Abstract
STAT1 mediates Interferon (IFN)-dependent positive and negative regulation of inflammatory gene expression in lung. In this study, we examined the effect of IFN-γ on the expression of SCGB3A1 which is thought to play crucial roles in inflammation and epithelial cell differentiation in lung. We found that expression of SCGB3A1 was down-regulated by IFN-γ in a time- and dose-dependent manner in the murine transformed Clara Cells (mtCC) line. IFN-γ induced the phosphorylation of STAT1, which binds to a STAT-binding element (SBE) in the SCGB3A1 gene promoter, leading to decreased transcriptional activation of this gene.
Keywords: SCGB3A1, Uteroglobin-related protein 2, IFN-γ
Secretoglobin (SCGB) 3A1, also known as uteroglobin-related protein 2 (UGRP2) [1] or HIN-1 (high in normal-1) [2], belongs to the Secretoglobin (SCGB) gene superfamily, based on its amino acid sequence and predicted structure. SCGB3A1 is predominantly expressed in the lung and trachea [3]. Although its function in vivo is not fully defined, SCGB3A1 appears to be involved in the regulation of local immune responses in the lung for the following reasons. First, in amino acid sequence, SCGB3A1 has similarity to Uteroglobin/Clara cell secretory protein (UG/CCSP), the prototypical protein of the SCGB gene superfamily, also called SCGB1A1, which is known to be a regulator of airway inflammation [3]. Second, the human SCGB3A1 gene is located at 5q31–q35, which has been suggested to be an asthma susceptibility region. Further, other studies have found a reduced level of expression of SCGB3A2, a member of the same gene family as SCGB3A1, in ovalbumin-induced airway inflammation in mice [4,5].
IFN-γ, a T helper 1 (Th1) cytokine, plays an important role in inducing and modulating an array of immune responses against many microbial pathogens, including bacteria, viruses, and fungi [6]. IFN-γ is the sole type-II IFN, and the functional IFN-γ receptor complex (IFNGR) is comprised of two receptor chains: IFNGR1, which is the primary ligand-binding chain, and IFNGR2 which serves an accessory role by recruiting the Janus tyrosine kinase, Jak2 [7]. The primary signaling of IFN-γ occurs through the Jak-Stat pathway, which involves sequential receptor recruitment and activation of members of the Janus family of kinases, specialy Jak1 and Jak2. IFN-γ induces the autophosphorylation and activation of Jak2, which promotes trans-phosphorylation of Jak1 [8], while activated Jak1 causes IFNGR1 phosphorylation to form two adjacent docking sites for the SH2 domains of STAT1 [9,10]. Receptor-recruited STAT1 is phosphorylated by Jak2, and phosphorylated STAT1 then homodimerizes and enters the nucleus, where it binds to promoter elements to initiate or suppress transcription of IFN-γ responsible genes [11–13].
SCGB3A1 gene expression is regulated by several growth factors and cytokines, including EGF/TGF-α, IL-4 and IL-13, in mouse transformed Clara Cells (mtCC) [14]. EGF/TGF-α stimulate both the ERK-MAPK and PI3K pathways via activation of EGF receptors, leading to up-regulation of SCGB3A1 gene expression [14]. Furthermore, the T helper 2 (Th2)-type cytokines, IL-4 and IL-13, stimulate expression of the SCGB3A1 gene through binding of STAT6 to the STAT-binding element (SBE) that resides –209 to –201 bp from the transcriptional start site of this gene [15]. In that study, we showed that the IL-4/IL-13-induced increase of SCGB3A1 gene expression was inhibited by IFN-γ. IFN-γ is a well known antagonist that inhibits the expression of several IL-4/IL-13-inducible genes, including IGF1, FcεRIIb (CD23), IgE, 15-lipoxygenase, IL-1R1 and IL-1RII, and the IL-1R antagonist [16]. Interestingly, IFN-γ inhibits not only IL-4/IL-13-induced SCGB3A1 gene expression, but also basal SCGB3A1 gene expression in mtCC cells [15].
The aim of this study was to investigate the mechanism that governs the negative regulation of SCGB3A1 gene expression by IFN-γ. We found that SCGB3A1 gene expression is down-regulated by IFN-γ via the binding of STAT1 to the SBE located in the SCGB3A1 gene promoter.
Materials and methods
Reagents and experimental animals
Pregnant C57BL/6 mice were purchased from Saitama Experimental Animals. Recombinat murine IFN-α, β and γ were purchased from R&D systems (Minneapolis, MN, USA). Antibodies against STAT1 and phospho-STAT1 were obtained from Cell Signaling Technology (Boston, MA, USA), HIN-1 (SCGB3A1) from R&D systems, and β-actin from Sigma-Aldrich (St. Louis, MO, USA). Horseradish peroxidase-conjugated secondary antibody was purchased from Santa Cruz (Santa Cruz, CA, USA).
Cell culture
Mouse transformed Clara cells (mtCC) were generated from tumor cells derived from transgenic mice expressing the SV40 large T Antigen gene under the control of the Uteroglobin/CCSP promoter [17]. mtCC and RAW264.7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (WAKO Pure Chemical Industries Ltd., Osaka, Japan) supplemented with 10% fetal bovine serum (Sigma-Aldrich).
RT-PCR
Total RNAs from cells were prepared using TRIzol solution (Invitrogen Life Technologies, Carlsbad, CA, USA). First-strand cDNA was synthesized using Superscript III (Invitrogen Life Technologies) and subjected to amplification with Taq polymerase (Sigma-Aldrich) using the following specific PCR primers:GAPDH, 5′-GAAGGTCGGTGTGAACGGATTTGGC-3′ and 5′-CATGTAGGCCATGAGGTCCACCAC-3′; mouse IFNGR1, 5′-CGAAAGACGTCTGTATCCCT-3′ and 5′-AAAAGCTGCGTCTTTGTGTC-3′; mouse IFNGR2, 5′-GATGATGTGCTCCAAACACC-3′ and 5′-AGGGGTCTCACATTTAAGCC-3′.
Northern blot analysis
Total RNAs (10 μg) were electrophoresed on a 1% agarose gel containing 2% formaldehyde and blotted onto nylon membranes (Hybond N, GE Healthcare UK Ltd., Buckinghamshire, UK). Filters were serially hybridized with mouse SCGB3A1 and GAPDH cDNA as a probe. Hybridization was performed in Perfect Hybridization solution (Sigma–Aldrich) at 65 °C overnight. The membranes were washed twice with 2 × SSC containing 0.5% SDS at 65 °C for 30 min, and once with 2 × SSC at 65 °C for 30 min, then exposed to STORM phosphoimager (Molecular Dynamics, Sunnyvale, CA, USA).
Western blot analysis
Total cell lysate (20 μg) was isolated, separated by 10% SDS-PAGE, and transferred onto Immobilon-P membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 3% BSA in TBST (150 mM NaCl, 20 mM Tris–HCl, pH 7.4 and 0.1% Tween 20), then subjected to immunostaining with antibodies. The membranes were developed using an ECL detection kit (ECL-plus, GE Healthcare UK Ltd.).
Luciferase reporter assay
mtCC cells were seeded in 24-well tissue culture plates, grown to 80% confluency, and transfected with pGL3 reporter plasmids containing various lengths of the mouse SCGB3A1 gene promoter using Lipofectamine 2000 (Invitrogen). Cells were cultured in 0.5% FBS-containing medium in the presence or absence of cytokine for 48 h and then lysed with passive lysis buffer (Promega, Madison, WI, USA). Luciferase activity was measured according to the technical manual for the Dual-Luciferase Reporter Assay System (Promega) and Lumat LB9507 (Berthhold Technologies, Bad Wildbad, Germany).
Electrophoretic mobility shift assay (EMSA)
Nuclear protein extracts were prepared from IFN-γ-treated RAW264.7 using a modification of the original method described by Schreiber et al. [18]. A double-stranded oligonucleotide (21 nt) prepared based on the DNA sequence present in the mouse SCGB3A1 gene promoter was used as a probe in EMSA. This oligonucleotide contains a STAT-binding element (SBE) that can bind STAT1 and STAT6 [15]. Binding reactions were performed as described previously [15].
Results
IFN-γ inhibits SCGB3A1 gene expression in a time- and dose-dependent manner
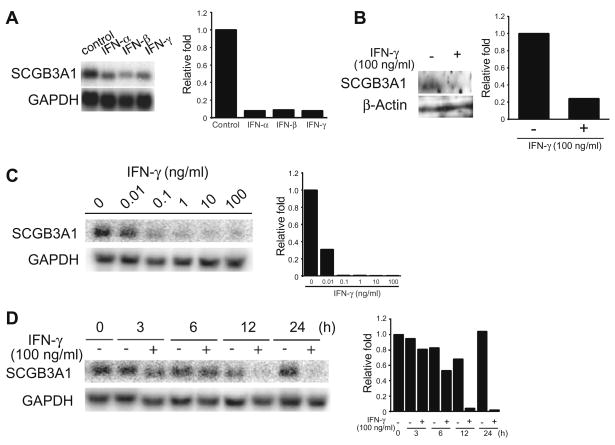
Our previous findings showed that SCGB3A1 gene expression is up-regulated by IL-4/IL-13, whereas it is down-regulated by IFN-γ [15]. Both IL-4 and IL-13 stimulated expression of the SCGB3A1 gene through the binding of STAT6 to the proximal SBE in the SCGB3A1 gene promoter. To investigate the mechanism of IFN-γ-mediated down-regulation of SCGB3A1 gene expression, we treated mtCC cells with either 100 ng/ml of type-I IFNs such as IFN-α and IFN-β, or IFN-γ (type-II IFN) for 12 h. All three IFNs markedly inhibited the expression of SCGB3A1 mRNA (Fig. 1A). Furthermore, Western blot analysis showed that SCGB3A1 protein expression was also decreased by IFN-γ treatment (Fig. 1B). These results indicate that all IFN-α, IFN-β, and IFN-γ down-regulate SCGB3A1 gene expression.
Fig. 1.
Regulation of SCGB3A1 gene expressions by IFNs. (A) mtCC cells were treated with 100 ng/ml of selected IFNs (IFN-α, IFN-β and IFN-γ) for 12 h, and mRNAs for SCGB3A1 and GAPDH as a control were examined by Northern blot analysis. (B) Effect on SCGB3A1 protein expression of IFN-γ. mtCC cells were treated with 100 ng/ml of IFN-γ, and proteins for SCGB3A1 and β-actin as a control were examined by Western blot analysis. (C) Dose effects of IFN-γ analyzed using Northern blots. mtCC cells were treated with 0.01, 0.1, 1, 10, or 100 ng/ml of IFN-γ for 24 h. (D) Time course analysis of IFN-γ effects by Northern blot analysis. mtCC cells were treated with 100 ng/ml of IFN-γ for 3, 6, 12, or 24 h. In each panel, band intensities were quantitated and the relative expression of SCGB3A1 vs GAPDH as a control is plotted on the right. Experiments were repeated at least twice and a representative result is shown.
The regulation of SCGB3A1 gene expression by IFN-γ was examined in more detail. When mtCC cells were treated with different concentrations of IFN-γ for 24 h, a significant decrease in the SCGB3A1 mRNA level was seen at a concentration of 0.01 ng/ml (Fig. 1C), while IFN-γ markedly inhibited SCGB3A1 mRNA expression at 0.1 ng/ml and higher concentrations. The time-dependent effect of IFN-γ on the suppression of SCGB3A1 mRNA was also examined using a fixed concentration of 100 ng/ml (Fig. 1D). A significant decrease was first detectable at 12 h after the addition of IFN-γ. These results demonstrate that IFN-γ suppresses the expression of SCGB3A1 mRNA in a dose- and time-dependent manner.
Activation of STAT1 in mtCC cells through IFNGRs
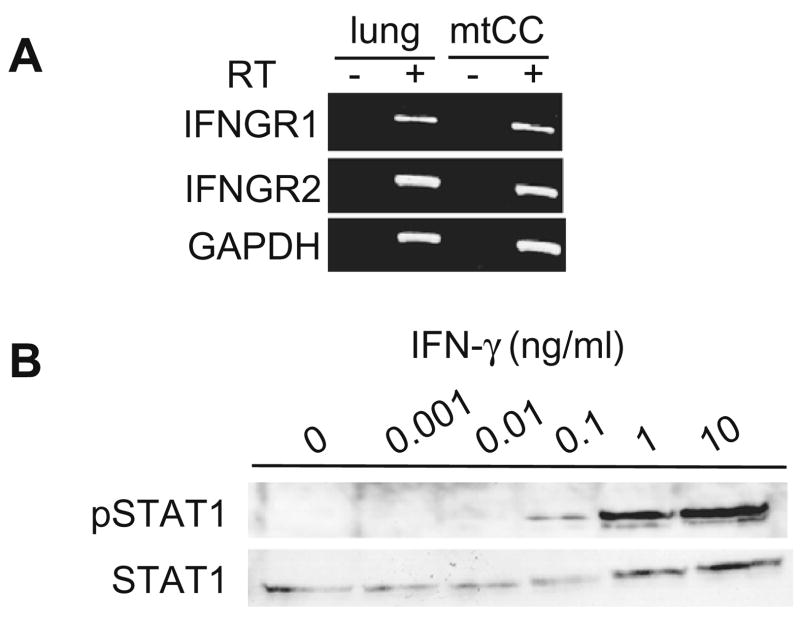
In order to study the mechanism of the IFN-γ-mediated decrease of SCGB3A1 mRNA expression, the presence of the IFN-γ receptors IFNGR1 and IFNGR2 was examined using RT-PCR analysis (Fig. 2A). The results clearly demonstrated that both receptors are present in mtCC cells. IFN-γ has been reported to activate STAT1, which is capable of binding to SBEs in the promoters of various IFN-γ responsive genes. To determine whether STAT1 is activated in mtCC cells upon IFN-γ treatment, phosphorylation of STAT1 was examined by Western blot analysis using antibodies against phospho-STAT1 and STAT1 (Fig. 2B). The levels of phosphorylated STAT1 were increased in the presence of as little as 0.1 ng/ml of IFN-γ, and increased in a dose-dependent manner. These results demonstrate that STAT1 is activated by IFN-γ in mtCC cells.
Fig. 2.
STAT1 is activated by IFN-γ in mtCC cells. (A) RT-PCR analysis of IFNGR1 and IFNGR2 using RNAs prepared from mtCC cells and adult mouse lung as a control. The sizes of PCR products are 426 bp for IFNGR1, 439 bp for IFNGR2, and 982 bp for GAPDH. (B) mtCC cells were treated with 0.001, 0.01, 0.1, 1, or 10 ng/ml of IFN-γ for 30 min. The level of activated-STAT1 was measured using a rabbit polyclonal anti-phospho-STAT1 (Tyr694) antibody.
Regulation of SCGB3A1 gene promoter through binding of STAT1 to proximal SBE
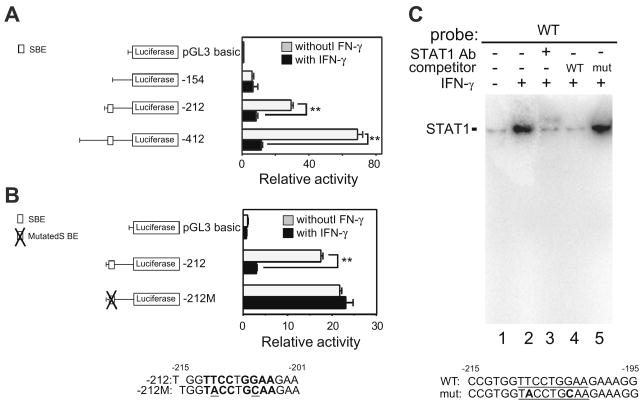
Three mouse SCGB3A1 gene promoter-luciferase constructs (pGL3-154, –212, and –412) [15] were prepared and transfected into mtCC cells, and the cells were then treated with or without IFN-γ. The basal reporter activity of the SCGB3A1 gene promoter without IFN-γ treatment was gradually increased up to –412 bp of the promoter sequence (Fig. 3A) [15]. In the presence of IFN-γ, the reporter activities of the constructs pGL3-212 and –412, but not –154 were down-regulated by the addition of IFN-γ as compared to those in the absence of IFN-γ. These results suggest that the element(s) responsible for the IFN-γ-induced suppression of SCGB3A1 gene promoter activity reside in the region from –154 and –212 bp upstream from the transcription start site of the SCGB3A1 gene. In fact, within this region, one SBE (5′-TTCCTGGAA-3′) is located at a position –209 to –201 bp from the transcription start site of the gene, which our previous study demonstrated is responsible for the IL-4/IL-13-induced increase of SCGB3A1 gene expression [15]. To determine whether this SBE may be responsible for the IFN-γ-induced decrease of SCGB3A1 promoter activity, mutations were introduced to modify two of its nucleotides (5′-TTCCTGGAA-3′ to 5′-TACCTGCAA-3′; mutations are underlined). We found that the construct pGL3-212M carrying these mutated nucleotides almost completely inhibits the IFN-γ induced decrease of SCGB3A1 gene promoter activity (Fig. 3B), suggesting that this SBE plays an important role in the suppression of SCGB3A1 promoter activity by IFN-γ. STAT1 binds to this SBE (–209 to –201 bp) in EMSA analysis using a 32P end-labeled double-stranded oligonucleotide spanning –215 through –195 bp of the mouse SCGB3A1 gene promoter (Fig. 3C) [15].
Fig. 3.
Identification of an IFN-γ-responsive region in the SCGB3A1 gene promoter. (A) Transfection analysis of a series of luciferase reporter plasmids containing various lengths (–412, –212 and –154/+52 bp) of the mouse SCGB3A1 gene promoter with or without IFN-γ. The proximal SBE (5′-TTCCTGGAA-3; –212 to –154 bp) previously shown to be involved in the IL-4/IL-13-induced increase of SCGB3A1 expression[15], is indicated in the illustration on the left. The SBE is characterized by a palindromic sequence shown in boldface. Luciferase assays were performed in mtCC cells with (black bar) or without (gray bar) stimulation with 100 ng/ml of IFN-γ. Results shown are the mean ± SE from three independent experiments. A significant difference was obtained with versus without IFN-γ treatment: **P < 0.01. (B) Mutation analysis. The SBE site and the mutated sequences (underlined) are shown in boldface. The mutation of the proximal SBE abrogates the IFN-γ-induced promoter activity of the –212 construct. (C) Electrophoretic mobility shift assays (EMSA) for the STAT1-binding site in the SCGB3A1 gene promoter. A SBE is underlined. In the mutated SBE oligonucleotide (mut), mutations were introduced by changing 2 nucleotides (boldfaced). Nuclear extract proteins (5 μg) from RAW264.7 cells stimulated with 100 ng/ml of IFN-γ for 30 min at 37 °C were incubated with labeled wild-type (WT) SBE oligonucleotide. For supershift assays, 2 μg of STAT1 antibody was added to probe-extract complexes. After incubations, DNA/protein complexes were resolved by 6% polyacrylamide gel electrophoresis.
Discussion
Previously, we demonstrated that the suppression of basal SCGB3A1 gene expression by IFN-γ can inhibit the induction of SCGB3A1 gene expression independently of IL-4 and IL-13 [15]. In this study, SCGB3A1 mRNA was found to be negatively regulated by IFN-γ through STAT1 activation. Activated STAT1 binds to the proximal STAT-binding element and inhibits the promoter activity of the SCGB3A1 gene.
SCGB3A1 mRNA expression was inhibited by IFN-α/β as well as IFN-γ (Fig. 1A). IFN-α/β are the type-I IFNs, which are well known to be central to the host defense against microbial pathogens including most viruses [19,20]. The IFN-α/β and IFN-γ signaling pathways exhibit intracellular cross-talk at multiple levels. Furthermore, the signaling pathways and target genes activated by type-I and -II IFNs partially overlap each other, which provides an opportunity for cross-talk within cells to synergize or antagonize particular functions [21]. Thus, it is possible that IFN-α/β regulates the expression of SCGB3A1 mRNA through STAT1 activation as seen with IFN-γ. In this regard, it is interesting that the expression of SCGB3A2, the gene from the same genefamily as SCGB3A1, is inhibited by IFN-α/β but not IFN-γ in mtCC cells (A. Yamada, unpublished observations).
Transcriptional analysis of the SCGB3A1 promoter–reporter constructs showed that the proximal SBE located between –209 and –201 bp from the transcription start site contains the elements that are responsible for the IFN-γ-induced inhibition of SCGB3A1 gene promoter activity (Fig. 3B). Previously, we demonstrated that IL-4- or IL-13-activated STAT6 binds to the same SBE in the SCGB3A1 gene promoter [15]. When cells were co-treated with IFN-γ and IL-4 or IL-13, binding of STAT6 to the SBE decreased apparently due to the binding of STAT1 to the same element. Ohmori et al. reported that the affinity of STAT1 is greater than the affinity of STAT6 for the SBE in the promoter of IRF-1 gene [22]. This may also be the case for the SCGB3A1 gene promoter. We do not know how the binding of STAT1 to the proximal SBE in the SCGB3A1 gene promoter suppresses SCGB3A1 promoter activity. Additional studies are required to address this issue.
Regulation of IFN-γ signal transduction and gene expression in airway epithelial cells is of critical importance in allergic diseases such as asthma because epithelial cells stimulated with IFN-γ express a multitude of inflammatory mediators that trigger and then maintain airway inflammation [23]. SCGB3A1 gene expression is up-regulated by retinoic acid, which induces mucinous differentiation in primary bronchial epithelial cells. The potential pathophysiological role of decreased SCGB3A1 expression induced by IFN-γ in the lung remains to be understood.
In conclusion, we demonstrated that IFN-γ suppresses expression of the SCGB3A1 gene in lung epithelial cells through activation of the STAT1 pathway.
Acknowledgments
We would like to thank Dr. J. Francesco DeMayo for providing mtCC cells. This work was supported in part by the ‘High-Tech Research Center’ Project for Private Universities matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Grants-in-Aid for Scientific research from the Japan Society for the Promotion of Science.
References
- 1.Reynolds SD, Reynolds PR, Pryhuber GS, Finder JD, Stripp BR. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am J Respir Crit Care Med. 2002;166:1498–1509. doi: 10.1164/rccm.200204-285OC. [DOI] [PubMed] [Google Scholar]
- 2.Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, Seth P, Kaelin CM, Rhei E, Bosenberg M, Schnitt S, Marks JR, Pagon Z, Belina D, Razumovic J, Polyak K. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:9796–9801. doi: 10.1073/pnas.171138398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niimi T, Copeland NG, Gilbert DJ, Jenkins NA, Srisodsai A, Zimonjic DB, Keck-Waggoner CL, Popescu NC, Kimura S. Cloning, expression, and chromosomal localization of the mouse gene (Scgb3a1, alias Ugrp2) that encodes a member of the novel uteroglobin-related protein gene family. Cytogenet Genome Res. 2002;97:120–127. doi: 10.1159/000064067. [DOI] [PubMed] [Google Scholar]
- 4.Chiba Y, Kusakabe T, Kimura S. Decreased expression of uteroglobin-related protein 1 in inflamed mouse airways is mediated by IL-9. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1193–1198. doi: 10.1152/ajplung.00263.2004. [DOI] [PubMed] [Google Scholar]
- 5.Chiba Y, Srisodsai A, Supavilai P, Kimura S. Interleukin-5 reduces the expression of uteroglobin-related protein (UGRP) 1 gene in allergic airway inflammation. Immunol Lett. 2005;97:123–129. doi: 10.1016/j.imlet.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira LK, Fonseca BP, Barboza BA, Viola JP. The role of interferon-gamma on immune and allergic responses. Mem Inst Oswaldo Cruz. 2005;100(Suppl 1):137–144. doi: 10.1590/s0074-02762005000900024. [DOI] [PubMed] [Google Scholar]
- 7.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 8.Briscoe J, Rogers NC, Witthuhn BA, Watling D, Harpur AG, Wilks AF, Stark GR, Ihle JN, Kerr IM. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. Embo J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 9.Heim MH, Kerr IM, Stark GR, Darnell JE., Jr Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi K, Garotta G, Ozmen L, Ziemiecki A, Wilks AF, Harpur AG, Larner AC, Finbloom DS. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J Biol Chem. 1994;269:14333–14336. [PubMed] [Google Scholar]
- 11.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 12.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 13.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interf Cytok Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 14.Yamada A, Kimura S. Induction of uteroglobin-related protein 2 (Ugrp2) expression by EGF and TGFalpha. FEBS Lett. 2005;579:2221–2225. doi: 10.1016/j.febslet.2005.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada A, Sheikh F, Niimi T, DeMayo FJ, Keegan AD, Donnelly RP, Kimura S. Induction of uteroglobin-related protein 2 (Ugrp2) gene expression by the Th2 cytokines IL-4 and IL-13. J Immunol. 2005;175:5708–5715. doi: 10.4049/jimmunol.175.9.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickensheets HL, Donnelly RP. IFN-gamma and IL-10 inhibit induction of IL-1 receptor type I and type II gene expression by IL-4 and IL-13 in human monocytes. J Immunol. 1997;159:6226–6233. [PubMed] [Google Scholar]
- 17.Magdaleno SM, Wang G, Jackson KJ, Ray MK, Welty S, Costa RH, DeMayo FJ. Interferon-gamma regulation of Clara cell gene expression: in vivo and in vitro. Am J Physiol. 1997;272:L1142–1151. doi: 10.1152/ajplung.1997.272.6.L1142. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis DB, Wilson CB. Gamma-interferon: an immunoregulatory lymphokine with immunotherapeutic potential. Pediatr Infect Dis J. 1990;9:642–651. [PubMed] [Google Scholar]
- 20.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 21.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 22.Ohmori Y, Smith MF, Jr, Hamilton TA. IL-4-induced expression of the IL-1 receptor antagonist gene is mediated by STAT6. J Immunol. 1996;157:2058–2065. [PubMed] [Google Scholar]
- 23.Heller NM, Matsukura S, Georas SN, Boothby MR, Rothman PB, Stellato C, Schleimer RP. Interferon-gamma inhibits STAT6 signal transduction and gene expression in human airway epithelial cells. Am J Respir Cell Mol Biol. 2004;31:573–582. doi: 10.1165/rcmb.2004-0195OC. [DOI] [PubMed] [Google Scholar]