Abstract
Very high-throughput sequencing technologies need to be matched by high-throughput functional studies if we are to make full use of the current explosion in genome sequences. We have generated a very large bacterial mutant pool, consisting of an estimated 1.1 million transposon mutants and we have used genomic DNA from this mutant pool, and Illumina nucleotide sequencing to prime from the transposon and sequence into the adjacent target DNA. With this method, which we have called TraDIS (transposon directed insertion-site sequencing), we have been able to map 370,000 unique transposon insertion sites to the Salmonella enterica serovar Typhi chromosome. The unprecedented density and resolution of mapped insertion sites, an average of one every 13 base pairs, has allowed us to assay simultaneously every gene in the genome for essentiality and generate a genome-wide list of candidate essential genes. In addition, the semiquantitative nature of the assay allowed us to identify genes that are advantageous and those that are disadvantageous for growth under standard laboratory conditions. Comparison of the mutant pool following growth in the presence or absence of ox bile enabled every gene to be assayed for its contribution toward bile tolerance, a trait required of any enteric bacterium and for carriage of S. Typhi in the gall bladder. This screen validated our hypothesis that we can simultaneously assay every gene in the genome to identify niche-specific essential genes.
Current nucleotide sequencing technologies are generating data at a phenomenal rate, allowing the generation of complete nucleotide sequences of a large number of genomes and facilitating the high-throughput identification of genes (Hall 2007; MacLean et al. 2009). One of the next important challenges for molecular biology is to use high-throughput techniques to help determine functions for these genes. The question of which genes are required for cellular viability is of fundamental importance to biology. Recent developments have seen estimates of the minimal gene set required for bacterial cell viability (Baba et al. 2006; Liberati et al. 2006; Gallagher et al. 2007; de Berardinis et al. 2008; French et al. 2008). Such studies performed in the laboratory, by their nature, identify those bacterial genes required for viability under specific laboratory growth conditions. Signature-tagged mutagenesis (STM) (Hensel et al. 1995) and transposon-site hybridization (TraSH) (Sassetti et al. 2001) are methods that make use of hybridization to identify genes disrupted by transposon insertions. They are also negative selection methods that have been used to identify “niche-specific” virulence genes in bacterial pathogens (for review, see Andrews-Polymenis et al. 2009). Most recent bacterial transposon mutant library screens have used a few thousand transposon mutants, which represent, on average, several insertions per gene (Salama et al. 2004; Glass et al. 2006; Liberati et al. 2006; Gallagher et al. 2007; Laia et al. 2009). A more recent method, transposon mediated differential hybridization (TMDH) (Charles and Maskell 2001), has been used to analyze approximately one million mutants to identify essential genes in Staphylococcus aureus (Chaudhuri et al. 2009). However, these approaches are all suboptimal, due to inaccuracy in the estimation of the transposon insertion site from microarrays, and because some genes, especially those which are smaller, will be missed by chance.
Illumina (formerly Solexa) sequencing technology generates short sequence reads (currently up to 100 bases) from very large numbers of DNA fragments. The millions of sequence reads generated allow a whole bacterial genome to be resequenced in one experiment (Hernandez et al. 2008; Holt et al. 2008). Normally, oligonucleotide linkers are ligated to randomly sheared genomic DNA fragments, all fragments in the pool are PCR amplified using universal primers, and sequencing-by-synthesis is performed using complementary oligonucleotide sequencing primers (Bentley et al. 2008). However, by adapting the sample preparation so that fragment pools are PCR amplified using one universal primer, and a second primer that is complementary to one end of the transposon, we have been able to generate simultaneously several million sequence reads adjacent to transposon insertion sites for a pool of ∼1.1 million transposon mutants.
This high-throughput approach, which we have called TraDIS (transposon-directed insertion-site sequencing), was used to investigate the essential gene set of the bacterium Salmonella enterica serovar Typhi (S. Typhi) under both standard laboratory and biologically relevant conditions. S. Typhi is the bacterial agent of typhoid fever and is estimated to cause over 22 million cases and 220,000 deaths every year, mainly in regions of the world where sanitation and clean supplies of drinking water are inadequate (Crump et al. 2004). An estimated 5% of typhoid patients continue to excrete S. Typhi for many years due to chronic infection of the gall bladder (Parry et al. 2002). Such typhoid carriers not only pose a significant health risk to others, but have a higher risk of developing cancers of the gall bladder, pancreas, and large bowel (Caygill et al. 1995; Dutta et al. 2000; Shukla et al. 2000). Carriers also provide a reservoir for S. Typhi, contributing significantly to the persistence of typhoid in endemic regions (Roumagnac et al. 2006). The gall bladder is the site of bile storage and therefore the long-term persistence of S. Typhi in this organ requires genes involved in bile resistance. Identification of such genes will enable a greater understanding of how enteric bacteria resist the toxic effects of bile in the human gut and will also contribute to the future development of therapeutics targeted to the treatment of S. Typhi carriage.
Results
Assaying one million transposon mutants
Using a derivative of Tn5, a transposon mutant library was generated for S. Typhi comprising a pool of an estimated 1.1 million individual mutants. We used the attenuated strain of S. Typhi Ty2, CVD908-htrA, which has deletion mutations in aroC, aroD, and htrA. Genomic DNA was extracted from this pool for nucleotide sequencing from the transposon into the adjacent sequences of the insertion sites. For each sequencing run, two lanes of the Illumina sequencing flow cell generated between seven million and 11 million nucleotide sequence reads, of which 75%–90% included an identical match to the 10 base transposon nucleotide sequence tag (Table 1; Supplemental Table S1). Of the tagged sequence reads, up to 70% (minimum 2.4 million per run) could be mapped unambiguously to the S. Typhi Ty2 chromosome sequence. This allowed the identification of between 200,000 and 300,000 individual transposon insertion sites; an average of one insertion site for every 15–20 base pairs (bp) (Table 1). Combining four sequencing lanes yielded over 370,000 insertion sites, an average of more than 80 inserts per gene (Supplemental Table S1). This is far in excess of the number of insertions achieved previously for bacterial transposon mutant libraries, which have reported an average of five or just over nine inserts per gene (Salama et al. 2004; Gallagher et al. 2007), and makes possible the assay of every gene in the genome.
Table 1.
Transposon inserts recovered from passage pools
One sequencing library was prepared for each sample from the 5′ end of the transposon, and sequenced on two lanes. Data are represented as the combined results of both lanes.
The distribution of mapped sequence reads across the whole genome is shown in Figure 1A. Their positions were compared with the genome sequence annotation to determine the number and precise position of sequence reads for each individual gene. As the number of sequence reads that map to a unique position within a gene is dependent upon gene length, we normalized the data by dividing the number of unique insertion sites for any given gene by the gene length to give an insertion index. A frequency distribution of the insertion index for all the annotated genes in the genome gives a clear bimodal distribution (Fig. 1B). The left-most peak represents genes in which transposon insertions significantly inhibit cellular growth, leading to the absence or greatly diminished representation of insertions in these genes. The right-most peak represents genes into which transposon insertion does not adversely affect cellular growth, so that large numbers of insertions can occur in these genes. Using this bimodal distribution, we calculated likelihood ratios (LR) to determine whether genes were more likely to be in the essential peak. A log2-LR of less than −2 was taken as the cutoff for essentiality (at which value a gene is at least four times more likely to be essential than not), and a log2-LR of greater than 2 was taken as the cutoff for nonessentiality (at which value a gene is at least four times more likely to be nonessential than essential).
Figure 1.
Essential genes in S. Typhi. (A) Frequency and distribution of transposon directed insertion-site sequence reads across the entire S. Typhi Ty2 genome for a pool of one million transposon mutants. The y-axis shows the number of mapped sequence reads within a window size of 3. ori and ter indicate the approximate positions of the replication origin and terminus, respectively. (B) Bimodal frequency distribution of insertion index (number of inserts per gene divided by gene length). Genes with insertion indices in the leftmost peak represent those that have none, or very few insertions (essential genes). (C–E) Detailed plots generated using Artemis (Rutherford et al. 2000) showing distribution of sequence reads across selected regions of the S. Typhi Ty2 genome. The y-axis shows the number of mapped sequence reads within a window size of three. The maximum number of sequence reads within each plot is shown (top right) and the position of annotated genes relative to the plotted sequence reads is indicated below the distribution plot; gray boxes represent genes, and white boxes pseudogenes. (C) The essential plsC gene and topoisomerase IV genes, parC and parE, showing the absence of transposon insertions. (D) Sequence reads mapping to regions between essential genes and the fabZ lpxA lpxB rnhB dnaE accA operon disrupted by insertions into rnhB (E) show that the Tn5-derived transposon is capable of generating many nonpolar mutations.
S. Typhi genes essential for growth
Three hundred fifty-six genes had a log2-LR of less than −2; thus, these genes are essential for growth under standard laboratory conditions based on the above criteria. Conversely, 4162 genes had a log2-LR greater than 2 and are thus nonessential. Nineteen genes had log2-LRs between the two cutoff values and so it is not possible to assign these as essential or nonessential with the same degree of confidence. In addition, the density of insertions across the genome is such that a 60 bp region without insertion has only a 1% chance of occurring randomly. Thus, we cannot make conclusions with confidence for very short genes (<60 bp) with no insertions, which may have been missed by chance. However, there are only two annotated genes that are less than 60 bp long and have no mapped insertion sites. Thus, we can effectively assay all but two very short annotated genes, and draw conclusions with statistical confidence for 4518 of 4537 (99.6%) annotated genes in the genome. The genomic locations of essential and nonessential genes are shown in Figure 2.
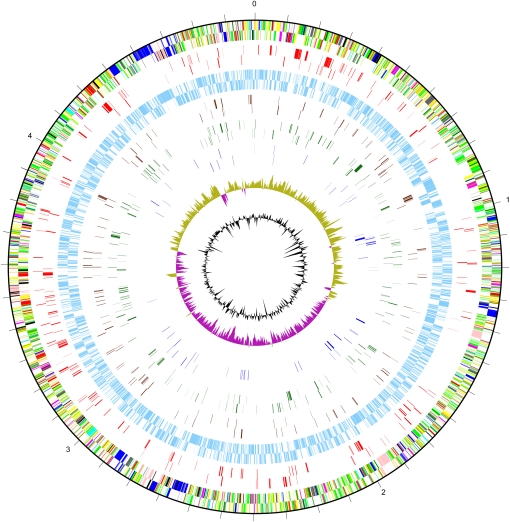
Figure 2.
Genetic map showing results of simultaneous assay of the whole S. Typhi Ty2 genome. The outer scale is marked in megabases. Circles range from 1 (outer circle) to 8 (inner circle) and represent genes on both forward and reverse strands. Circle 1, all genes (color-coded according to function: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, membranes/surface structures; cyan, degradation of macromolecules; purple, degradation of small molecules; yellow, central/intermediary metabolism; light blue, regulators; pink, phage/IS elements; orange, conserved hypothetical; pale green, unknown function; brown, pseudogenes); circle 2, essential genes (red); circle 3, nonessential genes (light blue); circle 4, genes involved in bile tolerance (brown); circle 5, genes advantageous for growth over six passages (dark green); circle 6, genes disadvantageous for growth over six passages (dark blue); circle 7, GC bias [(G – C)/(G + C)]; khaki indicates values > 1; purple < 1; circle 8, %(G + C) content.
Many of the 356 essential genes are required for fundamental biological processes, including cell division, DNA replication, transcription, and translation (Table 2). The full list is available in Supplemental Table S2. A few are worthy of note, including DNA polymerase III, a multimeric enzyme encoded by nine subunit genes, seven of which are identified as essential. The remaining two genes are holE, (log2-LR = 4.83), and holC (log2-LR = 5.36), which are unlikely to be essential. All the aminoacyl-tRNA synthetase genes were identified as candidate essential genes except for trpS (t4024) and trpS (t4557), which are both tryptophanyl-tRNA synthetases and therefore mutually redundant. Similarly, of the 11 genes that are involved in peptidoglycan biosynthesis, nine were assigned as essential, while ddlA and ddlB were assigned as nonessential; both these genes perform the same function. Of the 356 S. Typhi candidate essential genes identified by TraDIS, 256 (∼70%) are also essential in Escherichia coli (Baba et al. 2006), including 110 of the genes in Table 2. Of the 100 genes essential in S. Typhi but not E. coli, almost half are involved in energy metabolism or regulation of gene expression.
Table 2.
All known genes coding for fundamental biological processes in S. Typhi
Gene names in bold are also essential in E. coli (Baba et al. 2006).
aThis gene falls into the unassigned region, with a log2-LR of −1.8.
One particularly intriguing case is recA, mutants of which exist in E. coli, suggesting that this is not an essential gene in this bacterium (Baba et al. 2006). However, our data indicate that in S. Typhi, recA is a candidate essential gene (log2-LR = −11.5). In support of this, multiple attempts in our laboratory to generate a recA mutant in S. Typhi, using the suicide vector allelic-exchange method (Turner et al. 2006), have failed (see Supplemental material). During bacterial growth, RecA is involved in DNA replication and the reactivation of stalled replication forks. This occurs via the “restart” primosome, a multimeric enzyme complex made up of seven proteins encoded by dnaTBCG and priABC (Sandler and Marians 2000). In E. coli, priC mutants have little phenotypic effect on growth (Sandler et al. 1999), and in S. Typhi, priC is a pseudogene (Parkhill et al. 2001). However, without priC, there is only a priA-dependent pathway for replication fork restart and our results suggest that this is not viable in a recA mutant background.
The high density of insertions across the S. Typhi genome allows a clear demarcation between many candidate essential and nonessential genes. For example, topoisomerase IV, an essential enzyme for maintaining DNA supercoiling, is encoded by parC and parE and almost no insertion sites were identified for these genes, or for plsC, a lipid biosynthesis gene (Fig. 1C). It is of note that the genome coverage of the million mutant library is so great that insertions into small intergenic regions between essential genes such as pyrH, frr, and dxr can also be seen clearly (Fig. 1D). Elsewhere, the intergenic region between essential genes leuS and rlpB is only 14 bp, but we observed six sequence reads mapping to one insertion site here without any insertions into the adjacent coding sequence.
Transposon insertions into operons may produce polar mutations; in such cases an insertion into a nonessential gene in the operon may disrupt the expression of an essential gene downstream. This would indicate incorrectly that the gene with the insertion is essential. Thus, some caution should be exercised when making conclusions about the essentiality of genes in operons. These queries can only be confirmed when one gene is disrupted, leaving expression of the other intact. However, many instances can be observed where numerous transposon insertion sites occur immediately 5′ to an essential gene, indicating that many insertions for this transposon are nonpolar. For example, the fabZ lpxA lpxB rnhB dnaE accA operon includes candidate essential genes, with the exception of rnhB. Large numbers of transposon insertions are present in rnhB in the middle of this operon (Fig. 1E), suggesting that this does not disrupt expression of the downstream genes, at least in some of the mutants.
Genes advantageous or disadvantageous during growth
Transposon insertion into some genes severely reduces the growth rate or arrests growth completely and these genes will be identified as essential. However, there will be many other genes into which insertions reduce growth rate to a lesser degree, and it is likely that there is a continuum of growth rates for various mutants in the one million mutant pool. In order to see if other such genes could be identified, the one million mutant pool was passaged six consecutive times in nutrient-rich broth at 37°C. During these passages, the number of unique insertion sites identified decreased from over 370,000 prior to passaging to under 260,000 sites after the sixth passage (Fig. 3A), indicating a reduction in the number of different mutants in the pool. Following the first passage, we recovered very few insertion sites for 100 genes (log2-LR < −2). Previously, in the original mutant pool, 94 of these genes had a large number of insertion sites (log2-LR > 2), while the other six were from the 19 that previously could not be assigned as essential or not (log2-LR between −2 and 2). We termed these 100 genes “advantageous” for growth. By the third and sixth passages, a further 78 and 96 genes, respectively, fell into this category. Overall, in S. Typhi 356 candidate essential genes and 274 genes advantageous for growth in nutrient-rich broth at 37°C were identified using TraDIS (Fig. 2; Supplemental Table S2).
Figure 3.
Changes in composition of the mutant pool with passaging. (A) Frequency and distribution of transposon directed insertion-site sequence reads across the entire Ty2 genome in the original 1 million mutant pool (top) and following growth in broth of the pool through one, three, and six passages. The plot has a window size of 3. (B) Frequency and distribution of sequence reads across the flagella (fli) operon at passage 6 compared to the original mutant pool (passage 0) showing an increase in the number of flagella gene mutants following growth. (C) With genes divided into functional classes based on the S. Typhi CT18 annotation, the observed number of different transposon insertion sites per functional gene class is expressed as a proportion of the insertion sites expected if sites were distributed randomly for passages 0 and 6. Values greater than 1 indicate that insertions into the gene class are better tolerated, while values less than 1 indicate that insertions into the gene class are more poorly tolerated. (D) Passage 0 value minus Passage 6 value from C plotted to emphasize how much the proportion of expected differs between these passages. Larger values indicate a more important role during growth in broth for the gene class than smaller or negative values. Path./adapt./chap., pathogenicity, adaptation, chaperones; Mem./surface structures, membrane/surface structures; Deg., degradation; Cen./int. metabolism, central/intermediary metabolism; Cons. hypothetical, conserved hypothetical.
After the sixth passage, a greatly increased number of sequence reads mapped to a relatively small number of genes, compared to the original one million mutant pool. These included 30 genes involved in flagella biosynthesis and assembly (Fig. 3B; Supplemental Table S3). These may represent metabolically costly genes or genes that are disadvantageous for growth under laboratory conditions. Insertions within these genes appear to have given the bacteria a fitness advantage, allowing them to out-compete others, and leading to their overrepresentation in the mutant pool. Thus, in order to account for their persistence in the genome, there must be very strong selection for these genes during S. Typhi passage through natural environments.
Classifying genes by function gave a broad overview of the type of gene function for which insertions significantly affected cellular growth. We calculated the expected number of insertion sites per functional class (based on the average number of insertion sites across the whole genome and the cumulative length of all the genes in the functional class), and determined whether the observed number was higher or lower than the expected. Prior to passage, in the original mutant pool, more insertions than expected were recovered in pseudogenes, phage/IS elements, and pathogenicity/adaptation/chaperone genes, indicating that these gene classes are generally less important for growth in nutrient-rich laboratory conditions (Fig. 3C). Fewer insertions than expected were recovered for information transfer (replication, transcription, and translation) and energy metabolism genes in particular. This is unsurprising as these classes include the genes encoding essential proteins, such as ribosomal subunits, RNA/DNA polymerases, and tRNA synthetases. After six passages in nutrient-rich broth, the classes showing the most marked difference between the original pool and the sixth passage were energy metabolism, central and intermediary metabolism, degradation of macromolecules, and information transfer (Fig. 3D), all of which had even fewer insertions than expected when compared to the original pool.
Candidate genes involved in bile tolerance
The ability to resist high concentrations of bile is of particular importance for S. Typhi carriage in the gall bladder, but bile tolerance is also required for the many other species of bacteria that inhabit the mammalian alimentary tract. In order to identify many of these genes, and show that TraDIS can be used to assay almost every gene simultaneously to determine function in different niches, the S. Typhi mutant pool was grown in nutrient broth culture supplemented with 10% ox bile. We identified 169 genes in which the number of insertions showed a significant fold difference (P < 10−5) between cultures grown with and without ox bile (Fig. 4; Supplemental Table S4).
Figure 4.
Bile tolerance. (A) With genes divided into functional classes based on the S. Typhi CT18 annotation, the observed number of different transposon insertion sites per functional gene class is expressed as a proportion of the insertion sites expected if sites were distributed randomly for growth in the presence or absence of bile (abbreviations as in Fig. 3). Values greater than 1 indicate that insertions into the gene class are better tolerated, while values less than 1 indicate that insertions into the gene class are more poorly tolerated. (B) Data from A plotted to show how much the proportion of expected differs between LB (no bile) and bile (LB supplemented with 10% ox bile). A positive value indicates more insertions than expected in LB relative to bile; a negative value indicates fewer insertions than expected in LB relative to bile. (C) Detailed plot generated using Artemis (Rutherford et al. 2000), comparing the frequency and distribution of transposon directed insertion site sequence reads across the O-antigen biosynthesis (wba) genes following growth in the absence (top distribution) or presence (bottom distribution) of ox bile. The y-axis shows the number of mapped sequence reads within a window size of 3. The maximum number of sequence reads within each plot is shown top right. After growth in the presence of ox bile the number of transposon insertions is much reduced in this region. (D) Similar to (C), showing the frequency and distribution of sequence reads in and adjacent to the pagP gene.
All the genes from each growth condition were sorted by functional class and the number of insertions per class was compared to that expected if insertions were random (Fig. 4A). When compared to the LB growth condition, four functional classes contained fewer insertions in the presence of bile: energy metabolism, membrane/surface structures, degradation of macromolecules, and central/intermediary metabolism (Fig. 4B), indicating an important role for these gene classes during laboratory growth.
Among the 169 genes identified as being required for bile tolerance were those involved in the synthesis of lipopolysaccharide (LPS), which makes up the outer leaflet of the Gram-negative bacterial outer membrane. Of the 10 waa genes involved solely in LPS core biosynthesis, eight were required for growth in the presence of bile (the remaining two were essential). Mutants lacking waaL, waaK, and waaI genes showed the greatest significant fold difference (log2 fold changes of 139, 91, and 90, respectively) in the number of insertions between growth in the presence or absence of bile. Mutants for these genes probably show the greatest susceptibility to bile. All 4 wba genes involved solely in LPS core biosynthesis were also required for growth in the presence of bile, as were a number of wba genes involved in capsular polysaccharide biosynthesis (Fig. 4C). Several LPS biosynthetic genes have been identified previously as being involved in bile tolerance and it is known that mutants of E. coli and other Salmonella serovars defective in LPS biosynthesis are attenuated for colonization of the alimentary tract of animals (Nevola et al. 1985; Licht et al. 1996; Turner et al. 1998; Moller et al. 2003).
Other genes previously implicated in bile tolerance were also identified in this assay. The acrAB and tolC genes encode an efflux complex involved in the excretion of bile acids from the cell cytoplasm. S. Typhimurium with mutations in these genes was susceptible to bile (Prouty et al. 2004), and our acrAB deletion mutant of S. Typhi showed a minimum inhibitory concentration (MIC) of ox bile at least 60 fold less than the parental strain. In addition, rob, which is required for bile tolerance in E. coli, and whose product induces expression of acrAB (Rosenberg et al. 2003), was also identified in our assay. Other genes previously identified in S. Typhimurium and confirmed here as being required for bile tolerance in S. Typhi are seqA, dam (Prieto et al. 2007), phoP, and phoQ (van Velkinburgh and Gunn 1999).
Previously, none of the genes comprising the phoPQ regulon were found to be required for bile tolerance in S. Typhimurium (van Velkinburgh and Gunn 1999). However, our data show that the phoPQ induced pagP gene (encoding a lipid A acylation protein required for antimicrobial peptide resistance [Guo et al. 1998]) was required for bile tolerance (Fig. 4D). Another gene of note is hupA, which is required by S. Typhimurium for colonization of the alimentary tract of poultry (Turner et al. 1998), but was not known to have an effect on bile tolerance. Our assay also identified a number of genes encoding membrane-associated proteins, including mrcA and mrcB, penicillin binding proteins 1a and 1b, and sanA, which has been previously reported to play a role in the resistance of E. coli to vancomycin due to its barrier function in the cell envelope (Rida et al. 1996). Alongside these, there are also over 30 putative or hypothetical genes, which warrant further investigation into their role in bile tolerance (Supplemental Table S4).
Discussion
A variety of previous methods has identified a number of essential and niche-specific genes, but to do this effectively on a genome-wide scale has required the use of microarrays to indirectly assay the sites of transposon insertion. Microarrays have their drawbacks: resolution is limited, and distinguishing a positive from a negative signal for some microarray features can be difficult. With sequencing, the signal is of a “digital” nature; any sequence read that has the 10-bp transposon tag with adjacent genomic sequence is almost certainly an indication of the exact position of a transposon insertion site.
The combination of an extremely large transposon mutant pool and high-throughput Illumina sequencing from the transposon insertion sites has brought an unparalleled degree of resolution to a transposon mutagenesis screen. Indeed, the number of insertions was sufficiently great that a gap between insertion sites of 39 bp had a less than 5% probability of occurring by chance, indicating the resolution available from this approach. This has allowed us to distinguish between essential and nonessential genomic regions to within a few base pairs and to confidently assign 99.6% of all the genes in the genome as essential or nonessential. In addition, there are sufficient insertions to allow the assay of nearly every gene in the genome for a particular growth condition; only small genes, with few or no transposon insertions, cannot be assayed. Thus, TraDIS can be used for the accurate estimation of minimal gene sets, and as a very effective negative selection method.
Transposon insertion site bias is often cited as a limitation of transposon mutagenesis techniques. We detected a bias toward insertion in A + T-rich regions (Supplemental Fig. S1), but the frequency of insertion achieved by the number of transposon mutants ensured that even G + C-rich regions contained numerous insertions. Given that transposon insertion sites delimited essential genes, we have no reason to believe that transposon insertion bias has had any bearing upon our conclusions.
We have also used this technique in S. Typhimurium with a pool of one million transposon mutants. The sequence data from this library is currently being analyzed, but preliminary results indicate genome coverage is of an equivalent density to the S. Typhi library. This technique should work for any bacterium in which a large enough mutant library can be obtained, which is dependent upon transformation rates and a suitable transposon.
Passaging the full transposon library six times allowed the identification of genes that are advantageous for growth without being absolutely essential. More strikingly, a small set of genes were identified that are apparently disadvantageous during nutrient-rich growth under laboratory conditions. This is the first time, to our knowledge, that a transposon mutagenesis screen has been used successfully to determine genes costly to a growth condition, as well as those required for it.
It is of note that a small number of insertions were identified in some genes considered essential by our criteria. This is probably because such mutants, whilst unable to grow and divide on the initial selection plate, are still present and thus harvested, and therefore represented at very low frequency in the mutant pool. TraDIS is sufficiently sensitive to identify these sequences occasionally, confirming that insertions into essential genes do occur. The passage data show that this small number of insertions in some essential genes is lost over time.
We have identified at least 169 genes involved in bile tolerance including many not implicated previously. These include 30 putative or hypothetical genes and over 10 regulatory genes, the majority of which have not previously been linked to survival in bile. Taken together, our results highlight a number of possible new targets for therapies aimed at reducing S. Typhi carriage in the gall bladder. There are also wider implications for basic pathogen biology; the ability to identify genes that are costly and advantageous in addition to essential gene sets should reveal more about the functions necessary to support these bacteria throughout their entire disease cycle.
Methods
Strain
The S. Typhi strain used in these studies is WT26 pHCM1, a derivative of the attenuated Ty2-derived strain CVD908-htrA, which has deletion mutations in aroC, aroD, and htrA (Tacket et al. 1997). WT26 (Turner et al. 2006) has a point mutation in gyrA conferring reduced susceptibility to fluoroquinolone antibiotics and the multiple antibiotic resistance plasmid, pHCM1, has been introduced. These additions are intended to allow the transposon mutant library to be used for fluoroquinolone resistance and plasmid studies.
Preparation of transposomes
The transposon is a derivative of EZ-Tn5 < R6Kγori/KAN-2> (Epicentre Biotechnologies) with outward oriented T7 and SP6 promoters at each end, respectively, and with R6Kγori deleted. The transposon was amplified using oligonucleotides 5′-CTGTCTCTTATACACATCTCCCT and 5′-CTGTCTCTTATACACATCTCTTC with Pfu Ultra Fusion II, (Stratagene) and the amplicon was phosphorylated using polynucleotide kinase (New England Biolabs). Four hundred nanograms of this DNA were incubated with EZ-Tn5 transposase (Epicenter Biotechnologies) at 37°C for 1 h then stored at −20°C.
Preparation of bacterial cells for transformation
Bacterial cells for electrotransformation were grown in 2× TY broth to an OD600 of 0.3–0.5, then cells were harvested and washed three times in 1/2 vol 10% glycerol. Cells were finally resuspended in 1/1000 vol 10% glycerol and stored at −80°C. Sixty microliters of cells were mixed with 0.2 μL of transposomes and electrotransformed in a 2-mm electrode gap cuvette using a Bio-Rad GenePulser II set to 1.4 kV, 25 μF, and 200 Ω. Cells were resuspended in 1 mL of SOC medium (Invitrogen) and incubated at 37°C for 2 h then spread on L-agar supplemented with “aro mix” (40 μg/mL each of L-phe and L-trp, and 10 μg/mL each of p-aminobenzoic acid and 2,3-dihydroxybenzoic acid final concentration) and kanamycin at 7.5 μg/mL. After incubation overnight at 37°C, the number of colonies on several plates was estimated by counting a proportion of them, and from this the total number of colonies on all plates was estimated conservatively. Kanamycin resistant colonies were resuspended in sterilized deionized water using a bacteriological spreader.
Normally, 10 or more electrotransformations would be performed to generate one batch of mutants. The number of mutants in each batch ranged from estimates of 42,000–146,000. From the estimated total number of mutants and using the OD600 to estimate the cell concentration in each batch, volumes containing approximately similar numbers of mutants from 13 batches were pooled to create the mutant library mixture estimated to include 1.1 million mutants.
Transposon library passage
Approximately 2 × 108 viable mutants were inoculated into 500 mL of LB broth and grown overnight at 37°C with shaking. Subsequently, 1 mL of this culture was transferred to 500 mL of fresh LB broth and similarly grown overnight. This was continued for a total of six passages. Genomic DNA was extracted directly from cells harvested from 5 mL of each passage and from ∼5 × 109 cells of the original 1.1 million mutant pool, using tip-100-g columns and the genomic DNA buffer set from Qiagen. For the bile tolerance experiment, ∼3 × 107 viable mutants were added to 1 mL of LB-broth supplemented with aro mix and 0.02% (w/v) Oxgall (Oxoid). The cell suspension was incubated at 37°C for 50 min then transferred to 50 mL of fresh LB-broth supplemented with aro mix and 2% (w/v) Oxgall. This culture was growth overnight at 37°C with shaking after which 1 mL was transferred to 50 mL of fresh LB-broth supplemented with 10% (w/v) Oxgall and aro mix, and this was grown for 24 h at 37°C. Genomic DNA was extracted from cells harvested from 5 mL of the culture as above.
Nucleotide sequencing
Five micrograms of genomic DNA was fragmented to an average size of 300 bp by Covaris AFA (Quail et al. 2008) and Illumina DNA fragment library preparation was performed following the manufacturer's instructions, but using 1.5× the recommended reagent volumes in each step. Ligated fragments were run in a 12-cm 2% agarose gel in 1× TBE buffer, at 6 V/cm without the preceding column clean-up step. After 45 min, fragments corresponding to an insert size of 250–350 bp were excised, and DNA was extracted from the gel slice without heating (Quail et al. 2008). The DNA was quantified on an Agilent DNA1000 chip, following the manufacturer's instructions.
To amplify the transposon insertion sites, 22 cycles of PCR were performed using a transposon-specific forward primer and a custom Illumina reverse primer (see Supplemental material), and 100 ng of DNA fragment library per reaction. Amplified libraries were cleaned up with a QiaQuick PCR product purification column following the manufacturer's instructions, eluted in 30 μL of EB, and then quantified by qPCR (Quail et al. 2008). The amplified DNA fragment libraries were sequenced on paired or single end Illumina flow cells using an Illumina GAII sequencer, for 36 or 54 cycles of sequencing, using a custom sequencing primer and 2× hybridization buffer (see Supplemental material; Supplemental Fig. S2). This primer was designed such that the first 10 bp of each read was transposon sequence. All raw sequence data can be accessed at ftp://ftp.era.ebi.ac.uk/vol11/ERA000/ERA000097 and metadata files at ftp://ftp.era-xml.ebi.ac.uk/ERA000/ERA000097.
Analysis of nucleotide sequence data
Sequence reads from the Illumina FASTQ files were parsed for 100% identity to the last 10 bp of the transposon (TAAGAGACAG). Matching sequence reads were stripped of this transposon tag, converted to Sanger FASTQ format and mapped to the S. Typhi Ty2 chromosome using MAQ version MAQ-0.6.8 (Li et al. 2008). The output from the MAQ mapview command was used to determine the first nucleotide position to which each read mapped, giving a precise insertion site. The number and frequency of insertions mapping to each nucleotide in the S. Typhi genome were then determined for each growth condition. Comparison of these data with gene boundaries defined from the GenBank annotation (accession no. AE014613) enabled the number of sequence reads and the number of different insertion sites to be determined for every gene. Genes were grouped into functional classes based on the S. Typhi CT18 annotation (Parkhill et al. 2001). The number of insertions expected per functional class was calculated by dividing the total number of insertions recovered for a particular growth condition by the summed total of all gene lengths within that class.
Statistical analyses
Essential genes
As the number of insertion sites for any gene is dependent upon the gene length, the values were made comparable by dividing the number of insertion sites by the gene length to give an “insertion index” for each gene. The distribution of insertion indices is bimodal, corresponding to the essential (mode at 0) and nonessential models. For the original mutant pool and each passage condition, we fitted gamma distributions for the two modes using the R MASS library (http://www.r-project.org). Log2-likelihood ratios (LR) were calculated between the essential and nonessential models for each condition and we called a gene essential if it had a log2-LR of less than −2, indicating it was at least four times more likely according to the essential model than the nonessential model. Genes were assigned “nonessential,” if they had a log2-LR of greater than 2.
Comparison of culture passages and growth in the presence and absence of bile
For each pair of conditions tested (A,B), we calculated the log2 fold change ratio Sg,A,B in the number of observed reads ng,A, ng,B for every gene g as Sg,A,B = log2\frac(ng,A + 100)(ng,B + 100). The correction of 100 reads smoothes out the high scores for genes with very low numbers of observed reads. We fitted a normal model to the mode of distribution of SA,B, and calculated P-values for each gene according to the fit. After excluding essential genes from the original pool, we considered genes to be important/costly for a particular condition with a log2 fold change of at least two, which corresponds to a 10−5 P-value and a 2.5 × 10−4 false discovery rate according to the normal model.
We calculated the P-value for the distances between insertion sites using F = G/N, where G is the number of bases in the genome (4,791,961) and N is the number of unique insert sites (371,775). The P-value for at least X consecutive bases without an insert site is e(−X/F), giving a 5% cutoff at 39 bp and a 1% cutoff at 60 bp.
Acknowledgments
This work was supported by the Wellcome Trust. We thank Elizabeth Huckle, who helped with the design of the sequencing assay.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to European Nucleotide Archive (http://www.ebi.ac.uk/embl) under accession no. ERA000097.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.097097.109.
References
- Andrews-Polymenis HL, Santiviago CA, McClelland M. Novel genetic tools for studying food-borne Salmonella. Curr Opin Biotechnol. 2009;20:149–157. doi: 10.1016/j.copbio.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Sys Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill CP, Braddick M, Hill MJ, Knowles RL, Sharp JC. The association between typhoid carriage, typhoid infection and subsequent cancer at a number of sites. Eur J Cancer Prev. 1995;4:187–193. doi: 10.1097/00008469-199504000-00010. [DOI] [PubMed] [Google Scholar]
- Charles IG, Maskell DJ. Transposon mediated differential hybridisation. 2001 Patent no. WO2001/007651. [Google Scholar]
- Chaudhuri R, Allen A, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, et al. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH) BMC Genomics. 2009:10. doi: 10.1186/1471-2164-1110-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Sys Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta U, Garg PK, Kumar R, Tandon RK. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am J Gastroenterol. 2000;95:784–787. doi: 10.1111/j.1572-0241.2000.01860.x. [DOI] [PubMed] [Google Scholar]
- French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, III, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proc Natl Acad Sci. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laia ML, Moreira LM, Dezajacomo J, Brigati JB, Ferreira CB, Ferro MIT, Silva ACR, Oliveira JCF, Ferro JA. New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC Microbiol. 2009;9:12. doi: 10.1186/1471-2180-1189-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht TR, Krogfelt KA, Cohen PS, Poulsen LK, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D, Jones JDG, Studholme DJ. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat Rev Microbiol. 2009;7:287–296. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- Moller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA, Cohen PS. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun. 2003;71:2142–2152. doi: 10.1128/IAI.71.4.2142-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevola JJ, Stocker BA, Laux DC, Cohen PS. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun. 1985;50:152–159. doi: 10.1128/iai.50.1.152-159.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- Prieto AI, Jakomin M, Segura I, Pucciarelli MG, Ramos-Morales F, Garcia-Del Portillo F, Casadesus J. The GATC-binding protein SeqA is required for bile resistance and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:8496–8502. doi: 10.1128/JB.01156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Brodsky IE, Falkow S, Gunn JS. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology. 2004;150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rida S, Caillet J, Alix JH. Amplification of a novel gene, sanA, abolishes a vancomycin-sensitive defect in Escherichia coli. J Bacteriol. 1996;178:94–102. doi: 10.1128/jb.178.1.94-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, et al. Evolutionary history of Salmonella Typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ, Marians KJ. Role of PriA in replication fork reactivation in Escherichia coli. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ, Marians KJ, Zavitz KH, Coutu J, Parent MA, Clark AJ. dnaC mutations suppress defects in DNA replication- and recombination-associated functions in priB and priC double mutants in Escherichia coli K-12. Mol Microbiol. 1999;34:91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Singh H, Pandey M, Upadhyay SK, Nath G. Carcinoma of the gallbladder—is it a sequel of typhoid? Dig Dis Sci. 2000;45:900–903. doi: 10.1023/a:1005564822630. [DOI] [PubMed] [Google Scholar]
- Tacket CO, Sztein MB, Losonsky GA, Wasserman SS, Nataro JP, Edelman R, Pickard D, Dougan G, Chatfield SN, Levine MM. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect Immun. 1997;65:452–456. doi: 10.1128/iai.65.2.452-456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun. 1998;66:2099–2106. doi: 10.1128/iai.66.5.2099-2106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AK, Nair S, Wain J. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J Antimicrob Chemother. 2006;58:733–740. doi: 10.1093/jac/dkl333. [DOI] [PubMed] [Google Scholar]
- van Velkinburgh JC, Gunn JS. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect Immun. 1999;67:1614–1622. doi: 10.1128/iai.67.4.1614-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]