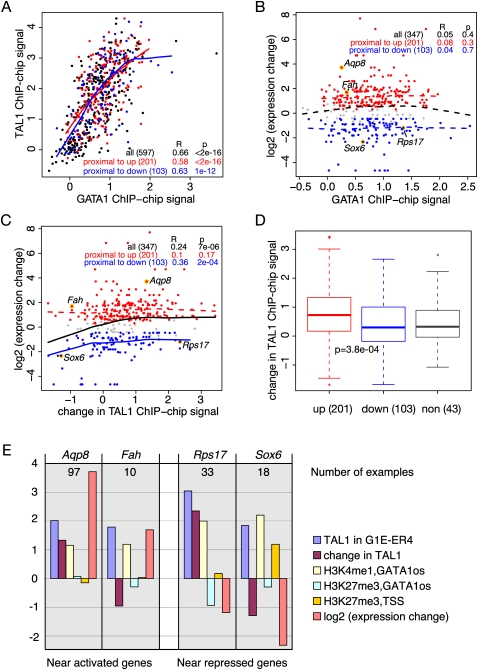
Figure 4.
Correlation among GATA1, TAL1, and changes in gene expression. (A) Scatterplot for the level of occupancy by GATA1 (x-axis) and TAL1 (y-axis) for all GATA1 OSs in the 66-Mb region of mouse chromosome 7, with the GATA1 OSs in the proximal neighborhood of up-regulated genes shown as red dots (increase in expression above the FDR threshold of 0.001), those in the proximal neighborhood of down-regulated genes shown as blue dots (decrease in expression exceeding the FDR threshold of 0.001), and all others as black dots. The ChIP-chip hybridization levels for the several probes in each interval covering a GATA1 OS were averaged and used as a proxy for occupancy for GATA1 and TAL1. The Pearson's correlation coefficient R and P-value are listed for three categories of GATA1 OS (all, and those in the proximal neighborhoods of up- or down-regulated genes), and the lowess line is drawn separately for each category of GATA1 OSs. The lowess lines for nonsignificant associations are broken whereas those for significant associations are solid. (B,C) Scatterplots for the relationship between the change in expression level for the genes in whose proximal neighborhood a GATA1 OS is found (y-axis) and GATA1 occupancy (x-axis in B) or the change in TAL1 occupancy between G1E-ER4 cells (GATA1 restored and activated) and G1E Gata1 knockout cells (x-axis in C). The largest difference in expression level (compared with that at time 0) at any point in the time course after activation is the expression change. (D) Boxplots of the distributions of values for the change in TAL1 occupancy in GATA1 OSs associated with genes in three expression categories (non, nonresponsive; up, induced; down, repressed). The differences between the up- and down-regulated categories are significant by a Student's t-test (P = 3.3 × 10−5). (E) Examples illustrating the range of features observed at GATA1 OSs in the neighborhoods of two induced genes and two repressed genes (right). The bars are the mean ChIP-chip signals for the probes in the interval for each GATA1 OS (see key) or the log (base 2) of the expression change. The number of GATA1 OSs that fit the pattern shown is given in each graph (see Table 1).
Despite the overall correlation between GATA1 and TAL1 occupancy, with our large data set we confirm previous observations (Tripic et al. 2009) that co-occupancy of GATA1 OS by TAL1 is associated with induced expression and the absence or reduction of TAL1 levels is associated with repression. For this analysis, we examine not only the direction of the change in expression but also the amount of change, e.g., the largest difference (compared with time zero) that occurs during the time course of the expression analysis. The largest differences are most frequently seen at the later time points (21 h and 30 h, Fig. 1A); thus, our occupancy measurements at 24 h roughly coincide with or occur before the maximal change. We also did the analyses using the expression change at 21 h rather than the maximal change, and very similar results were obtained (see Supplemental material). The amount of induction or repression is not dependent on the level of GATA1 occupancy within the proximal neighborhood of genes (Fig. 4B). However, the change in gene expression is positively correlated with the change in TAL1 occupancy upon restoration and activation of GATA1-ER (Fig. 4C). This association is driven primarily by the decrease in TAL1 levels for strongly repressed genes (bottom left of the graph). GATA1 OSs in the vicinity of both strongly and weakly induced genes show similar ranges for the changes in TAL1 levels (Fig. 4C). The distribution of values for the change in TAL1 occupancy is significantly higher for GATA1 OSs in induced genes than for those in repressed genes, with a substantial number of the latter showing decreases in TAL1 (Fig. 4D). For example, a GATA1 OS close to the induced gene Aqp8 shows increased TAL1 upon activation of GATA1-ER, while one near the repressed gene Sox6 shows decreased TAL1 (Fig. 4E). While these are distinctive properties of GATA1 OSs near induced versus repressed genes as a group, they are not universal, as illustrated by the opposite trends seen for GATA1 OSs near the induced gene Fah and the repressed gene Rps17 (Fig. 4E).
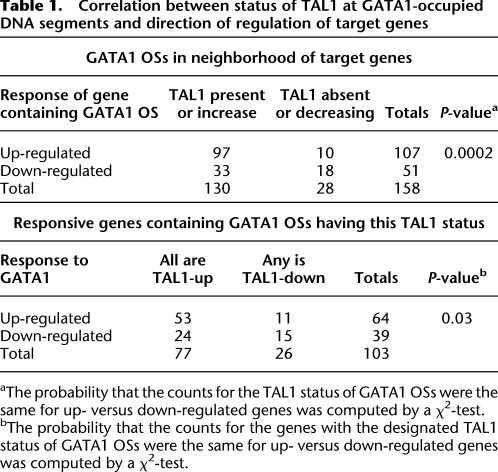
We then examined co-occupancy by GATA1 and TAL1 for whole genes. Each GATA1 OS in the proximal neighborhood of a gene was classified as TAL1-up if it was co-occupied by TAL1 or as TAL1-down if it showed an absence of or decline in TAL1 upon activation of GATA1. (Details and thresholds are in Supplemental material.) The results strongly support the association of induction with co-occupancy by GATA1 and TAL1, and the counterassociation of repression with no or decreased TAL1 at GATA1 OSs (Table 1). Considering GATA1 OSs, 75% of those co-occupied by TAL1 are in the proximal neighborhood of induced genes, while 64% of those lacking TAL1 or exhibiting a decline in TAL1 are in repressed genes. Considering the genes, 83% of up-regulated genes show co-occupancy by TAL1 at all GATA1 OSs, whereas a greater fraction of repressed genes (38%) than induced genes (17%) have a TAL1-down GATA1 OS. These associations are highly significant by a χ2-test (P = 0.0002 for the GATA1 OS comparison; P = 0.03 for the gene comparison).
Table 1.
Correlation between status of TAL1 at GATA1-occupied DNA segments and direction of regulation of target genes
aThe probability that the counts for the TAL1 status of GATA1 OSs were the same for up- versus down-regulated genes was computed by a χ2-test.
bThe probability that the counts for the genes with the designated TAL1 status of GATA1 OSs were the same for up- versus down-regulated genes was computed by a χ2-test.
However, it is also clear that the amount of TAL1 on GATA1 OSs does not decrease for a substantial number of down-regulated genes. This indicates that more than one mechanism may be used for GATA1-dependent repression in erythroid cells. We note that for the induced genes, the distinctive feature is co-occupancy by TAL1 and GATA1 (Table 1), not an increase in TAL1 upon activation by GATA1 (Fig. 4C).
The Polycomb mark H3K27me3 is placed on a subset of genes repressed by GATA1
Trimethylation of lysine 27 of histone H3 (H3K27me3) is a chromatin modification associated with repression; it is catalyzed by the Polycomb protein complex PRC2 (Muller et al. 2002). Monomethylation of lysine 4 of histone H3 (H3K4me1) is strongly associated with enhancers (Heintzman et al. 2007). In G1E-ER4 cells with activated GATA1-ER, we examined the levels of these two histone modifications throughout the 66-Mb region of chromosome 7 (Zhang et al. 2009), expecting them to distinguish active from inactive genes and perhaps GATA1-induced from GATA1-repressed genes. The overall results reveal abundant histone modifications in large regions that are also bound by transcription factors GATA1, TAL1, and LDB1 and contain GATA1-responsive genes, separated by large regions depleted for histone modifications and transcription factor binding and containing no GATA1-responsive genes (Fig. 1B). The latter appear to be “dead zones” with respect to marks of functional chromatin, and they may represent chromosomal regions with genes absent or stably repressed. Although both histone modifications are found in the same broad regions when viewed at low resolution (Fig. 1B), as expected they often occur in a mutually exclusive manner when examined at higher resolution.
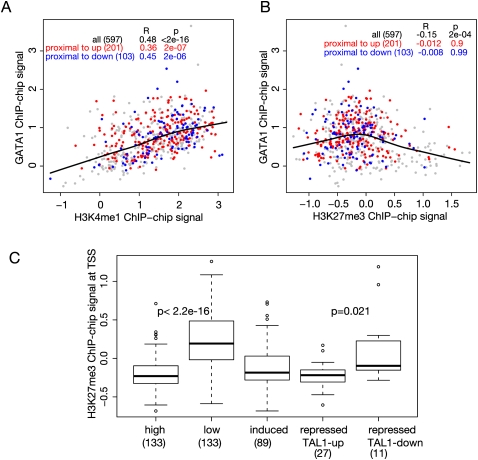
Occupancy by GATA1 is positively correlated with the level of H3K4me1, but most GATA1 OSs have low levels of H3K27 trimethylation (Fig. 5A,B). In fact, the histone modification status is the best predictor of occupancy by GATA1, considerably better than any short DNA sequence motif or motif combination (Zhang et al. 2009). The correlation of occupancy with H3K4 monomethylation is not significantly different whether the GATA1 OS is present within up-regulated or down-regulated genes. Thus, while H3 modification status is an important determinant of occupancy by GATA1, it does not distinguish the directions of response of the presumptive target genes.
Figure 5.
Correlations of histone modifications with occupancy and transcriptional status. (A,B) Scatterplots showing the correlation of GATA1 occupancy (proxied by mean ChIP-chip signal on the y-axis) with the levels of monomethylation of histone H3K4 (H3K4me1, x-axis in A) or with levels of trimethylation of histone H3K27 (H3K27me3, x-axis in B) in each GATA1 OS. The histone modifications were determined in G1E-ER4 cells treated with estradiol. Colors of the dots for GATA1 OSs are red for those associated with up-regulated genes, blue for those associated with down-regulated genes, and gray for all other GATA1 OSs in the 66-Mb region of chromosome 7. The lowess line is for all the data points, and correlations for the different expression categories are given in the inset table in each graph. (C) Boxplot comparing the distributions of H3K27me3 around the TSS of genes in the indicated expression categories. The mean levels of the histone modification around the TSS for each gene in a category were computed. The distributions are significantly different when comparing the high (top quartile of expression levels from the transcriptome analysis) versus low expressed genes (bottom quartile) (P < 2.2 ×10−16) and repressed genes distinguished by co-occupancy between GATA1 and TAL1 (TAL1-down vs. TAL1-up, P = 0.021), using a single-tailed t-test. The numbers of genes in each category are given in parentheses.
In contrast, the level of H3K27me3 around the TSS of the presumptive target gene does distinguish active from inactive genes, and it separates down-regulated genes into two classes. We aggregated the ChIP-chip signal for H3K27me3 around the TSS for each gene in the 66-Mb region and examined the distributions of the mean signals for genes in five response categories. As shown in Figure 5C, genes with the highest expression levels in G1E-ER4 cells (top quartile) are depleted of H3K27me3 around the TSS, while the opposite pattern is seen for chromatin around the TSSs of genes with low or no gene expression (bottom quartile). The distributions of H3K27me3 levels for both induced and repressed genes are lower than that for the low-expression genes (Fig. 5C). However, when the down-regulated genes are partitioned by the TAL1 status of GATA1 OSs in their proximal neighborhood (TAL1-up and TAL1-down, see above), then a separation by level of H3K27me3 is observed. The TSSs of the TAL1-down class of repressed genes have significantly more H3K27me3 than do the TSSs of the TAL1-up class (Fig. 5C).
Thus, genes repressed by GATA1 fall into at least two categories based on the TAL1 status of GATA1 OSs within their proximal neighborhood. One category, containing 38% of the down-regulated genes that could be classified in the 66-Mb region, has at least one GATA1 OS with no TAL1 or in which the level of TAL1 decreases when GATA1 is restored. Notably, this is the category of repressed genes that also has a significant accumulation of the repressive chromatin modification H3K27me3 around the TSS; an example is the Sox6 gene (Fig. 4E). In contrast, the other category of down-regulated genes have GATA1 OSs that are co-occupied by TAL1, and their TSSs accumulate significantly less H3K27 trimethylation than the TAL1-down group, illustrated by Rps17 (Fig. 4E). These differences in transcription factor occupancy and in histone modifications suggest that the mechanism for GATA1-mediated repression differs for the genes in these two categories.
Constraint on GATA1 binding site motifs is associated with induction by GATA1
Consistent with our previous work on the 66-Mb region (Cheng et al. 2008), almost all (87%) of the GATA1 OSs detected genome-wide contain a match to the canonical binding site motif for GATA1, WGATAR. The small minority lacking the motif also have a significantly lower level of occupancy compared with the GATA1 OSs with a WGATAR (Fig. 6B; P < 2.2 × 10−16 using a two-sided Mann-Whitney U test). The low occupancy level coupled with the absence of a WGATAR motif suggests that the interaction between GATA1 and these DNA segments may be indirect, e.g., through a different DNA-binding protein that interacts with GATA1.
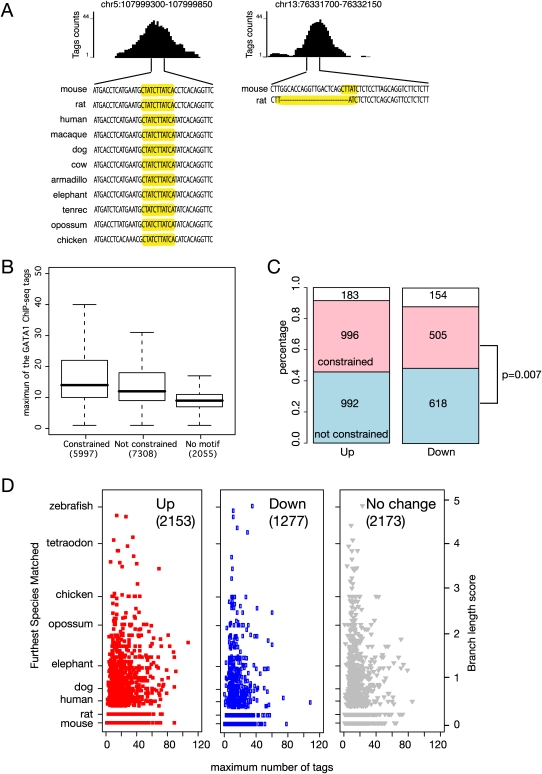
Figure 6.
Correlation of evolutionary constraint on the WGATAR binding site motif and level of occupancy and induction of target genes. (A) Examples of GATA1 OSs with equal occupancy by GATA1, with deep preservation of the WGATAR motif on the left but a rodent-specific motif on the right. (B) Boxplot comparing the distributions of occupancy level in GATA1 OSs after partitioning them by evidence of purifying selection (constraint) on the binding site motif or absence of the motif. Analyses in this figure use all GATA1 OSs in the proximal neighborhood of genes throughout the mouse genome. (C) Bar graphs presenting the percentages (y-axis) and the numbers (in each box) of GATA1 OSs in the proximal neighborhoods of up-regulated and down-regulated genes, again partitioning them by evidence of constraint (red) or not (blue) on the binding site motif. The numbers of GATA1 OSs with no WGATAR motif are given in the white boxes. The P-value is for a χ2-test on motif constraint and direction of regulation. (D) The ranges of constraint on the most deeply conserved WGATAR binding site motif in each GATA1 OS (expressed on the y-axis as the branch length score from mouse to the most distant species to which the motif is preserved) and of occupancy (expressed as the maximum number of sequence tags in the ChIP-seq data). The results are shown for GATA1 OSs in the proximal neighborhood of up-regulated, down-regulated, and nonresponsive genes. Along the left side, the branch length score is calibrated by the comparison species representing the major clades.
The GATA1 OSs containing a WGATAR motif were partitioned by a feature indicative of evolutionary constraint on the motif, which is preservation of the binding site motif in multiple lineages of mammals. The WGATAR motif in some GATA1 OSs is preserved across a considerable phylogenetic depth, whereas others have the motif only in mouse or rodents (Fig. 6A). Preservation of the motif in at least one mammalian lineage outside rodents is strongly indicative of evolutionary constraint (Cheng et al. 2008). While the level of occupancy, as measured by the number of GATA1 ChIP-seq tags, can be the same for constrained and nonconstrained motifs (such as those shown in Fig. 6A), the distribution of occupancy levels is significantly higher in the GATA1 OSs with a constrained motif (Fig. 6B; P < 2.2 × 10−16 by a two-sided Mann-Whitney U test).
Since evolutionary constraint on the WGATAR motif in GATA1 OSs is strongly associated with the ability to enhance gene expression in transfected mammalian erythroid cells (Cheng et al. 2008), we reasoned that constraint on the WGATAR motif could be associated with positive regulation. Indeed, the fraction of GATA1 OSs showing evidence of constraint on the binding site motif is higher for those in the proximal neighborhood of induced genes than for those proximal to repressed genes (46% compared with 40%) (Fig. 6C; P = 0.007 by a χ2-test).
Given that constraint on the motif has a positive association with both level of occupancy and up-regulation, we compared the profiles of occupancy (determined by number of GATA1 ChIP-seq tags) and depth of preservation of the WGATAR motif, measured as the branch length score to the most distant species aligning and preserving the motif (Kheradpour et al. 2007; King et al. 2007; Cheng et al. 2008). While GATA1 OSs within all three expression categories of genes (up-regulated, down-regulated, and nonresponsive) show wide ranges for both features, more of the GATA1 OSs in up-regulated genes have high occupancy and preservation of the motif through mammals (Fig. 6D). Considering preservation of the motif in mammals as reflecting purifying selection and a threshold of 18 reads (which is the top 25% of the GATA1 OSs) as indicating high occupancy, then a larger fraction of GATA1 OS in up-regulated genes (44%) are in this category than are those in down-regulated (14%) or nonresponsive (7%) genes. Thus, we see a strong tendency for GATA1 to bind to constrained motifs in up-regulated genes. However, it is not a feature exclusive to up-regulation; examples of occupancy of WGATAR motifs preserved through fish can be found in down-regulated genes.
Discussion
The data presented here provide a genome-wide view of the relationships between occupancy by a tissue-specific transcription factor, GATA1, and the response of target genes in their level of expression. The G1E cell system is ideal for studying these relationships because both occupancy and expression can be examined after genetic complementation of a critical nuclear factor that induces cellular maturation through a complex, concerted network of transcriptional events. Thus, the responses in gene expression are consequences of restoration of the transcription factor.
Much of the previous work on erythroid genes has emphasized long-range regulation of gene expression by distal CRMs, and thus it might be expected that occupied segments are interspersed over long distances relative to the response genes. Instead, the GATA1-responsive genes tend to cluster in regions with many DNA segments occupied by GATA1, and responsive genes are significantly closer to the GATA1 OSs than are nonresponsive genes. These results focus attention on more local regulation. To be sure, the previously characterized “distal” erythroid regulatory sequences of the HBB and HBA clusters and Gata2 (Grosveld et al. 1987; Vyas et al. 1992; Grass et al. 2006; Wang et al. 2006) are within the 70-kb region that we consider “local” in the present context. The new insight is that CRMs located 100 kb to 1000 kb away either are not common or they are located close to other regulated genes.
Genes whose expression is induced upon activation of GATA1 differ from repressed genes in at least five diagnostic features. GATA1 tends to bind close to the TSS, most often in the first intron but frequently in the proximal 5′ flanking region; this is the case for over 80% of the activated genes. Activated genes have multiple GATA1 OSs within and around them, and they have more GATA1 OSs than do repressed genes. Almost invariably, the GATA1 OSs in the proximal neighborhood of up-regulated genes are also occupied by TAL1. The GATA1-binding site motif within GATA1 OSs shows evidence of evolutionary constraint more frequently for induced genes than for repressed genes. Finally, the region around the TSS of induced genes is depleted of the Polycomb mark H3K27me3, which is associated with repression.
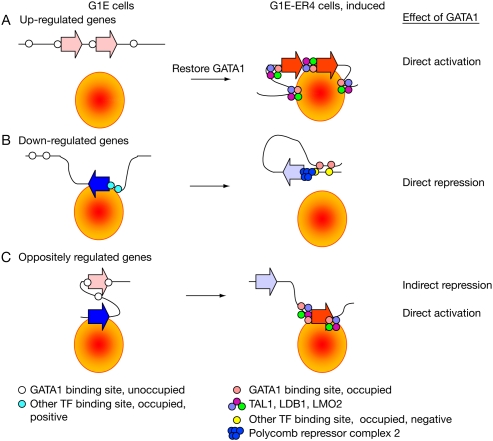
Thus, GATA1-dependent activation of transcription is associated with multiple binding sites for the multiprotein complex containing GATA1, TAL1, LDB1, and LMO2 (Wadman et al. 1997; Anguita et al. 2004; Wozniak et al. 2008). Gene activation in response to restoring GATA1 can be explained by this transcription factor within the multiprotein complex binding to sequences in the proximal neighborhood of the induced gene (Fig. 7A). This in turn can recruit coactivators such as CREBBP and EP300 (Hung et al. 1999; Blobel and Weiss 2001) and increase the frequency of association with the transcriptional machinery or with a transcription factory (Osborne et al. 2004).
Figure 7.
Direct activation and direct versus indirect repression of genes. (A) Up-regulation via direct activation. (B) Down-regulation via direct repression. (C) Down-regulation as a consequence of up-regulation. Gene transcription is diagrammed as occurring in a transcription factory (orange disk with red center); genes not in contact with the factory are not expressed. Genes are shown as boxed arrows, with a bright solid fill indicating active transcription and a light fill indicating no transcription (red for induced, blue for repressed genes). Circles along the line (representing the DNA fiber) are transcription factor binding sites. Open circles indicate a lack of occupancy, and solid colors indicate occupancy; the color code is in the key. The situations prior to and subsequent to restoring GATA1 are on the left and right, respectively. Repressor proteins can recruit the Polycomb repressor complex 2 to methylate histone H3K27, but the chromatin structure is not shown explicitly.
The resulting enhancement of transcriptional activity appears to be subject to purifying selection. GATA1-activated genes are distinguished from repressed genes by more frequent evolutionary preservation of the binding site motif in their GATA1 OSs, reaffirming our previous work showing that negative selection on enhancer activity leads to preservation of the GATA1 binding site motif (Cheng et al. 2008). Similarly, constrained binding site motifs for the glucocorticoid hormone receptor are occupied more frequently around hormone-induced genes than repressed genes (So et al. 2008). These results indicate that constraint on binding site motifs occurs more frequently for positive regulation (enhancement) than for repression. Possible explanations include gene repression being altered in specific lineages more frequently than gene activation or gene repression being subject to less severe purifying selection than gene activation.
Our results on occupancy and response suggest more than one mechanism for repression after restoring GATA1. The CRMs for down-regulated genes studied previously tend to be regulatory switches, activating transcription when occupied by GATA2 but repressing transcription when occupied by GATA1 (Grass et al. 2006; Wozniak et al. 2008). Tripic et al. (2009) discovered several examples of GATA1-bound CRMs associated with down-regulated genes that all lack co-occupancy by TAL1, and they suggest that this is a characteristic of GATA1-dependent down-regulation. We find many additional examples that fit this model and further show that the GATA1-dependent changes in gene expression are significantly associated with the changes in TAL1 occupancy at GATA1 OSs, especially for decreases in TAL1 at repressed genes. Thus, the associations discovered in Tripic et al. (2009) are robust and strongly supported statistically. In the subset of repressed genes with TAL1 reduced or absent from their GATA1 OSs, the regions around the TSSs accumulate the Polycomb repressive histone mark H3K27me3. Presumably, at this class of repressive CRMs, GATA1 recruits transcriptional repressors and corepressors such as PRC2 (shown in the model of Fig. 7B). This supports the independent results from our laboratory (Yu et al. 2009) implicating Polycomb group repressors in GATA1-dependent down regulation.
However, a significant number of down-regulated genes are closely linked to DNA segments co-occupied by GATA1 and TAL1. One possible explanation is that GATA1 and TAL1 co-occupying these DNA segments recruit repressors and corepressors, i.e., carrying out the opposite function to that invoked for up-regulation. TAL1 has been shown to recruit coactivators such as EP300 (Huang et al. 1999) and corepressors such as SIN3A (Huang and Brandt 2000) in erythroid cells.
Another model for GATA1-dependent repression is that the down-regulation occurs as a consequence of up-regulation of other genes, i.e., through indirect effects. If one assumes that the transcriptional capacity is close to fully engaged prior to restoring GATA1 and that no increase in the levels of transcriptional activators and the transcriptional machinery accompanies the activation of new genes, then the up-regulation of previously unexpressed genes by GATA1 will necessarily reduce the access of previously expressed genes to the transcriptional apparatus (Fig. 7C). Such indirect repression could account for an appreciable number of the down-regulated genes. In this case, the GATA1 OSs co-occupied by TAL1 in down-regulated genes are not involved directly in repression, but rather their role in activation of one set of genes leads to down-regulation of other, previously active genes.
Limiting access to the transcriptional machinery would lead to down-regulation, but that leaves open the question of why it would affect specific genes. As diagrammed in Figure 7, the transcriptional apparatus could be localized in the nucleus, in transcription factories (Pombo et al. 2000). Individual genes are preferentially transcribed at particular transcription factories (Osborne et al. 2004, 2007). Genes engaged at a particular transcription factory would be negatively affected by activation of other genes using that factory. Thus, it is possible that genes down-regulated as a consequence of up-regulation of other genes are in proximity in three-dimensional space in the nucleus. This can be tested in future work by exploring interactions of up- and down-regulated genes within the nucleus. While we have discussed the model in terms of access to transcription factories, the model is more general and applies to access to the transcriptional machinery, regardless of whether it is localized in a factory or dispersed through the nucleus.
In addition to the new insights into GATA1-dependent positive and negative regulation in erythroid cells, our data provide an important resource for further investigation of many issues in erythroid differentiation and gene regulation. Thus, we have made all the raw and processed data available both in the Gene Expression Omnibus (Edgar et al. 2002) and in a custom browser (http://main.genome-browser.bx.psu.edu/) based on the UCSC Genome Browser (Kent et al. 2002).
Methods
Gene expression measurements on microarrays
Growth conditions for the cells are described in the Supplemental material. G1E-ER4 cells were induced for 0, 3, 7, 14, 21, and 30 h with 10−7 M beta-estradiol in three independent experiments. RNA from G1E-ER4 cells was extracted using the TRIzol reagent and processed for hybridization to GeneChip Mouse Genome 430 2.0 Arrays (MOE430v2) from Affymetrix. These results are available from the Gene Expression Omnibus (submission GSE18042). Analysis was conducted using the Bioconductor Package in the R project for statistical computing. Hybridization signals on each microarray were first normalized by RMA methods. M versus A plots for the expression arrays (Supplemental material) show that the normalizations were appropriate. The log2 transformed signal intensities were averaged, and the mean value was used to compute the fold change. Pairwise t-tests between 0 h and different times of induction were computed with the Limma package with the BH-FDA adjustment. K-means clustering was performed with kmeans function in the R package after normalization with the standard score on each row. The probes that passed a threshold of twofold enrichment when compared with the zero time point were subgrouped according to their profile of expression over time using the Ordered Restricted Inference for Ordered Gene Expression (ORIOGEN) 5 package (Peddada et al. 2005).
ChIP assay
The ChIP assay was conducted as described previously (Welch et al. 2004). Two different cells were used in this assay: the parental G1E Gata1 knockout cell line and the G1E-ER4 subline (GATA1 restored as a hybrid protein with the hormone binding domain of the estrogen receptor) after activation of the GATA-1-ER hybrid with estradiol. Purified ChIP DNA (10 ng) was further amplified with the WGA amplification kit (Sigma) to obtain enough material to hybridize to the high-density tiling microarray. Amplified material was checked by qPCR on positive and negative control DNA regions to ensure high quality. NimbleGen high-density tiling arrays were hybridized with 4 μg of amplified ChIP DNA for the single microarray covering the 66 Mb in chr 7 or 60 μg for the whole-genome HD2 tiling array set. Peak calling on the ChIP-chip data is described in detail in the Supplemental material.
For the ChIP-seq analysis, an Illumina sequencing library was prepared from a 10-ng sample of GATA1 ChIP DNA from induced G1E ER4, using the ChIP-seq Sample Preparation Kit provided by Illumina. DNA fragments were repaired to generate blunt ends, and a single A nucleotide was added to each end. Double-stranded Illumina adaptors were ligated to the fragments. Ligation products were amplified by 18 cycles of PCR, and the DNA between 200 and 400 bp was gel purified. Completed libraries were quantified with a Quant-iT dsDNA HS Assay Kit. The DNA library was sequenced on the Illumina Genome Analyzer II. Cluster generation, linearization, blocking, and sequencing primer reagents were provided in the Solexa Cluster Amplification kits. The resulting 36-nucleotide sequence reads were mapped to the mouse genome (mm8 assembly) using the program Eland from the Illumina software suite. Only the reads with a unique position in mouse genome were kept for the following analysis. About three-fourths of the reads mapped uniquely to the mouse genome. For GATA1 ChIP-seq, 32,329,253 reads were obtained, of which 23,858,147 mapped uniquely to the mouse genome, and for the input DNA, 20,711,007 reads were obtained, of which 15,651,823 mapped uniquely to the mouse genome. MACs (Zhang et al. 2008) was used to call peaks for GATA1 occupancy, using the parameters mfold = 15, bandwidth = 125.
Peaks of GATA1 occupancy were tested for validation using quantitative PCR as detailed in the Supplemental material.
Acknowledgments
This work was supported by NIH grants DK065806 (R.C.H., M.J.W., and G.A.B.), DK54937 and DK58044 (G.A.B.), HG002238 (W.M.), and HG004718 (Y.Z.). S.C.S. is supported by the Gordon and Betty Moore Foundation. M.J.W. is a Leukemia and Lymphoma Society Scholar.
Footnotes
[Supplemental material is available online at http://www.genome.org. The gene expression data and the Illumina ChIP-seq sequencing read data from this study have been submitted to NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE18042 and GSE18164, respectively. The results on occupancy, chromatin modifications, and expression are available at http://main.genome-browser.bx.psu.edu/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.098921.109.
References
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Weiss MJ. Nuclear factors that regulate erythropoiesis. In: Steinberg MH, et al., editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge University Press; Cambridge, UK: 2001. pp. 72–94. [Google Scholar]
- Cantor A, Orkin S. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Cheng Y, King DC, Dore LC, Zhang X, Zhou Y, Zhang Y, Dorman C, Abebe D, Kumar SA, Chiaromonte F, et al. Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 2008;18:1896–1905. doi: 10.1101/gr.083089.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR. Establishing the transcriptional programme for blood: The SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim SI, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves D, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes & Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol Cell Biol. 2000;20:2248–2259. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Qiu Y, Stein RW, Brandt SJ. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: Combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes & Dev. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DC, Taylor J, Zhang Y, Cheng Y, Lawson HA, Martin J, ENCODE groups for Transcriptional Regulation and Multispecies Sequence Analysis. Chiaromonte F, Miller W, Hardison RC. Finding cis-regulatory elements using comparative genomics: Some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- MacQeen JB. Some methods for classification and analysis of multivariate observations. In: Le Cam LM, Neyman J, editors. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability. University of California Press; Berkeley: 1967. pp. 281–297. [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A, Jacob F, Monod J. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase by E. coli. J Mol Biol. 1959;1:165–178. [Google Scholar]
- Peddada S, Harris S, Zajd J, Harvey E. ORIOGEN: Order restricted inference for ordered gene expression data. Bioinformatics. 2005;21:3933–3934. doi: 10.1093/bioinformatics/bti637. [DOI] [PubMed] [Google Scholar]
- Pombo A, Jones E, Iborra FJ, Kimura H, Sugaya K, Cook PR, Jackson DA. Specialized transcription factories within mammalian nuclei. Crit Rev Eukaryot Gene Expr. 2000;10:21–29. [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci. 2008;105:5745–5749. doi: 10.1073/pnas.0801551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR. Cis-acting sequences regulating expression of the human α-globin cluster lie within constitutively open chromatin. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz G, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NL1 proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang Y, Cheng Y, Zhou Y, King DC, Taylor J, Chiaromonte F, Kasturi J, Petrykowska H, Gibb B, et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Keles S, Lugus JJ, Young KH, Boyer ME, Tran TM, Choi K, Bresnick EH. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol Cell Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Xu J, Cheng Y, Hardison RC, Weiss MJ, Orkin SH, et al. Involvement of Polycomb Repressor Complex 2 in GATA-1 mediated gene silencing during erythroid maturation. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.11.002. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Poisson approximation for significance in genome-wide ChIP-chip tiling arrays. Bioinformatics. 2008;24:2825–2831. doi: 10.1093/bioinformatics/btn549. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu W, Cheng Y, King DC, Harris RS, Taylor J, Chiaromonte F, Hardison RC. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucl. Acids Res. 2009 doi: 10.1093/nar/gkp1747. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Barrera LO, Ren B, Wu YN. ChIP-chip: Data, model, and analysis. Biometrics. 2007;63:787–796. doi: 10.1111/j.1541-0420.2007.00768.x. [DOI] [PubMed] [Google Scholar]