Abstract.
This review focuses on the known factors required for selenocysteine (Sec) incorporation in eukaryotes and highlights recent findings that have compelled us to propose a new model for the mechanism of Sec incorporation. In light of this data we also review the controversial aspects of the previous modelspecifically regarding the proposed interaction between SBP2 and eEFSec. In addition, the relevance of two recently discovered factors in the recoding of Sec are reviewed. The role of the ribosome in this process is emphasized along with a detailed analysis of kinkturn structures present in the ribosome and the L7Ae RNA-binding motif present in SBP2 and other proteins.
Key words. Selenocysteine, SBP2, eEFSec, L7Ae, L30
Selenium and selenocysteine
Most trace elements function as cofactors for various enzymes and are thus essential for their activity. Unlike other cofactors which interact with their cognate protein through post-translational modifications, selenium is incorporated into proteins co-translationally. Selenium is unique because it is biologically active in the form of the amino acid selenocysteine (Sec), which is incorporated into at least 25 selenoproteins in humans. Despite its exclusion from many textbooks, Sec is widely considered to be the 21st amino acid and exists in all three lineages of life. The significance of the properties that it imparts on selenoenzymes, such as the glutathione peroxidases, is found in the simple fact that the pKa of Sec is much lower than that of Cys, allowing a much greater nucleophilic activity at physiological pH [reviewed in ref. 1]. Sec has captured the attention of researchers in the protein synthesis field because unlike other amino acids, its insertion into proteins requires a translational recoding event that occurs at select UGA codons, which usually signal translation termination. While the mechanism governing insertion of Sec in prokaryotes has been fairly well deciphered [reviewed in ref. 2], the mechanism in eukaryotes has proven more difficult to discern. The topic of selenoprotein biology and the general mechanisms of eukaryotic Sec incorporation have been thoroughly reviewed in recent years [3–8]. This work will focus on the biochemical details regarding each of the known protein factors involved in Sec incorporation, assembling the latest findings into the larger picture of how the ribosome switches languages midstream.
The factors
SECIS elements
Common to both the prokaryotic and the eukaryotic system are the two known cis-acting factors required for Sec incorporation: the UGA codon and a stable stemloop structure referred to as the selenocysteine insertion sequence (SECIS) element. Likewise, both systems utilize a SECIS-binding protein and a Sec-specific elongation factor-functions embodied in a single factor in bacteria (SelB) and by two factors in eukaryotes (SBP2 and eEFSec/mSelB, respectively). Eukaryotic SECIS elements are exclusively found in the 3′ untranslated region (UTR) of every selenoprotein mRNA. In the case of selenoprotein P, the 3′ UTR contains two SECIS elements in tandem, the concerted action of which is probably required for the efficient decoding of the ten Sec codons found in mammalian Sel P. Part of the SECIS element is strikingly similar to an RNA structural motif recently dubbed the kink-turn [9]. As such, it contains a motif that is found in several RNA species including rRNA, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). Figure 1 illustrates the SECIS element found in the selenoprotein glutahione peroxidase 4 (GPX4) next to the consensus kink-turn sequence derived from those found in the crystal structure of the large ribosomal subunit derived from Haloarcula marismortui [9]. While the kink-turn motif within the SECIS element (refered to as the SECIS core) clearly functions as the SBP2-binding site [10], the apical stem and loop, which contain the conserved AAR sequence (see fig. 1), have no known function. As discussed below, they make an attractive target for the controversial idea that eEFSec physically interacts with the SECIS element at the terminal loop region.
Figure 1.
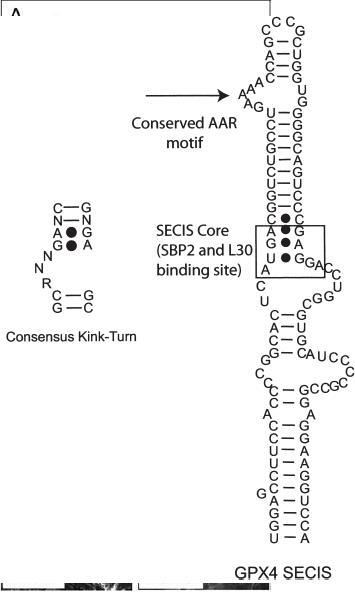
Comparison of the consensus kink-turn motif (left) and the GPX4 SECIS element (right). The SECIS core motif is boxed, emphasizing the SBP2- and rpL30-binding site. The conserved AAR motif is also noted within the apical bulge.
SECIS-binding factors
The most widely studied protein required for Sec incorporation is SECIS-binding protein-2 (SBP2). Rat SBP2 contains 846 amino acids and has three domains which were identified by structure/function analysis as well as sequence homology [11]. The three identified functions for SBP2 include Sec incorporation in vitro, binding to the SECIS core, and binding to the ribosome. The N-terminal 400 amino acids of this protein have no known function, and this domain is not conserved in the SBP2 found in lower eukaryotes (i.e., insects and worms). Since this domain possesses a predicted nuclear localization signal, nuclear retention may somehow contribute to or regulate SBP2 function. The current data do not support that idea, but are limited to the detection of strictly cytoplasmic SBP2 in transiently transfected rat hepatoma cells [12]. In addition, C-terminal SBP2 and full-length SBP2 have nearly identical Sec incorporation activities in vitro. The C-terminal 450 amino acids of this protein are sufficient for all three of the aforementioned functions. Amino acids 399–517 contain a functional domain required for Sec incorporation and amino acids 517–777 contain the SECIS-binding domain. Within this SECIS-binding domain is an L7Ae RNA-binding motif found in several proteins including the ribosomal protein L30 (rpL30) and the eukaryotic translation termination release factor 1 (eRF1). SBP2 and the SECIS element are believed to provide specificity for the incorporation of Sec at select UGA codons as opposed to the UGAs at the end of coding regions. Exactly how this specificity is achieved remains unclear but is likely to rely on the concerted action of all of the cis- and trans-acting factors described, and since the system has not been reconstituted in vitro, other as yet undiscovered factors could be required as well.
eEFSec/mSelB
The Sec-specific elongation factor was independently identified by two groups nearly simultaneously and termed either mSelB [13] or eEFSec [14]. This protein bears strong similarity to the canonical translation elongation factor eEF1A but contains a C-terminal extension that has been reported to interact with SBP2 but only when expressed as an isolated fragment [15]. Interestingly, bacterial SelB also contains a C-terminal extension relative to EF-Tu that is known to be required for binding to the bacterial SECIS element [16]; however, the bacterial and eukaryotic C-terminal extensions are not related [7]. In addition, the archaeal SelB protein lacks a C-terminal extension and does not possess SECIS element-binding activity [17]. eEFSec has been shown to specifically bind the Sec-tRNA[Ser]Sec and is predicted to act independently of a guanine nucleotide exchange factor based on relatively high intrinsic GTP affinity [13, 14]. Whether eEFSec interacts with the SECIS element and/or SBP2 is addressed in detail below.
Multimerization state of SBP2
Based on evidence of complex formation in glycerol gradients, SBP2 was hypothesized to self-associate and function as a homomultimer [11]. The fact that SBP2 has only one canonical RNA-binding domain but is known to interact with both the SECIS element and rRNA supported this idea, such that in a head-to-head dimer configuration, SBP2 would have two RNA-binding domains available to interact simultaneously with both components and provide a means for communication between the ribosome and SECIS elements downstream in the 3′ UTR.
Recent findings have challenged several aspects of this notion. Using a combination of gel filtration and pulldown assays, it was found that SBP2 did not self-associate [18]. Self-association and complex formation were attributed to a tag-tag interaction resulting from the Streptagged version of SBP2 which was shown to form a large salt-sensitive complex. Furthermore, a sucrose cushion assay demonstrated that SBP2 could not simultaneously interact with the ribosome and the SECIS element in vitro. The presence of a 2.5-fold excess of a wild-type SECIS element to ribosome, but not a mutant lacking the SECIS core, was able to effectively compete some of the SBP2 off of the ribosome. 32P-labeled SECIS element was present in supernatant fractions and not in the pellet fractions, suggesting that the competition observed was due to SBP2/SECIS interactions. Furthermore, when SECIS was incubated with ribosomes in the absence of SBP2, no pelleting of SECIS elements was observed, verifying that the SECIS element cannot independently associate with purified ribosomes.
These results are consistent with the fact that SBP2 is monomeric and contains only one canonical RNA-binding domain and further support the idea that the SBP2/ribosome interaction occurs at a kink-turn motif on the ribosome. The indication is that SBP2 function is dynamic and that the exchange of SBP2 between the ribosome and the SECIS element during translation may be important for the incorporation of Sec. If true, this would support the idea that the SBP2/ribosome interaction is somehow making ribosomes competent for the incorporation of Sec.
Kink-turns and SBP2
The most under-studied contributor of the Sec recoding mechanism is the ribosome. This lack of attention stems largely from the fact that although SBP2 has been shown to bind to the ribosome at the level of 28S rRNA [11], the significance of this interaction for the incorporation of Sec is unknown. SBP2 is known to interact stably and quantitatively both in vivo and in vitro with the ribosome [11,18]. This interaction is likely to occur at the level of the recently classified rRNA structures called kink-turns, described above (fig. 1). This strong structural resemblance provides the best explanation for an interaction between the SBP2 SECIS-binding domain and both the SECIS element and rRNA.
The kink-turn was described as a ‘new’ structural motif in 2001 [9] at about the same time as it was described as the ‘GA motif’ [19] by a separate group. These reports should not be confused with the original descriptions of GA base pairing which date back to the early 1980s [20, 21]. In fact, evidence for a kinked structure associated with sheared GA base pairs was provided by chemical and enzymatic analysis of SECIS elements in 1996 [22]. Rather, the more recent studies provide definitive structural data on multiple iterations of the motif. All of the known non-ribosomal kink-turn motifs have corresponding binding partners that possess a conserved binding motif (see below). Nine of the 28 large subunit ribosomal proteins in H. marismortui interact with at least some part of the six 23S kink-turns [9], but only one is bound by a protein possessing the conserved motif (L7Ae). This raises the intriguing possibility that the ribosome may support interactions with non-ribosomal proteins using exposed kink-turns as binding sites.
As mentioned above, the current evidence suggests that SBP2 may interact with a kink-turn on the ribosome. Indirect support of this idea is the fact that SBP2 is one member of a small family of proteins that possess a so-called L7Ae RNA-binding motif. This motif was first identified in 1994 in a purely computational study in which the RNA-binding function was proposed but not tested [23]. Table 1 lists the members of this group that are found in humans, illustrating both the similarities and differences in function associated with this motif. All of these factors seem to play a role in translation or splicing and most are somehow associated with ribosome or rRNA interactions. The structure of the 15.5-kDa U4 snRNA-binding protein complexed with its target RNA has been solved by X-ray crystallography [24]. In addition, the crystal structure of L7Ae has been solved both on the ribosome [25] and in association with Box C/D snoRNAs [26], and the solution structure of rpL30 in complex with its own mRNA has been solved by nuclear magnetic resonance, (NMR) spectroscopy. Figure 2 shows an alignment of human sequences from SBP2, rpL30, and L7Ae along with information derived from crystallographic data regarding the specific RNA/protein contacts. In addition, based on the work of Allmang et al. [27], the residues that have been mutated to alanine in human SBP2 are also noted by either red (mutations that resulted in little or no SECIS element binding) or green (resulting in between 46–64% binding). None of the mutant proteins bound as well as the wild-type control in that study. Together, these data paint a fairly clear picture of how SBP2 might interact with both the SECIS element and the ribosome, but there are a few inconsistencies that should be pointed out. First, SBP2 binding has been reported to be magnesium intolerant. Initial reports indicated an inhibitory concentration of about 4 mM [28], while a more recent study put that figure at about 20 mM [29]. Both SBP2 and rpL30 will bind the SECIS element in the absence of magnesium, and the former has been shown to be insensitive to high concentrations (>5 mM) of EDTA [28]. Recent work indicates that the kink-turn RNA structure is regulated by divalent cation concentration, where increasing magnesium concentrations resulted in stabilization of the kinked structure [30]. If this is the case, then SBP2 appears to favor the low-magnesium, unkinked state. Since all of the kink-turns described within the H. marismortui and Thermus thermophilus rRNA are by definition in the kinked conformation, it is perhaps unlikely that SBP2 is interacting with one of these unless crystallization procedures induced stable kinks that are flexible in vivo. Interestingly, the presence of kink-turns in the H. marismortui ribosome appears to be magnesium independent because subsequent analysis of the positions of all divalent cations in the structure showed no indication of magnesium near the kink-turns [31]. This is consistent with the idea that the kink is actually protein induced as has been shown for the L7Ae/snoRNA interaction [26]. Thus, SBP2 may interact with a helix that contains potential G:A base pairs distinct from those identified in the archaeal large ribosomal subunit, and the kinked structure may be induced by SBP2 binding — an attractive scenario considering that our model of SBP2 functionrelies on ribosome conformational changes.
Table 1.
List of human proteins containing the L7Ae RNA-binding motif.
| Common name | Accession (GI) | Size (kDa) | Function | Reference |
|---|---|---|---|---|
| rpL7Ae | 4506661 | 30 | core ribosomal protein, contacts 28S helix 15 | 26 |
| rpL30e | 63100761 | 13 | core ribosomal protein, autoregulation of splicing, targets: 28S helix 34 and rpL30 mRNA | 40 |
| rpS12e | 14277700 | 15 | core ribosomal protein, contacts unknown | 23 |
| 15.5 kDa | 72086684 | 10 | spliceasome component, binds U4 snRNA and Box C/D snoRNA | 24 |
| RNAse P (p38) | 13124513 | 32 | tRNA processing | 41 |
| NHP2 | 68565945 | 17 | rRNA modification, ribosome biogenesis | 42 |
| Gadd45β | 120751 | 18 | DNA damage induced factor, JNK pathway inhibitor | 43 |
| eRF1 | 4759034 | 49 | translation termination | 44 |
| SBP2 | 21359955 | 95 | Sec incorporation | 4 |
| SLP (KIAA0256) | 59803102 | 122 | unknown function | 11 |
Figure 2.
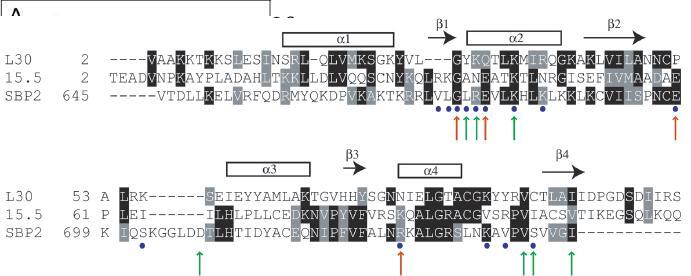
Alignment of the core L7Ae binding motif in the human versions of the 15.5-kDa snRNA-binding protein, rpL30 and SBP2. Residues shaded in black represent identities, those shaded in gray represent conservative substitutions. The secondary-structure motifs, derived from the 15.5-kDa co-crystal analysis [24] are illustrated above the sequence. Red arrows indicate alanine substitutions in SBP2 that inactivated SECIS element-binding activity. Green arrows indicate alanine substitutions in SBP2 that had a minimal affect on SECIS element binding [27].
Another apparently unique feature of the known SECIS element interactions is the fact that a much larger piece of RNA is required for optimal binding than has been suggested by the co-crystal data. Both SBP2 and rpL30 prefer to bind to a SECIS element with an extended stem that totals 105 nucleotides of the GPX4 SECIS [28, 29]. This structure is considerably larger than that used in the co-crystal and NMR studies, perhaps suggesting that SECIS element interactions are fundamentally different than those that occur on rRNA or snRNAs. Indeed, this phenomenon combined with the implications surrounding magnesium sensitivity may be the basis for the ability of SBP2 to discriminate between the ribosome and the SECIS element during incorporation (see below).
In sum, the body of information regarding the L7Ae family of proteins likely provides some information about how SBP2 interacts with its target RNAs, but little, if any, information about how SBP2 actually works to support Sec incorporation. One intriguing aspect of this story is that the translation termination factor eRF1 also possesses an L7Ae RNA-binding domain. While the function of this domain with respect to the termination reaction is not known, it is an obvious target for future investigation, since at some level, SBP2 and eRF1 may compete for the same binding site.
The SBP2/ribosome interaction. One observation that supports the idea that the SBP2/ribosome interaction is essential is the fact that the ribosomal binding site for SBP2 appears to be conserved. We have observed that SBP2 also binds Escherichia coli ribosomes and yeast ribosomes [S. A. Kinzy and P. R. Copeland, unpublished observation], the latter being an organism that does not synthesize selenoproteins. Unfortunately, the interaction between SBP2 and the ribosome is not as straightforward as one would initially predict. Although truncation analysis reveals that the SECIS-binding domain of SBP2 is necessary for its interaction with the ribosome, a single point mutant in the L7Ae RNA-binding motif (G669R) had no effect on ribosome binding. This same mutation resulted in a dramatic loss of SBP2 binding to the SECIS element which indicates that the SECIS-binding and ribosome-binding domains overlap but are not identical. In addition, truncations within the functional domain of SBP2 also affect ribosome binding while leaving SECIS element binding intact [11]. Together these results paint a convoluted picture that goes beyond a simple competitive model where SBP2 either binds to 28S rRNA via a kinkturn or to the SECIS element. Clearly, the interaction is more complex, and in that complexity most likely lies the heart of SBP2 function in promoting Sec incorporation, a concept in limbo until formal proof of the requirement for an SBP2/ribosome interaction is provided. To determine definitively whether the SBP2/ribosome interaction is essential requires identifying a point mutant with properties opposite to that of the G669R mutant described above. That is, a mutant of SBP2 that fails to bind the ribosome yet retains its SECIS-binding capability. Such a mutant version of SBP2 has not yet been reported.
New factors
Until recently, the UGA recoding machinery was thought to be comprised of the two cis-acting factors and the three trans-acting factors described above. As previously mentioned, Sec incorporation in vivo is likely to be an efficient event, but to date all studies of efficiency using transfection systems or in vitro translation of reporter mRNAs have produced values in the range of 1–10% [12, 32, 33]. These studies suggest that other factors may be required for efficient Sec incorporation.
Numerous cases of programmed frameshifting allowing stop codon read-through have been shown, and in many cases, frameshifting efficiency is determined by mRNA elements near the stop codon [34]. In an effort to learn how similar elements within the coding region of selenoprotein mRNAs may affect Sec incorporation, Howard et al. [35] sought to identify other elements that may be involved in read-through in selenoprotein mRNAs. Using a combination of comparative sequence analysis and site-directed mutagenesis, they discovered an element in the vicinity of the selenoprotein N UGA codon. When cloned into a luciferase construct, this stem-loop was found to increase read-through in the absence of a SECIS element. Interestingly, several mutants that changed base identity but not overall structure failed to restore read-through, indicating the importance of sequence identity and hinting at the potential for the existence of a factor that binds in a sequence-dependent manner. The addition of a SECIS element to the 3′ UTR further stimulated read-through in an additive fashion. As pointed out by the authors, a major limitation to these findings is the fact that Sec incorporation per se was not analyzed and the effect was not specific for UGA, since a UAG codon also supported read-through. The absence of this redefinition element in selenoprotein P is also troubling since it would seem to be a logical recipient of the aid of a potential efficiency element. The contribution of this element in the context of Sec incorporation awaits further research to clarify its role in read-through mediated by the insertion of Sec at the UGA codons in selenoprotein mRNAs.
Chavatte et al. [29] recently showed that rpL30 can bind to a SECIS element in vitro an in vivo and enhance the incorporation of Sec in transfected cells. The involvement of rpL30 is not entirely surprising since, like SBP2, it also contains an L7Ae RNA-binding motif. The rpL30/SECIS element interaction is specific, however, since several other proteins containing the L7Ae motif were shown not to bind under the same conditions. The authors propose that the SECIS element is acting as a molecular switch that binds to rpL30 under conditions where it forms a kink-turn. Alternatively, rpL30 has been suggested to function as a tether for communication between the ribosome at the upstream in-frame UGA codon and the SECIS element found far downstream [36]. The latter is interesting in the context of our recent observation that SBP2 does not appear to be interacting simultaneously with the ribosome and the SECIS element, but both rpL30 and SBP2 seem to be capable of providing communication with the ribosome. The potential role of rpL30 for the incorporation of Sec is reviewed below in the context of our revised model.
Interactions
The most controversial aspect of all the current models pertaining to Sec incorporation is the proposed interaction between the Sec-specific elongation factor eEFSec and both SBP2 and the SECIS element. First, with regard to the eEFSec/SECIS interaction, the papers often cited for evidence of a lack of an interaction between eEFSec and the SECIS element actually show an interaction in the absence of other factors. In one case, the binding persisted, albeit at a lower level, when mutations were present in the SECIS core [13], while in the other case, a mutation in the SECIS core had no effect on SECIS element binding except when SBP2 was added to the reaction, in which case no binding was observed at all [14]. A mutation in the conserved AAR motif in the apical loop has not been tested but the motif seems like a reasonable binding site since no factors have been identified that bind there, including SBP2, and mutations in this motif are sufficient to prevent Sec incorporation [37].
The contribution of other cellular factors to the eEFSec/SECIS interaction was investigated by analyzing binding in the presence of HeLa whole-cell extracts [13]. The authors suggest that the binding of eEFSec is enhanced in the presence of a factor that anchors eEFSec to the SECIS element, as demonstrated by a supershift during elecrophoretic mobility shift assays (EMSAs). Interestingly, when the experiment was repeated using a SECIS element with a point mutation in the SECIS core, the extent of super shifting observed was significantly reduced, although some complexes were still detected. From these data, the authors conclude that this anchor may be SBP2, however, the involvement of an as yet unidentified factor was not ruled out. An alternative possibility is that this other factor and eEFSec simultaneously but independently bind the SECIS element.
In keeping with the idea that Sec incorporation requires the tethering of elongation and SECIS binding as is the case in bacteria, initial analysis of an SBP2/eEFSec interaction by co-immunoprecipiation from cells overexpressing both factors indicated that the two proteins do interact but that complex formation was RNA dependent (i.e. RNAse A sensitive [14]). As described above, both eEFSec and SBP2 have been shown to independently bind the SECIS element. The expectation, then, was that if these two components were simultaneously binding the SECIS element, then a super shift would be observed. Unfortunately, EMSA assays containing both SBP2 and eEFSec did not clearly demonstrate that these factors were bound to the SECIS element at the same time since no super shifting could be observed [14], but further investigation will be required to settle the question definitively. Together, these results indicate that eEFSec does bind the SECIS element independently of SBP2, but whether this accounts for the co-immunoprecipitation of the two proteins from cell extracts remains to be determined.
An important consideration that had not been addressed was the contribution, if any, of the Sec-tRNA[Ser]Sec to the eEFSec/SBP2 interaction. Recently, however, it was shown that SBP2 and eEFSec can be co-immunoprecipitated in HEK-293 cells overexpressing these two proteins, an interaction that was significantly enhanced when Sec-tRNA[Ser]Sec was overexpresed as well [15]. This interaction could not be replicated in rabbit reticulocyte lysate unless truncations of eEFSec were utilized. The authors suggest that the reticulocyte was limiting for some cellular factor that allowed for this interaction and they predicted that this factor was Sec-tRNA[Ser]Sec. The binding of Sec-tRNA[Ser]Sec to eEFSec was hypothesized to alter the conformation of the C-terminal extension of eEF-Sec such that it could interact with SBP2. However, a major limitation to this idea is that rabbit reticulocyte is only limiting for SBP2 and is not limiting for Sec-tRNA[Ser]Sec [12]. The inability to immunoprecipitate SBP2 and eEFSec in rabbit reticulocytes could be attributed to the poor expression of SBP2 seen in those reactions [15]; however, even when SBP2 is overexpressed relative to eEFSec in rabbit reticulocyte lysate, co-immunoprecipitation is not observed [P. R. Copeland, unpublished data]. The contradictory data derived from transfected cells and in vitro translation remain an unresolved issue, but strongly suggest that the interaction between SBP2 and eEFSec is not direct.
The cumulative data suggest that eEFSec does, in fact, bind the SECIS element, but biochemical detail is lacking to address the location of binding or whether the C-terminal extension in eEFSec is responsible for the interaction as it is in the case of bacterial SelB. As for the eEFSec/SBP2 interaction, more work needs to be done to clarify the conflicting data described above. The function of eEFSec outside its role as an elongation factor awaits further investigation to determine precisely where and when it acts, and whether these putative interactions are essential for the recoding of UGA to Sec.
Current models for the mechanism of Sec incorporation
The incorporation of Sec into a distinct class of essential proteins requires a modification of the standard translation machinery such that UGA ceases to function as a stop codon. Currently, two distinct models have been proposed to describe Sec incorporation. The most commonly cited model speculates that SBP2 is bound to the SECIS element prior to translation. In this scenario, the SBP2-SECIS complex recruits eEFSec/Sec-tRNA[Ser]Sec to the SECIS element. Consequently, the Sec-tRNA[Ser]Sec is delivered to the ribosomal A-site (fig. 3A).
Figure 3.
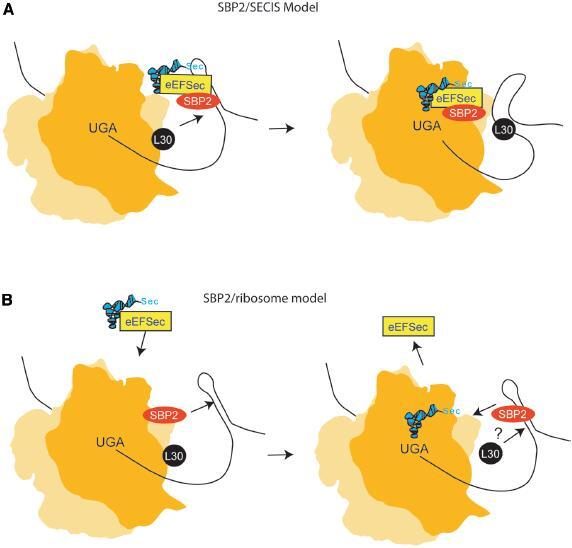
Contrasting models for Sec incorporation. (A) A common model for Sec incorporation. An SBP2-bound SECIS element recruits the eEFSec/Sec-tRNA[Ser]Sec complex. After association with the ribosome, SBP2 is exchanged for rpL30 resulting from a kinked SECIS element. This conformational change in the SECIS element is hypothesized to trigger the release of the Sec-tRNA[Ser]Sec and GTP hydrolysis. (B) A model of Sec incorporation based on the topics discussed in this review. SBP2 is pre-bound to the ribosome. Upon arrival at a UGA codon, ribosome stalling allows for a distant SECIS element to find and interact with SBP2. Movement of SBP2 may trigger conformational changes in the ribosomal A-site that favor the binding of eEFSec/Sec-tRNA[Ser]Sec and the subsequent insertion of Sec. After incorporation, rpL30 may function to displace SBP2 off the SECIS element and back to its original position on the ribosome.
The recent finding that rpL30 specifically binds the SECIS element has stimulated discussion regarding its role in Sec incorporation. With this finding, an updated version of this model has been offered in which rpL30 was proposed to function as an anchor between the SECIS element and the ribosome. It has been further suggested that rpL30 displaces SBP2 from the SECIS element during the recoding event, but biochemical data with regard to the mechanism of rpL30 function during Sec incorporation are thus far lacking [29].
As described above, the current data do not support many features of the model shown in figure 3A. We propose an alternative model for the insertion of Sec, where SBP2 remains ribosome bound and only interacts with the SECIS element during active translation or perhaps only during Sec incorporation (fig. 3B). SBP2 has been shown to stably and quantitatively bind to ribosomes, which indicates that there is no cytoplasmic pool of SBP2 that is interacting with the SECIS element in the absence of translation. The fact that SBP2 binds to ribosomes suggests that it is selecting a subset of ribosomes and making them competent for Sec insertion. Although a SECIS element was able to compete for SBP2 binding to the ribosome, it could not do so completely even when SBP2 was preincubated with SECIS prior to the addition of ribosomes [K. Caban and P. R. Copeland, unpublished observation]. Further studies will be required to determine the binding affinity for SBP2/SECIS interactions versus SBP2/ribosome binding to determine whether this phenomenon is due to preferential binding as opposed to a purely mass action event due to the abundance of ribosomes compared to SECIS-containing mRNA. Assuming that the SBP2/ribosome interaction is essential, a major function of this interaction may be to prevent release factor binding and/or to increase the stalling time at the UGA to give time for the SBP2/SECIS interaction to take place. Alternatively (or additionally), the SECIS-induced movement of SBP2 off the ribosome may allow conformational changes within the ribosomal A-site that provide an optimal environment for eEFSec/Sec-tRNA[Ser]Sec binding, thus allowing Sec-tRNA[Ser]Sec the freedom to act like a true suppressor tRNA.
The role of eEFSec in delivering the Sec-tRNA[Ser]Sec is clear, but how it receives the specificity information it needs to find the right UGA codon is not obvious. It does seem that an indirect eEFSec/SBP2 interaction is mediated by an RNA component, and although preliminary evidence points to a role for tRNA in linking eEFSec with SBP2 [15], whether the link is provided by tRNA, rRNA, the SECIS element, or even simply selenoprotein mRNA has not been examined thoroughly. A formal possibility is that eEFSec does not require any interaction with the SBP2/SECIS complex since, as an elongation factor, its affinity for the ribosomal A-site is inherent. In this scenario, SECIS element-dependent specificity is conferred solely on the ribosome which undergoes an A-site conformation change that accommodates eEFSec/SectRNA[Ser]Sec.
Although the role of rpL30 is entirely speculative for either model, the fact that it can compete with SBP2 for SECIS element binding suggests it may displace SBP2 off the SECIS element allowing it to reassociate with the ribosome so that the ribosome remains competent for subsequent recoding events. In combination with the circular mRNA model of translation initiation [38], which may provide optimal SBP2/SECIS proximity, SBP2 cycling between the ribosome and SECIS element may allow for processive Sec incorporation. Despite recent criticism levied at this ‘closed loop’ model of translation initiation [39], the overwhelming dataset that supports it provides a good model for how a ribosome could be ‘primed’ to decode UGA as Sec during the initiation event. This is an important concept considering the fact that selenoprotein P contains multiple Sec residues and would require ribosomes to remain competent after recoding of the first UGA to Sec.
Conclusions
The question whether SBP2 is pre-bound to, and thus modifying, a subset of ribosomes or whether it preferentially binds the SECIS element and serves as a docking site for other trans-acting factors may seem too fine a point, but the sequence of events may turn out to have very important implications in determining the processivity and hence the efficiency UGA recoding.
As is usually the case, this review has dredged up many more questions than answers, but the basic requirements for Sec incorporation are taking shape. A complete understanding of this system will enable manipulations designed to maximize the benefits of selenoprotein function — a goal that remains elusive but one that is beginning to seem achievable.
Footnotes
Received 2 September 2005; received after revision 28 September 2005; accepted 12 October 2005
References
- 1.Stadtman T. C. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 2.Bock A. Biosynthesis of selenoproteins — an overview. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll D. M., Chavatte L. Finding needles in a haystack. In silico identification of eukaryotic selenoprotein genes. EMBO Rep. 2004;5:140–141. doi: 10.1038/sj.embor.7400080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll D. M., Copeland P. R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 5.Hatfield D. L., Gladyshev V. N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gladyshev V. N., Kryukov G. V., Fomenko D. E., Hatfield D. L. Identification of trace element-containing proteins in genomic databases. Annu. Rev. Nutr. 2004;24:579–596. doi: 10.1146/annurev.nutr.24.012003.132241. [DOI] [PubMed] [Google Scholar]
- 7.Copeland P. R. Regulation of gene expression by stop codon recoding: selenocysteine. Gene. 2003;312:17–25. doi: 10.1016/S0378-1119(03)00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland P. R. Making sense of nonsense: the evolution of selenocysteine usage in proteins. Genome Biol. 2005;6:221. doi: 10.1186/gb-2005-6-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein D. J., Schmeing T. M., Moore P. B., Steitz T. A. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher J. E., Copeland P. R., Driscoll D. M., Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 11.Copeland P. R., Stepanik V. A., Driscoll D. M. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta A., Rebsch C. M., Kinzy S. A., Fletcher J. E., Copeland P. R. Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tujebajeva R. M., Copeland P. R., Xu X.-M., Carlson B. A., Harney J. W., Driscoll D. M., et al. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:1–6. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavacki A. M., Mansell J. B., Chung M., Klimovitsky B., Harney J. W., Berry M. J. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol. Cell. 2003;11:773–781. doi: 10.1016/S1097-2765(03)00064-9. [DOI] [PubMed] [Google Scholar]
- 16.Kromayer M., Wilting R., Tormay P., Bock A. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol. 1996;262:413–420. doi: 10.1006/jmbi.1996.0525. [DOI] [PubMed] [Google Scholar]
- 17.Rother M., Wilting R., Commans S., Bock A. Identification and characterisation of the selenocysteine-specific translation factor SelB from the Archaeon Methanococcus jannaschii . J. Mol. Biol. 2000;299:351–358. doi: 10.1006/jmbi.2000.3756. [DOI] [PubMed] [Google Scholar]
- 18.Kinzy S. A., Caban K., Copeland P. R. Characterization of the SECIS binding protein 2-dependent selenocysteine incorporation complex. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler W. C., Grundy F. J., Murphy B. A., Henkin T. M. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/S1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982;10:2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walczak R., Westhof E., Carbon P., Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin E. V., Bork P., Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidovic I., Nottrott S., Hartmuth K., Luhrmann R., Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/S1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 25.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 26.Suryadi J., Tran E. J., Maxwell E. S., Brown B. A., 2nd The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry (Mosc) 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- 27.Allmang C., Carbon P., Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/S1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Copeland P. R., Driscoll D. M. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 29.Chavatte L., Brown B. A., Driscoll D. M. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 30.Goody T. A., Melcher S. E., Norman D. G., Lilley D. M. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein D. J., Moore P. B., Steitz T. A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berry M. J., Harney J. W., Ohama T., Hatfield D. L. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon: codon mutations. Nucleic Acids Res. 1994;22:3753–3759. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasim M. T., Jaenecke S., Belduz A., Kollmus H., Flohe L., McCarthy J. E. Eukaryotic selenocysteine incorporation follows a nonprocessive mechanism that competes with translational termination. J. Biol. Chem. 2000;275:14846–14852. doi: 10.1074/jbc.275.20.14846. [DOI] [PubMed] [Google Scholar]
- 34.Baranov P. V., Gesteland R. F., Atkins J. F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/S0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 35.Howard M. T., Aggarwal G., anderson C. B., Khatri S., Flanigan K. M., Atkins J. F. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry M. J. Knowing when not to stop. Nat. Struct. Mol. Biol. 2005;12:389–390. doi: 10.1038/nsmb0505-389. [DOI] [PubMed] [Google Scholar]
- 37.Berry M. J., Banu L., Harney J. W., Larsen P. R. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachs A. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Translational Control of Gene Expression. Cold Spring Harbor, N. J.: Cold Spring Harbor Laboratory Press; 2000. pp. 447–466. [Google Scholar]
- 39.Kozak M. How strong is the case for regulation of the initiation step of translation by elements at the 3′ end of eukaryotic mRNAs? Gene. 2004;343:41–54. doi: 10.1016/j.gene.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Mao H., White S. A., Williamson J. R. A novel looploop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- 41.van Eenennaam H., Jarrous N., van Venrooij W. J., Pruijn G. J. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life. 2000;49:265–272. doi: 10.1080/15216540050033113. [DOI] [PubMed] [Google Scholar]
- 42.Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gelugne J. P., Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papa S., Zazzeroni F., Pham C. G., Bubici C., Franzoso G. Linking JNK signaling to NF-kappaB: a key to survival. J. Cell Sci. 2004;117:5197–5208. doi: 10.1242/jcs.01483. [DOI] [PubMed] [Google Scholar]
- 44.Kisselev L., Ehrenberg M., Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]


