Abstract
Death-associated protein kinase (DAPK), a mediator of apoptotic systems, is silenced by promoter hypermethylation in lung and breast tumors. This gene has a CpG island extending 2500 bp from the translational start site; however, studies characterizing its transcriptional regulation have not been conducted. Two transcripts for DAPK were identified that code for a single protein, while being regulated by two promoters. The previously identified DAPK transcript designated as exon 1 transcript was expressed at levels 3-fold greater than the alternate exon 1b transcript. Deletion constructs of promoter 1 identified a 332 bp region containing a functional CP2-binding site important for expression of the exon 1 transcript. While moderate reporter activity was seen in promoter 2, the region comprising intron 1 and containing a HNF3B-binding site sustained expression of the alternate transcript. Sequencing the DAPK CpG island in tumor cell lines revealed dense, but heterogenous methylation of CpGs that blocked access of the CP2 and HNF3B proteins that in turn, was associated with loss of transcription that was restored by treatment with 5-aza-2′-deoxycytidine. Prevalences were similar for methylation of promoter 1 and 2 and intron 1 in lung tumors, but significantly greater in promoter 2 and intron 1 in breast tumors, indicative of tissue-specific differences in silencing these two transcripts. These studies show for the first time dual promoter regulation of DAPK, a tumor suppressor gene silenced in many cancers, and substantiate the importance of screening for silencing of both transcripts in tumors.
Introduction
Transcriptional silencing of death-associated protein kinase (DAPK) occurs in many diverse types of human cancers through promoter hypermethylation at prevalences ranging from 7% in liver tumors to 84% in B-cell non-Hodgkin's lymphoma (1,2). Methylation of DAPK in two of the most commonly diagnosed cancers, lung and breast, occurs at prevalences of 25–45% and 9–45%, respectively (3–7). Methylation of DAPK has been seen in the bronchial epithelium of some smokers (8). The ubiquitous silencing of this gene implies a critical role for it in cancer development. DAPK is a 16 kDa Ca+/calmodulin-regulated serine threonine kinase with a conserved death domain that is a positive mediator of a wide array of apoptotic systems (9–14). DAPK suppresses c-myc- and E2F-induced oncogenic transformation by p19ARF-dependent activation of the p53 apoptotic pathway (15). The reintroduction of DAPK into highly metastatic mouse lung carcinoma cells delayed tumor growth and strongly reduced their metastatic capacity, substantiating a link between this gene and cancer (16). Moreover, DAPK was methylated in 90% of brain metastasis derived from melanoma, lung, breast, ovarian and colon cancers (17). Methylation of DAPK in lung cancer has been associated with poor prognosis and advanced pathological stage and reduced expression of this gene predicted for reduced survival and recurrence in breast cancer patients (3,4,18). Thus, loss of expression of DAPK appears to confer a selective growth advantage for cancer cells that may drive tumor aggressiveness and progression.
There have been no extensive studies to characterize how DAPK is regulated transcriptionally. The DAPK gene lacks a defined TATA box but contains other positive regulatory elements located within 1500 bp of the translational start site in exon 2. These include multiple Sp1- and AP2-binding sites, an E box, a CAAT box and a number of other consensus binding sites [e.g. AP1, nuclear factor-kappaB (NF-κB) and E2F]. A CpG island of 590 bp containing 46 CpG dinucleotides is located directly upstream of the translational start site. An additional 100 CpGs, distributed evenly, are located 1000 bp upstream of this region. The published genomic and messenger RNA sequences (9,19) identified a transcribed region extending from −1194 to −966 upstream of the translational start site; however, additional transcriptional start sites below this region are also predicted. Adding further complexity to the regulation of this gene, bisulfite sequencing of a 659 bp fragment (−1411 to −752) that included exon 1 and contained 74 CpG dinucleotides in lung cancer cell lines revealed two methylation hotspots each containing 8–10 methylated CpGs that correlated with reduced to absent gene expression (20). Whether important transcription factors bind to these regions is not known or have studies addressed methylation in the 590 bp CpG island immediately upstream of the translational start site.
The purpose of the current study was to identify the transcriptional start sites, the minimal promoter region and critical transcription factors for the DAPK promoter and to assess methylation of CpG dinucleotides within the two CpG islands of the DAPK gene and the relationship to expression in lung and breast cancer cell lines. Prevalence for methylation across this entire region was also determined in primary lung and breast tumors.
Materials and methods
Cell lines and tumors
Twelve lung cancer cell lines (Calu-6, Calu-1, SKLU-1, A549, H2009, H1568, H358, H596, H522, H23, H125 and H460) and three breast cancer cell lines (MCF-7, T47D and MDA-MB-231) were obtained from the American Type Culture Collection (Manassas, VA). Human bronchial epithelial cells isolated from cancer-free smokers who underwent bronchoscopy (n = 3) and keratinocytes isolated from foreskin were used in expression studies for comparison with cancer cell lines. Cells were maintained at 37°C in a humid atmosphere containing 5% CO2 in American Type Culture Collection-recommended media or as described (8). A total of 35 frozen lung adenocarcinomas from smokers and 38 breast ductal carcinomas were obtained from tumor banks at Johns Hopkins and University of New Mexico, respectively. All persons providing tissue specimens signed informed consent and the Institutional Review Board of Lovelace Respiratory Research Institute approved this study.
Transcriptional start site identification
The transcriptional start sites of the DAPK gene were identified by RNA ligase-mediated rapid amplification of 5′ complementary DNA (cDNA) ends (RLM-RACE GeneRacer Kit; Invitrogen, San Diego, CA). RNA was isolated from human lung tumor-derived cell lines that express DAPK (H522, H1568 and Calu-6) using the Stratagene Strataprep Total RNA Miniprep Kit (Stratagene, LaJolla, CA). RNA (3 μg) was treated with calf intestinal phosphatase to remove the 5′ phosphates from partial transcripts while leaving capped RNA intact. RNA was then treated with tobacco acid pyrophosphate to remove the cap from capped messenger RNA to allow for ligation of the GeneRacer RNA Oligo (provides annealing site for universal GeneRacer primers) with T4 RNA ligase. Following generation of cDNA, 5′ and 3′ ends were amplified using the universal primer and DAPK-specific primers located in exon 2 (available upon request). PCR products were ligated into the pCR 4-TOPO vector using TA cloning kit (Invitrogen) and five clones from each sample were sequenced in both directions (University of New Mexico Center for Genetics in Medicine, Albuquerque, NM).
Quantitation of DAPK transcription
Cell lines were harvested in TRI reagent (Sigma, St. Louis, MO), and RNA was isolated following TRI reagent instructions. DNA was removed by digestion with DNase I (Life Technologies, Gaithersburg, MD) followed by phenol extraction and ethanol precipitation. Total RNA (1 μg) was reverse transcribed using the SuperScript™ First-Strand Synthesis System for reverse transcription–polymerase chain reaction (RT–PCR; Invitrogen) according to the cDNA synthesis protocol from Invitrogen®. Product sizes for exon 1–exon 2 and exon 1b–exon 2 transcripts were 250 and 180 bp, respectively. Quantitative RT–PCR analysis was performed on an ABI 7500 real-time system (Applied Biosystems, Foster City, CA). β-Actin was used as an internal control. PCR was performed using a SYBR Green PCR Master Mix Reagent Kit (Appliced Biosystems). Relative gene expression was determined based on the threshold cycles (Ct) of the DAPK exon 1 and exon 1b transcripts and the internal reference gene. The messenger RNA levels of each transcript were expressed as the ratio of each transcript to β-actin. The average Ct value of the β-actin gene was subtracted from the average Ct value of the DAPK transcripts and fold change in expression was calculated. All results were reproduced in three separate experiments. Primer sequences and PCR conditions are available by request.
Promoter analysis
The DAPK genomic sequence was obtained from GenBank (accession no. DQ436495). Full-length constructs for the luciferase reporter assay were generated by PCR amplification of the DAPK genomic region that included promoter 1 or promoter 2, exon 1b and intron 1 (Figure 1A). Primers (5′) that annealed progressively closer to the 3′ end of these two DNA fragments were designed to generate the deletion constructs. The primers were designed to contain unique MluI (5′) or BglII (3′) restriction sites (New England Biolabs, Beverly, MA) allowing for directional cloning of the PCR products. For additional mapping of reporter activity, several constructs of various sizes (described in Results) were created that were localized to different regions within the promoter 1 or the promoter 2–exon 1b–intron 1 sequence. All DNA fragments were directionally subcloned into the pGL2-basic luciferase reporter vector (Promega, Madison, WI) upstream of the luciferase coding sequence. A small amount of each ligation reaction was used to transform INVαF’ chemically competent cells (TA cloning kit; Invitrogen). Plasmid was purified from individual bacterial colonies using a Wizard DNA Miniprep kit (Promega) and samples were sequenced to confirm the presence of the insert as well as the correct nucleotide sequence (Sequetech, Mountain View, CA). Primer sequences and PCR conditions are available by request.
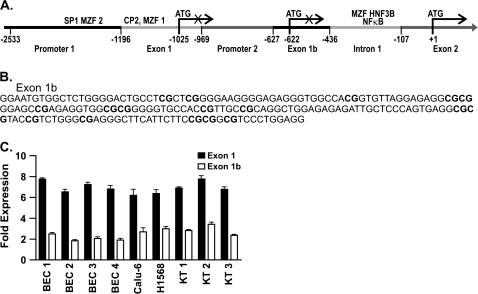
Fig. 1.
DAPK genomic structure, exon 1b sequence, and expression of exon 1 and 1b transcripts. (A) Schematic diagram depicting the location of promoter 1, exon 1 and 1b, promoter 2 and intron 1 in relation to the translational start site within exon 2 is shown. Shaded areas and numbers depict the boundaries and location of each region. The location of the ATGs in exon 1 and 1b that are not translational start sites due to stop codons in these exons are shown as are the location of transcription factor-binding sites that affect reporter activity. (B) Sequence of the 186 bp region designated exon 1b with the 17 CpG dinucleotides bolded is shown. (C) Quantitative expression of DAPK exon 1 and 1b transcripts in H1568, Calu-6, BEC and keratinocyte (KT) cell lines. Fold expression between exon 1 and 1b transcripts is compared.
Site-directed mutagenesis
The transcription factor-binding sites within the promoter were located by means of the ‘public domain’ version of MatInspector software, which uses a library of matrix descriptions to locate binding sites and assigns quality ratings to the resulting matches within a sequence (21). MatInspector software was also used to determine the essential nucleotides within the Sp1-, MZF-, CP2-, NF-κB- and hepatocyte nuclear growth factor 3B (HNF3B)-binding sites. At least two essential nucleotides were selected for mutation within each site. The selected mutations were introduced into the minimized promoter 1 and intron 1 DAPK/pGL2 luciferase constructs using the Stratagene Quik Change 2 mutagenesis kit. Briefly, plasmids were denatured, a primer containing the desired mutations was annealed to the single-stranded plasmid, the double-stranded plasmid was regenerated with the addition of polymerase and ligase and the new plasmid was amplified in repair-deficient bacteria. The amplified plasmids were purified and used to transform TOP10 chemically competent cells (TA cloning kit). Plasmid was purified from individual colonies (five colonies per mutation) and mutations were confirmed by sequencing (Sequetech).
Transient transfection and luciferase assay
Human lung tumor-derived cell lines (Calu-6, H1568 and H522) that express both DAPK transcripts were used for transient transfections. Cells (5 × 105) were plated in six-well dishes and transfected the following day. Approximately 1 μg of plasmid DNA that included 0.5 μg of pGL2 fragment plasmid and 0.5 μg of pSV-β-galactosidase (Promega) as an internal standard to normalize for differences in transfection efficiency was transfected into cells using Lipofectamine Plus transfection reagent (Life Technologies, Rockville, MD). A promoter-less pGL2-basic vector and the pGL2-control vector, which contains the SV40 promoter, were used as negative and positive controls, respectively. Forty-eight hours after transfection, cells were harvested and lysed. Immediately after lysing, cell extracts were assayed in a luminometer for luciferase activity using the Luciferase Assay System (Promega). β-Galactosidase activity in cell lysates was measured using the Galacto-Star Reporter Gene Assay System (Tropix, Bedford, MA). Promoter activity was calculated as the ratio of the activity of luciferase and β-galactosidase. Transfections were done in duplicate in three independent experiments.
Chromatin immunoprecipitation and siRNA
Chromatin immunoprecipitation (ChIP) was done using the ChIP Assay kit (Upstate, Charlottesville, VA). Antibodies for CP2, HNF3B and IgG were purchased from Upstate Chemicon (Temecula, CA) and used to capture protein–DNA complexes. ChIP PCR analysis was performed using 3 μl of DNA and primers spanning the region −1217 to −1060 and −156 to −6 (with respect to ATG) of the DAPK promoter 1 and intron 1, respectively, were used. Quantitative RT–PCR (TaqMan assays) was used to quantify the expression of the CP2 and HNF3B transcription factors in normal bronchial epithelial cell lines and cancer lines in which ChIP studies were conducted. Results were repeated in three separate experiments. siRNA sequence (available upon request) to HNF3B or a scrambled control was transiently transfected into Calu-6 cells and effect on expression of exon 1 and exon 1b transcripts was quantified 48 h after transfection.
DNA extraction and modification
DNA was isolated from cell lines and tumor samples using a standard phenol–chloroform extraction method. Genomic DNA was modified using the EZ DNA Methylation-Gold Kit™ (ZYMO Research, Orange, CA) as described by the manufacturer, and 100 ng of modified DNA was used per PCR for methylation-specific polymerase chain reaction (MSP) or bisulfite sequencing.
Bisulfite sequencing and MSP
The DAPK genomic sequence that included promoter 1, exon 1, promoter 2, exon 1b and intron 1 was amplified in three separate PCR reactions from bisulfite-modified DNA from lung and breast cancer cell lines. Promoter 1 and exon 1 [−1500 to −1076 bp (reference to translational start site in exon 2)] was amplified in PCR fragment 1 and contained 49 CpG dinucleotides. PCR fragment 2 included promoter 2 and exon 1 alternate (−925 to −490 bp) and contained 42 CpG dinucleotides. Finally, PCR fragment 3 included intron 1 (−450 to −1 bp) and contained 30 CpG dinucleotides. PCR products were cloned into the pCR®II cloning vector (Invitrogen), and five clones were sequenced per sample (University of New Mexico Genetics).
Three sets of MSP primers were designed to anneal to regions within promoter 1–exon 1, promoter 2 and intron 1 that were the most densely methylated by bisulfite sequencing. All primer sequences and conditions for PCR for sequencing and methylation assays are available by request. The MSP assay can detect one methylated allele in a background of 1000 unmethylated alleles (8).
DAPK gene re-expression
Cell lines (Calu-6, H2009, H23 and T47D) with unmethylated or methylated DAPK regions were treated with 1 μM 5-aza-2′-deoxycytidine (DAC) for 3 days, with media changes and fresh DAC added every 24 h. After 72 h, cells were harvested in TRI reagent for isolation of RNA. Following RNA isolation and generation of cDNA as described above, exon 1 and exon 1b transcripts, were amplified by PCR. A portion of the β-actin gene was amplified to control for cDNA quality.
Results
Identification of an alternative exon 1 for DAPK
Three lung tumor-derived cell lines (H522, H1568 and Calu-6) that express the previously identified DAPK transcript were used to determine if additional transcriptional start sites were present. RNA ligase-mediated rapid amplification of 5′ cDNA ends identified the expected transcriptional start site at −1196 bp (reference to the translational start site in exon 2); however, a second transcriptional start site was mapped to −622 bp (Figure 1A). Sequencing of this RACE product identified a 186 bp exon (designated exon 1b; Figure 1B). This CpG-rich exon has 17 CpG dinucleotides and an observed to expected CpG ratio of 0.86 indicative of a small CpG island according to the criteria of Takai et al. (22).
Quantitative RT–PCR was performed to determine the relative abundance of the exon 1 and 1b transcripts in the Calu-6 and H1568 cell lines. A 2.4-fold higher level of expression for exon 1 compared with exon 1b transcript was seen (Figure 1C). A similar difference in expression (2.3- to 3.6-fold) was also seen in normal keratinocytes and bronchial epithelial cells (Figure 1C). Finally, to assess whether any other sequence differences exist between the two DAPK transcripts, the entire cDNA of the DAPK gene was amplified from Calu-6 cells using primers specific to exon 1 and 1b and a common primer to exon 26 and sequenced. No differences other than the sequence of exon 1b were seen (data not shown). There is an ATG at −627 of exon 1b, however, initiating translation at this ATG ends with a stop codon after 23 amino acids. This scenario is similar to that observed when trying to initiate translation using the ATG present in exon 1. Therefore, it appears that translation of both transcripts begins in exon 2, consistent with the detection of only the 16 kDa protein by western blot (data not shown).
Analysis of DAPK promoters and intervening sequence
The sequences upstream of exon 1 and 1b were designated promoter 1 and 2, respectively (Figure 1A). Promoter 1 (−2533 to −1025) and seven segments representing 5′ deletions of this 1508 bp promoter were ligated into a luciferase expression vector and transfected into H522, H1568 and Calu-6 cells (Figure 2A). In general, 80–100% of full-length luciferase activity was seen across cell lines in the initial four deletion constructs up to nucleotide position −1357, the exception being the H522 cells that showed a progressive decrease in promoter activity. However, luciferase activity was reduced 65–75% in the −1278 to −1025 or −1208 deletion constructs (Figure 2A). Reporter activity was virtually ablated with further deletion (−1135 to −1025; Figure 2A). To characterize the region where critical transcription factors are probably binding, three additional constructs beginning at −1278 and extending 79, 135 and 222 bp were constructed. None of these constructs demonstrated reporter activity that exceeded 20% of the full-length activity, suggesting that the entire region between −1357 and −1025 (minimized promoter) is required for full promoter activity.
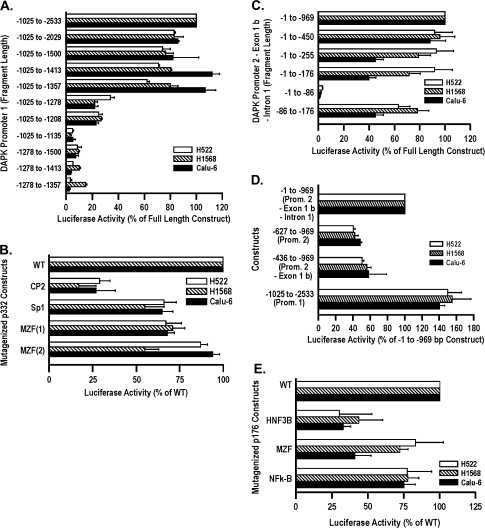
Fig. 2.
Characterization of promoter activity and identification of transcription factors for the DAPK gene. (A) Individual sequences comprising different portions of the DAPK promoter 1 were amplified by PCR (numbers indicate the size and location of the DNA fragment relative to the translational start site), ligated into a luciferase expression vector and then transfected into H522, H1568 and Calu-6 cells to identify the minimized region supporting reporter activity. Luciferase activity is expressed as the percentage of the full-length construct. All transfections were conducted in duplicate and repeated three times. Error bars represent the average of values obtained ± SD. (B) CP2-binding site is critical for DAPK promoter 1 activity. The effect of mutagenizing four specific transcription factor-binding sites, CP2, Sp1, MZF (1) and MZ1 (2), within the minimized DAPK promoter 1 (332 bp fragment) on luciferase reporter activity was determined. (C) Individual sequences comprising different portions of the DAPK promoter 2–exon 1b–intron 1 were amplified by PCR, ligated into a luciferase expression vector and transfected into H522, H1568 and Calu-6 cells to identify the minimized region supporting reporter activity. (D) Comparison of reporter activity between different genomic regions of DAPK. Luciferase activity of reporter constructs containing promoter 1, promoter 2 or promoter 2–exon 1b was compared with activity of the 969 bp construct that included promoter 2–exon 1b–intron 1. (E) HNF3B-binding site is critical for DAPK intron 1 activity. The effect of mutagenizing three specific transcription factor-binding sites, HNF3, NF-κβ and MZ1, within intron 1 (176 bp fragment) on reporter activity was determined.
There are several putative binding sites for transcription factors in the minimized promoter. The three most common transcription factors identified were CP2, Sp1 and MZF (two putative binding sites). Mutation of the CP2-binding site (−1184) had a dramatic effect, reducing activity ≥65% in all cell lines (Figure 2B). In contrast, reporter activity was reduced by 25% with mutation of the Sp1- (−1301) or MZF- (−169) binding site. Mutation of the second MZF-binding site (−1269) only affected luciferase activity in the H1568 cells (Figure 2B).
Initial studies to characterize the region responsible for expression of the exon 1b transcript found only modest activity within the nucleotide sequence −969 to −627 (promoter 2) located between exon 1 and 1b (Figure 1A, data not shown). Several studies demonstrate that introns can enhance promoter activity (23–25). Inspection of DAPK intron 1 identified multiple transcription factor-binding sites suggesting this region could contribute to transcription of this alternate transcript. Therefore, we evaluated luciferase activity in the sequence extending from −969 to −1 that included promoter 2, exon 1b and intron 1 and created four deletion constructs (Figure 2C). There was no effect on luciferase activity in the three cell lines following deletion of promoter 2 and exon 1b (−450 to −1 construct). Furthermore, reporter activity was largely unchanged in the H522 and H1568 cells in the next two deletion constructs but was totally abolished in the construct that included only 86 nt (Figure 2C). Interestingly, a decline in reporter activity in the Calu-6 cell line was seen in the 255 and 176 bp constructs, suggesting that additional regulatory factors present in this cell line are modulating luciferase activity in the full-length construct. The generation of a −176 to −86 construct restored 50–80% of reporter activity in these cell lines, suggesting that the most important transcription factors for sustaining reporter activity were binding within this region.
Reporter activity of promoter 1 (−2533 to −1025) was 40–50% higher than promoter 2–exon 1b–intron 1 (−969 to −1) consistent with the lower expression of the exon 1b transcript (Figure 2D). Additional constructs were made to compare reporter activity of the 969 bp sequence to promoter 2 (−969 to −627) and to promoter 2 and exon 1b (−969 to −436). Luciferase activity for promoter 2 was 50% less than that seen with the 969 bp fragment in all cell lines (Figure 2D). The addition of exon 1b had minimal effect on reporter activity. The fact that the 450 bp fragment containing only intron 1 has similar luciferase activity to the full-length construct suggests there is a repressor within promoter 2 (Figure 2C) and that activity within intron 1 probably contributes substantially to expression of the exon 1b transcript. Therefore, we focused on identifying the transcription factors within intron 1, specifically the 90 bp region from −176 to −86 because of its high reporter activity. The three most common transcription factors identified in this region were HNF3B, MZF and NF-κB. Mutation of the HNF3B-binding site reduced luciferase activity by 60–70% in all cell lines, whereas activity was reduced by ∼25% when either the MZF or NF-κB-binding sites were mutagenized (Figure 2E).
Relationship between methylation of DAPK promoters and expression of transcripts
The methylation of DAPK promoter 1, promoter 2, exon 1, exon 1b and intron 1 was assessed by bisulfite sequencing and MSP in 12 and 3 cell lines derived from lung and breast tumors, respectively. The relationship between methylation density by sequencing and MSP (yes or no) to expression of the exon 1 and 1b transcripts was determined (Table I). Sequencing evaluated 49, 42 and 30 CpG dinucleotides in promoter 1–exon 1, promoter 2–exon 1b and intron 1, respectively. Five cell lines (H2009, Calu-1, MDA-MB-231, MCF-7 and T47D) showed methylation of 51–91% of the CpGs in the promoter 1–exon 1 region and this was associated with complete loss of transcription of exon 1 (Figure 3A, Table I). The common areas for dense methylation were localized to CpGs 13–23 and 35–45. The transcriptional start site and the major transcription factor for promoter 1, CP2, are localized within the region that includes CpGs 35–45 (Figure 3A). The remaining cell lines showed no to sparse methylation of promoter 1–exon 1 and expressed the exon 1 transcript (Table I).
Table I.
Relationship between methylation of DAPK promoters, intron 1 and expression of transcripts
| Cell line | Prom. 1 methyl. status |
Exon 1 expr. | Prom. 2 methyl. status |
Intron 1 methyl. status |
Exon 1b expr.b | |||
| Seq. (%)a | MSP | Seq. (%)a | MSP | Seq. (%) | MSP | |||
| Calu-6 | 0 | U | + | 0 | U | 0 | U | + |
| H2009 | 59 | M | − | 71 | M | 57 | M | − |
| H1568 | 0 | U | + | 0 | U | 0 | U | + |
| H358 | 0 | U | + | 0 | U | 0 | U | + |
| A549 | 0 | U | + | 0 | U | 3 | U | + |
| SKLU-1 | 4 | U | + | 0 | U | 40 | M | − |
| H596 | 0 | U | + | 0 | U | 27 | M | − |
| H522 | 0 | U | + | 0 | U | 0 | U | + |
| H23 | 0 | U | + | 2 | U | 50 | M | − |
| H125 | 4 | U | + | 2 | U | 10 | U | + |
| H460 | 12 | U | + | 12 | M | 43 | M | − |
| Calu-1 | 55 | M | − | 26 | M | 17 | M | − |
| MDA-MB-231 | 82 | M | − | 86 | M | 67 | M | − |
| MCF-7 | 90 | M | − | 93 | M | 97 | M | − |
| T47D | 51 | M | − | 10 | U | 40 | M | − |
Expr, expression; M, methylated; Methyl, methylation; Prom, promoter; Seq, sequence; U, unmethylated.
Values are the percent of methylated CpGs among the five clones.
Expression of exon 1b was greatly reduced in the H2009 and T47D cell lines and absent in other cell lines as indicated by –.
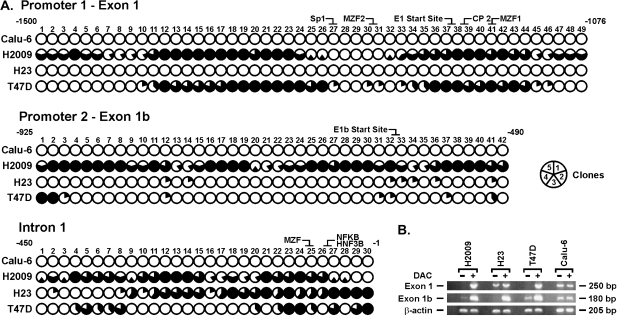
Fig. 3.
Methylation patterns within promoter 1–exon 1, promoter 2–exon 1b and intron 1 silence expression of specific transcripts. (A) Genomic regions containing 49, 42 and 30 CpG sites within promoter 1–exon 1, promoter 2–exon 1b and intron 1, respectively, were amplified and sequenced (five clones per sample). Each circle is divided into five equal parts (see key), representing the methylation status of one clone for the specified CpG. Filled regions, methylated clones; open regions, unmethylated clones. (B) DAC treatment restores expression of exon 1 and 1b transcripts. In Calu-6 cells where no methylation is present, DAC treatment has no effect on expression. In contrast, DAC treatment restores or increases expression of both transcripts in H2009 and T47D and exon 1b transcript in H23.
There was high concordance between being methylation positive in promoter 2–exon 1b and in intron 1. Four cell lines (H2009, Calu-1, MDA-MB-231 and MCF-7) showed methylation of 26–91% and 17–97% of the CpGs located in the promoter 1–exon 1b region and intron 1, respectively and this was associated with loss of expression (Figure 3A, Table I). Interestingly, little to no methylation in promoter 2–exon 1b was seen in four cell lines (SKLU-1, H596, H23 and T47D), whereas 27–50% of CpGs were methylated in intron 1 and this was associated with loss of expression of the exon 1b transcript (Table I). Methylation within intron 1 was localized to CpGs 23–30 in these cell lines, the location for binding of the three transcription factors that modulate reporter activity for this intron (Figure 3A). Both transcripts were expressed in the remaining cell lines (e.g. H358) where no methylation was seen in these CpG islands.
Primers for MSP were designed to areas of the highest density for methylation in promoter 1–exon 1, promoter 2 and intron 1 seen with sequencing. MSP assays in cell lines were 100% concordant with sequencing results and expression status of both transcripts (Table I). Cell lines with methylated and unmethylated DAPK promoters 1 and 2 and intron 1 were treated with the cytosine demethylating agent, DAC, to confirm the cause and effect relationship between CpG island hypermethylation and loss of gene expression. In Calu-6 cells that express both transcripts, treatment with DAC did not change expression levels, whereas expression of both transcripts was restored or increased in H2009 and T47D cells (Figure 3B). Expression of the exon 1b transcript that is silenced in H23 cells was restored following DAC treatment (Figure 3B).
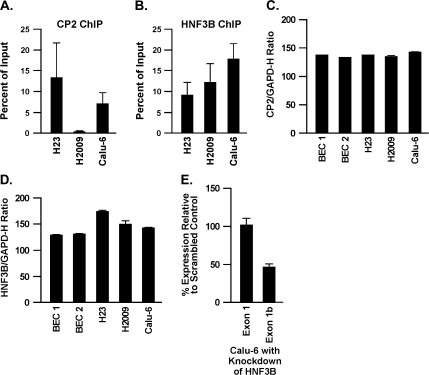
ChIP was conducted to assess the relationship between methylation, expression and binding of the CP2 and HNF3B proteins to promoter 1–exon 1 and promoter 2–exon 1b, respectively. Both proteins bound their respective sites in Calu-6 cells that are completely unmethylated across the two DAPK promoters and express both transcripts (Figure 4). In contrast, no binding of CP2 was seen for H2009 cells that are densely methylated around this transcription factor-binding site and in turn show no expression of the exon 1 transcript (Figures 3A and B and 4A). Binding of CP2 was readily seen in H23 cells that are unmethylated within promoter 1 and express the exon 1 transcript. Finally, binding of HNF3B was reduced to 35 and 50% in H2009 and H23 cells, respectively, as compared with Calu-6 and this also generally correlated with methylation density and expression of the exon 1b transcript (Figures 3A and B and 4B). Quantitative RT–PCR revealed similar levels of expression of the CP2 and HNF3B transcripts in Calu-6, H2009 and H23 compared with normal bronchial epithelial cell lines (Figure 4C and D). Thus, the reduced binding of these transcription factors detected by ChIP in the specific lung cancer cell lines was not due to differences in endogenous expression of these genes across cell lines. To further substantiate a role for HNF3B in regulating expression of the exon 1b transcript, siRNA experiments were performed. Transient transfection of siRNA to HNF3B in Calu-6 cells reduced expression of this transcription factor by 60%. This resulted in a 54% reduction in expression of the exon 1b transcript and no effect on expression of the exon 1 transcript compared with scrambled control (Figure 4E).
Fig. 4.
Binding of CP2 and HNF3B proteins to DAPK promoter 1 and 2 and expression in lung cancer cell lines. Antibodies to CP2 (A) and HNF3B (B) proteins were used to assess binding of these proteins to the DAPK promoters using EZ ChIP™. Binding of these proteins is compared between the Calu-6, H2009 and H23 cell lines. Densitometry compared the intensity of the PCR products between cell lines and values are the average ± SD from three separate PCRs. All values were normalized using input DNA. Expression of CP2 (C) and HNF3B (D) in two normal bronchial epithelial (BEC) cell lines, Calu-6, H2009 and H23 is depicted as the ratio of the fluorescence intensity value for each gene to that of GAPD-H, multiplied by 100. siRNA to HNF3B selectively reduces the expression of exon 1b transcript (E) in Calu-6 cells. Values are the average ± SD from three separate PCRs.
DAPK methylation in primary lung and breast tumors
Promoters 1 and 2 and intron 1 were methylated in 46, 74 and 66% of lung adenocarcinomas, respectively (Table II). Thirteen tumors were methylated in all three regions, whereas methylation of at least one region was seen in 87% of tumors. The prevalence for methylation of promoter 2 (39%) and intron 1 (55%) was significantly greater than promoter 1 (13%) in breast tumors. Only five breast tumors were methylated in all three regions, whereas 58% of tumors showed hypermethylation within at least one region. There was close to a 100% concordance for methylation in promoter 1 predicting for methylation in either promoter 2 or intron 1 for both lung and breast tumors (data not shown). In contrast, the concordance between methylation in promoter 2 and intron 1 was 77 and 50% in lung and breast tumors, respectively (data not shown).
Table II.
Prevalence for methylation of promoter 1 and 2 and intron 1 in lung and breast tumors
| Tumor type | Prevalence (%) of methylation for the region |
|||
| Promoter 1 | Promoter 2 | Intron 1 | All three regions | |
| Lung | 16/35 (46) | 26/35 (74) | 23/35 (66) | 13/35 (37) |
| Breast | 5/38 (13) | 15/38 (39)a | 20/38 (55)a | 5/38 (13) |
P < 0.05 when compared with promoter 1.
Discussion
This study identified two transcripts for the DAPK gene that code for a single protein while being regulated by two distinct active promoters and transcription factors. A strong correlation was seen between gene silencing of exon 1 and 1b transcripts and methylation of CpGs localized to regions where the CP2 and HNF3B transcription factors bound, respectively. Finally, the prevalence for silencing of the exon 1b transcript by promoter hypermethylation was significantly greater than for exon 1 in primary breast tumors.
Our studies have revealed significant associations and tissue-specific differences with respect to silencing of these transcripts by methylation. Methylation of promoter 1 and silencing of the exon 1 transcript were always associated with methylation of promoter 2 and/or intron 1 and silencing of the exon 1b transcript in both lung and breast tumors. This could stem from the initiation of methylation within promoter 1 and subsequent spreading to the transcriptional start sites, a scenario seen for the RUNX3 gene promoter (26). In contrast, our studies also clearly demonstrate that expression of the exon 1 transcript does occur in the absence of expression of the exon 1b transcript and this is associated with differences in methylation across the DAPK CpG island. Interestingly, there appears to be tissue-specific differences in coordinate silencing of the DAPK transcripts as evident by a similar versus 4-fold greater prevalence for methylation within intron 1 compared with promoter 1 in lung and breast tumors, respectively. Another important tumor suppressor gene whose transcription is regulated by two promoters and is silenced in cancer is hypermethylated in cancer-1 (HIC1; 27). HIC1 is transcribed from two promoters to produce two non-coding exons. However, in contrast to DAPK, dense methylation of either HIC1 promoter was associated with complete loss of transcription (28).
Deletion constructs of DAPK promoter 1 identified a 332 bp region that appeared most critical for reporter activity. Reporter activity was markedly reduced by deletion of the CP2 transcription factor-binding site located in the 5′-untranslated region of exon 1 and ChIP confirmed the binding of this protein within promoter 1. CP2 is a member of the NTF-like (neurogenic element-binding transcription factor) family of transcription factors. CP2 binds to human c-fos, ornithine decarboxylase, c-myc and DNA polymerase promoters and activates transcription in vitro and its activity can be modulated by cell growth signals (29,30). The methylation of CpGs 38 and 39 that reside within the CP2-binding site (Figure 3A) was strongly associated with loss of expression of the exon 1 transcript and blocked the binding of the CP2 protein, supporting an essential role for CP2 in regulating expression of this transcript.
A different paradigm was seen with respect to regulation of transcription initiating within promoter 2. While modest reporter activity was seen in the region designated as promoter 2, the region comprising intron 1 showed activity equivalent to that seen for constructs that included promoter 2 and intron 1. In fact high reporter activity was localized to a 90 bp sequence (−176 to −86) within intron 1 suggesting that an enhancer present within this region influences promoter 2 activity. Enhancers within the first intron of the osteopontin and cathepsin L genes significantly increase promoter activity and gene expression (23,24). Site-directed mutagenesis of the HNF3B-binding site within intron 1 identified this transcription factor as a major regulatory protein for transcription initiating within promoter 2. HNF3B, also called FOXA2, is a member of the winged helix/forkhead family of transcription factors that play a role in early development, organogenesis and in metabolism and homeostasis (31). For example, HNF3B is an important positive regulator of mucin MUC2 expression in the small intestine (32). Pertinent to our findings, HNF3B regulates expression of the microtubule-associated protein 1A gene through a binding site within an intronic region upstream of exon 3 (33). The importance of this transcription factor for expression of exon 1b was evident by the association between methylation of CpGs 23–30 around this transcription factor-binding site and loss or reduced expression of this transcript. The fact that expression of the exon 1b transcript is reduced or completely absent in cell lines that show minimal to no methylation within promoter 2, but dense methylation around the HNF3B-binding site, suggests that while transcription is probably being initiated in promoter 2, elongation is being blocked due to lack of access of HNF3B to its binding site. This premise is supported by our ChIP assays showing reduced binding of HNF3B in H2009 and H23 cells. Further support for the inhibition of transcription by intragenic methylation was shown using a transfected transgene methylated specifically in a region downstream of the promoter. RNA polymerase II was depleted in the methylated region that had also adopted a closed heterochromatin structure (34).
Our studies have shown for the first time dual promoter regulation of DAPK, a tumor suppressor gene silenced in many diverse types of cancer. Extensive bisulfite sequencing of the two DAPK CpG islands revealed dense, albeit heterogenous methylation of CpGs that blocked access of transcription factor-binding proteins requisite for gene expression. Thus, assessment for functional inactivation of this gene in tumors will necessitate evaluation of specific regions within the two CpG islands regulating the transcription of this gene. Methylation of DAPK has also shown promise as a molecular marker in sputum for identifying incident lung cancer, and future studies should assess both regions for silencing (35).
Funding
National Institute of Health (R01 ES008801).
Acknowledgments
Conflict of Interest Statement: S.A.B. is a consultant to Oncomethylome Sciences. Under a licensing agreement between Lovelace Respiratory Research Institute and Oncomethylome Sciences, nested MSP was licensed to Oncomethylome Sciences, and the author is entitled to a share of the royalties received by the Institute from sales of the licensed technology. The Institute, in accordance with its conflict of interest policies, is managing the terms of these arrangements.
Glossary
Abbreviations
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- DAC
5-aza-2′-deoxycytidine
- DAPK
death-associated protein kinase
- MSP
methylation-specific polymerase chain reaction
- NF-κB
nuclear factor-kappaB
- PCR
polymerase chain reaction
- RT–PCR
reverse transcription–polymerase chain reaction
References
- 1.Lehmann U, et al. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin. Cancer Res. 2009;11:3654–3660. doi: 10.1158/1078-0432.CCR-04-2462. [DOI] [PubMed] [Google Scholar]
- 2.Katzenellenbogen RA, et al. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- 3.Tang X, et al. Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small cell lung cancer. J. Natl Cancer Inst. 2000;92:1511–1516. doi: 10.1093/jnci/92.18.1511. [DOI] [PubMed] [Google Scholar]
- 4.Kim DH, et al. Promoter methylation of DAP-kinase: association with advanced stage in non-small cell lung cancer. Oncogene. 2001;20:1765–1770. doi: 10.1038/sj.onc.1204302. [DOI] [PubMed] [Google Scholar]
- 5.Belinsky SA, et al. Predicting gene promoter methylation in lung tumors by evaluating sputum and serum. Br. J. Cancer. 2007;96:1278–1283. doi: 10.1038/sj.bjc.6603721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann U, et al. Promoter hypermethylation of the death-associated protein kinase gene in breast cancer is associated with the invasive lobular subtype. Cancer Res. 2002;62:6634–6638. [PubMed] [Google Scholar]
- 7.Dulaimi E, et al. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin. Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 8.Belinsky SA, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 9.Deiss LP, et al. Identification of a novel serine-threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 10.Inbal B, et al. Death associated protein kinase-related protein 1, a novel serine-threonine kinase involved in apoptosis. Mol. Cell. Biol. 2000;20:1044–1054. doi: 10.1128/mcb.20.3.1044-1054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen O, et al. DAP-kinase participates in TNF-α- and Fas-induced apoptosis and its function requires the death domain. J. Cell Biol. 1999;146:141–148. doi: 10.1083/jcb.146.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelled D, et al. Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. J. Biol. Chem. 2002;277:1957–1961. doi: 10.1074/jbc.M104677200. [DOI] [PubMed] [Google Scholar]
- 13.Jang CW, et al. TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 2002;4:51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 14.Cohen O, et al. DAP-kinase: from functional gene cloning to establishment of its role in apoptosis and cancer. Cell Death Diff. 2001;8:6–15. doi: 10.1038/sj.cdd.4400794. [DOI] [PubMed] [Google Scholar]
- 15.Raveh T, et al. DAP-Kinase activates a p19ARF/p53-mediated apoptotic checkpoint to suppress oncogenic transformation. Nat. Cell Biol. 2001;3:1–7. doi: 10.1038/35050500. [DOI] [PubMed] [Google Scholar]
- 16.Inbal B, et al. DAP-kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Gomez P, et al. Frequent death-associated protein-kinase promoter hypermethylation in brain metastases of solid tumors. Oncol. Rep. 2003;10:1031–1033. [PubMed] [Google Scholar]
- 18.Lévy D, et al. Death-associated protein kinase loss of expression is a new marker for breast cancer prognosis. Clin. Cancer Res. 2004;10:3124–3130. doi: 10.1158/1078-0432.ccr-03-0213. [DOI] [PubMed] [Google Scholar]
- 19.Livingston RJ, et al. Homo sapiens Death-associated Protein Kinase 1 (DAPK1) Gene, Complete cds. Seattle, WA: Department of Genome Sciences; 2004. NIEHS ES15478. [Google Scholar]
- 20.Toyooka S, et al. Epigenetic down-regulation of death-association protein kinase in lung cancers. Clin. Cancer Res. 2003;9:3034–3041. [PubMed] [Google Scholar]
- 21.Quandt K, et al. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai D, et al. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giacopelli F, et al. The first intron of the human osteopontin gene contains a C/EBP-beta-responsive enhancer. Gene Expr. 2003;11:95–104. doi: 10.3727/000000003108748991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charron M, et al. The cathepsin L first intron stimulates gene expression in rat sertoli cells. Biol. Reprod. 2007;76:813–824. doi: 10.1095/biolreprod.106.057851. [DOI] [PubMed] [Google Scholar]
- 25.Kolb A. The first intron of the murine beta-casein gene contains a functional promoter. Biochem. Biophys. Res. Commun. 2003;306:1099–1105. doi: 10.1016/s0006-291x(03)01104-5. [DOI] [PubMed] [Google Scholar]
- 26.Homma N, et al. Spreading of methylation within RUNX3 CpG island in gastric cancer. Cancer Sci. 2006;97:51–56. doi: 10.1111/j.1349-7006.2005.00133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerardel C, et al. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J. Biol. Chem. 2001;276:3078–3089. doi: 10.1074/jbc.M008690200. [DOI] [PubMed] [Google Scholar]
- 28.Chen WY, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat. Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 29.Volker JL, et al. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 1997;11:1435–1446. doi: 10.1101/gad.11.11.1435. [DOI] [PubMed] [Google Scholar]
- 30.Shirra MK, et al. LSR and NTF-1 share a conserved DNA recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J. Biol. Chem. 1998;273:19260–19268. doi: 10.1074/jbc.273.30.19260. [DOI] [PubMed] [Google Scholar]
- 31.Friedman JR, et al. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Sluis M, et al. Forkhead box transcription factors Foxa1 and Foxa2 are important regulators of Muc2 mucin expression in intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2008;369:1108–1113. doi: 10.1016/j.bbrc.2008.02.158. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama A, et al. Characterization of two promoters that regulate alternative transcripts in the microtubule-associated protein (MAP) 1A gene. Biochim. Biophys. Acta. 2001;1518:260–266. doi: 10.1016/s0167-4781(01)00173-7. [DOI] [PubMed] [Google Scholar]
- 34.Lorincz MC, et al. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 35.Belinsky SA, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high- risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]