Abstract
Three human cases having mutations in the glycine N-methyltransferase (GNMT) gene have been reported. This enzyme transfers a methyl group from S-adenosylmethionine (SAM) to glycine to form S-adenosylhomocysteine (SAH) and N-methylglycine (sarcosine) and is believed to be involved in the regulation of methylation. All three cases have mild liver disease but they seem otherwise unaffected. To study this further, gnmt deficient mice were generated for the first time. This resulted in the complete absence of GNMT protein and its activity in livers of homozygous mice. Compared to WT animals the absence of GNMT resulted in up to a 7-fold increase of free methionine and up to a 35-fold increase of SAM. The amount of SAH was significantly decreased (3 fold) in the homozygotes compared to WT. The ratio of SAM/SAH increased from 3 in WT to 300 in livers of homozygous transgenic mice. This suggests a possible significant change in methylation in the liver and other organs where GNMT is expressed.
Keywords: enzyme activity, glycine N-methyltransferase, knockout, mouse, S-adenosylhomocysteine, S-adenosylmethionine
Introduction
S-adenosylmethionine is a universal donor of methyl groups in biological methylation in all organisms and is considered as a major component in regulation of methionine function (Finkelstein & Martin, 1986). It is synthesized from methionine and is used as substrate of numbers of methyltransferases (Clarke & Banfield, 2001). The most abundant methyltransferase in mammalian liver is GNMT, comprising about 1% of the soluble protein in rat liver (Takusagawa et al., 1999). GNMT is also present in large amounts in the exocrine pancreas and the prostate, but is present also in other tissues in certain specific cell types. There is only a single gene that codes for GNMT.
GNMT structure and enzyme properties have been studied in detail using rat liver and recombinant proteins (Takusagawa et al., 1999). The GNMT genes and cDNAs from several sources have been cloned and sequenced (Ogawa et al., 1993). The proteins are about 90% similar (Ogawa et al., 1993) regarding amino acid sequences and crystal structures (Fu et al., 1996; Luka et al., 2002; Pakhomova et al., 2004).
The biological function of glycine N-methyltransferase is still not fully understood. First found in the guinea pig this enzyme was rediscovered as a major folate binding protein in rat liver cytosol (Suzuki & Wagner, 1980; Cook & Wagner, 1984). It was subsequently found that a specific form of folate, 5-methyl-tetrahydrofolate pentaglutamate (5-methyl-THF-G5) is a potent inhibitor of GNMT enzyme activity (Wagner et al., 1985) and is tightly, but not covalently, bound to the enzyme in vivo (Cook & Wagner, 1984). This has established the role of GNMT as an important factor in the control of one-carbon metabolism (Balaghi et al., 1993).
The importance of GNMT is to maintain a constant ratio of SAM/SAH, which is considered to be an indicator of methylation capacity in the cell (Cantoni et al., 1978) and serve as an alternate route to convert SAM to SAH in addition to the 40–50 other methyltransferases. Indeed, the identification of several human individuals with persistent hypermethioninaemia resulted in discovery of natural mutations of GNMT as the reason for those metabolic abnormalities (Mudd et al., 2001; Augoustides-Savvopoulou et al., 2003). Although these individuals have evidence of mild liver disease they appear to be otherwise quite normal aside from their abnormal pattern of methionine metabolites. The individuals are still quite young and problems may develop as they get older. In order to investigate the role of GNMT and the regulation of methyl group metabolism in the liver, which is the major metabolic organ, we developed the mouse model with the loss of the gnmt gene. The results of the basic changes in the livers of such animals are reported in this work.
Methods, results and discussion
This study was begun with cloning and sequencing of the mouse gnmt gene since it was not cloned at that time. Cloning and sequencing of gnmt was done by using standard gene cloning procedure (Sambrook et al., 1989) from a mouse129 Svj gene library. The latter was prepared from mouse spleen DNA in Lambda FIX II vector by insertion into NotI restriction site (Stratagene, La Jolla, CA, USA).
At the first step of cloning the size of the mouse gnmt gene was determined by PCR amplification of the fragment of the mouse gene by using primers complementary to the 5′- and 3′-sequences of known sequence of mouse cDNA (Aida et al., 1997). The sequences of the primers were 5′- (5′-ATGGTGGACAGCGTGTACCGT) and 3′- (5′-GTCTGTCTTCTTGAGCACATG). The fragment with a molecular size of about 3.2 kb was synthesized by PCR, which meant that mouse gnmt is about the size of the rat gene.
The PCR amplified DNA was used as a probe for screening the mouse genomic library for the gnmt. After several screening cycles 8 positive clones with the strongest signals were isolated. From these, 3 identical clones containing the full size gnmt were found by PCR analysis. Using one of these clones the sequence of entire gene was determined containing a 5′-fragment upstream of the first ATG codon in first exon and a 3′-downstream fragment. This has been submitted to Gene Bank (Accession Number AF 325352).
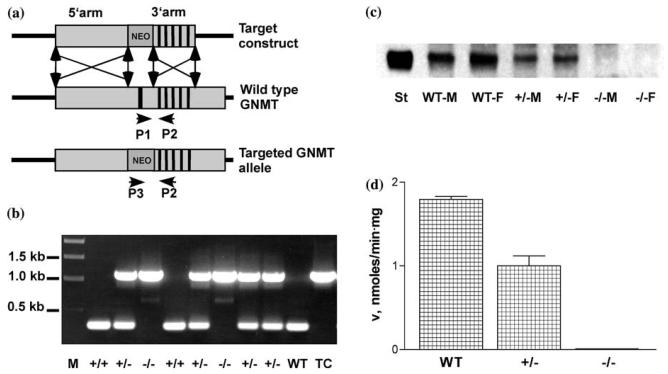
The target construct was prepared in a gene targeting vector pPNT (Tybulewicz et al., 1991). To get a convenient plasmid for restriction analysis and sequencing, the fragment of the mouse genomic DNA containing the gnmt was subcloned from phage DNA into pGEM-5Zf(+) vector into a unique NotI site. The strategy for preparation of target construct was to delete the first exon and potential promoter region from the gnmt, which would prevent gene expression. This was achieved in two steps. At the first step the 3′ BamHI-KpnI fragment of the GNMT gene containing exons 2–6 with a 3′ sequence of about 3 kb was inserted into BamHI-KpnI sites of pPNT, which generated the plasmid pL1. In the second step the 5′-fragment of the gene of about 9 kb was excised with NotI–XhoI and inserted into NotI–XhoI sites of the plasmid pL1. This cloning resulted in the preparation of the target construct pLT. The target construct is shown in Figure 1a. Position and orientation of all essential elements of the target construct was confirmed by restriction analysis and sequencing.
Figure 1.
Gnmt gene targeting. (a) Knockout strategy for replacing the first exon and promoter region of gnmt by the neomycin resistance gene (neo) from the targeting vector pPNT. The vertical bars represent exons 1–6. (b) Genotyping by PCR with P1, P2 and P3 primers of the mice from F2 generation after crossing gnmt+/− mice. M, molecular size markers; WT, wild type mouse; TC, target construct. (c) Levels of GNMT in liver crude extracts. Western blotting was performed after separation of 15 μg of total proteins from liver crude extracts. Rabbit antibodies against rat GNMT were used in the first step and goat IgG conjugated with peroxidase used as secondary antibodies. St, GNMT marker; M, male; F, female animals. (d) Enzyme activity. Values are means +/− S.E.M. Each group was significantly different from the others.
Gene targeting was done by IngenKO Company at Monash University (Clayton, Australia). Details of the procedure are discussed below.
To measure GNMT activity, the level of free methionine and perform Western blotting crude extracts of livers were prepared. This was done by homogenization of about 100 mg of liver in Homogenization Buffer containing 0.1 M Tris, pH 7.5–0.15 M NaCl-5 mM EDTA-10 mM DTT-2 mM PMSF. The homogenate was centrifuged at 20,000 g for 30 min at 2°C and supernatant was collected. Protein concentration in crude extract was determined by BCA Protein Assay Kit (Pierce Biotechnology Inc.) with bovine serum albumin as the standard.
GNMT activity was performed using the charcoal adsorption method described earlier (Wagner et al., 1985).
To determine GNMT protein expression Western blotting was performed. The membrane was incubated with primary rabbit antibodies against rat GNMT and secondary peroxidase conjugated goat anti-rabbit IgG and visualized by using SuperSignal West Femto Maximum Sensitivity Substrate Kit (Pierce Biotechnology Inc.).
SAM and SAH were determined in livers by a previously published method (Capdevila & Wagner, 1998). A piece of the frozen tissue of about 30 mg was homogenized in 0.7 ml of 10% TCA and the precipitate was removed by centrifugation.
Statistical analysis was carried out using the Kruskal–Wallis test for 3 independent variables. This showed that there are differences between groups (p<0.0001). The Wilcoxon–Mann–Whitney test was then used to compare each group with each other. In every case n=4. Differences were considered significant when p<0.05.
The concentration of free amino acids in the crude liver extract was determined by high performance liquid chromatography equipped with a scanning fluorescence detector at the Center for Molecular Neuroscience Core Facility at Vanderbilt University Medical Center.
Gnmt−/− deficient mice were generated by homologous recombination as shown in Figure 1a. The first step involved screening by geneticin (G418) of 400 selected clones of electroporated ES cells. Twenty clones were selected as having undergone the targeting events. From those 20 clones, 4 clones contained correctly targeted gnmt as confirmed by PCR and Southern analysis. Two of those clones were microinjected into C57BI/6J blastocytes, which resulted in generation of a high percentage of chimeric animals. Chimeric male animals were crossed with wild type C57BI/ 6J female animals, which resulted in obtaining gnmt+/− F1 animals. In one case all 12 pups were agouti, which indicated germ line transmission. Among them, 4 animals were heterozygous (2 male and 2 female). By crossing heterozygous animals 8 pups were born with classic Mendelian segregation of 2 animals of gnmt+/+ 4 animals of gnmt+/− and 2 mice of gnmt−/−. Genotyping of those animals was done by PCR using three primers: P1 (GTACCGCAGAGTACAAGGCG), P2 (CAATCGCAGGAGGAACAGCGC) and P3 (CTGAATGAACTGCAGGACGAG) and genomic DNA prepared from tails of the animals. This is shown in Figure 1b. PCR amplification with P1– P2 and P3–P2 primers resulted in expected 330 and 1151 bp fragments respectively. The heterozygous and the homozygous mice were no different from the WT mice in appearance.
Since GNMT is most abundant in mammalian liver this organ was used to determine loss of gnmt. As shown in Figure 1c and d the targeting strategy resulted in complete absence of the protein in the liver from homozygotes, as determined by activity assay and by Western Blotting. Activity of GNMT in the liver of heterozygous animals (1.0 nmol/ min · mg of total protein) exhibits of about 50% of the activity of WT animals (1.8 nmol/min mg), (p<0.05).
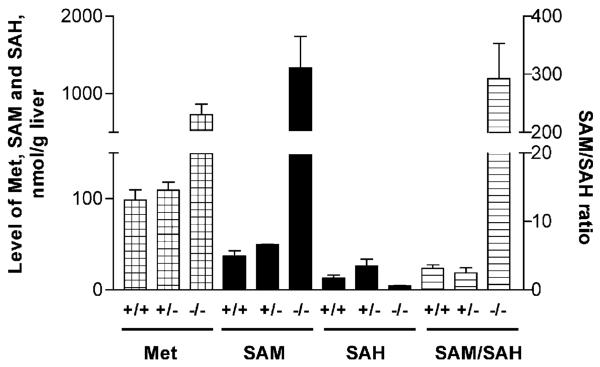
Absence of GNMT in the liver caused the highly elevated levels of free methionine and SAM as shown in Figure 2. The level of methionine in gnmt−/− mice increased from 100–700 nmol/g liver (p<0.05). The level of SAM in gnmt−/− animals was 1334 nmoles/g liver compared to 37 (p<0.05) in WT animals. The level of SAH in gnmt−/− animals (3–5 nmol/g liver) was slightly but significantly (p<0.05) decreased compared to WT mice (12–15 nmol/g). As a result the ratio of SAM/SAH increased 100-fold from about 3–300. It is noteworthy that the levels of the metabolites, methionine, SAM and SAH are quite close in the +/+ and the +/− animals in spite of the significant reduction in enzyme activity (Figure 1d). This is probably due to the action of the other methyltransferases in the liver that also convert SAM to SAH.
Figure 2.
Levels of methionine, SAM, SAH and ratio of SAM/SAH in mouse livers. Levels of methionine, SAM and SAH were determined in the crude extracts prepared from livers of WT, gnmt+/− and gnmt−/− mice and normalized to weight of the liver. The number of animals in each group was 4. In each case the values for the −/− group were significantly different from the other two groups but the +/− group was not significantly different from the +/+ group. Error bars indicate the S.E.M.
The increased level of methionine deserves some mention since MAT I and III, the isozymes responsible for SAM synthesis in the liver, are not reversible and elevated methionine cannot be a result of equilibrium effects. MAT I activity is inhibited by SAM and MAT III is stimulated by SAM. The net effect on the concentration of methionine will depend on the relative amounts of MAT I and MAT III and their response to elevated SAM in the livers of the mouse. In the three human mutations in GNMT all had elevated plasma methionine levels. In humans excess methionine (as in cases of cystathionine-ß-synthase deficiency) is disposed of by a transamination pathway.
This gnmt targeting project was undertaken in order to get a mouse model lacking a key enzyme involved in methionine, SAM and one-carbon unit metabolism. Human GNMT deficiency due to mutations resulted in mild liver disease characterized by hepatomegaly, in some cases, and elevated levels of enzymes characteristic for liver disease in the plasma. All had highly elevated levels of methionine and SAM in plasma (Mudd et al., 2001; Augustides-Savvopoulou et al., 2003). Impaired utilization of SAM has been implicated in liver disease (Mato et al., 1990).
Our knockout strategy resulted in complete absence of GNMT protein and obvious lack of GNMT activity. Liver tissue was used for characterization of the transgenic mice in comparison with WT animals because that tissue is characterized by the highest expression of GNMT and therefore the effect of absence of that enzyme should be more easily recognized. As we anticipated, in gnmt−/− mice the level of GNMT substrate, SAM, is highly elevated while the level of SAH is even lower that in WT mice (Figure 2). Methylation in cells depends not only on the SAM concentration but also on the level of the product of reaction, SAH. Therefore the ratio SAM/SAH is considered as an index of methylation capacity (Cantoni et al., 1978). This parameter also is highly elevated in gnmt−/− mice compared to WT mice as shown in Figure 2.
Acknowledgements
The authors thank Professor B. Shane of the University of California (Berkeley) for providing targeting vector pPNT and helpful initial discussion on the gnmt gene targeting strategy. Support was provided in part from grant # DK15289 of the U.S. Public Health Service and the Department of Veterans Affairs, USA.
Abbreviations
- GNMT
glycine N-methyltransferase
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
References
- Aida KM, Tawata Negishi MM, Onaya T. Mouse glycine N-methyltransferase is sexually dimorphic and regulated by growth hormone. Horm Metab Res. 1997;29:646–649. doi: 10.1055/s-2007-978982. [DOI] [PubMed] [Google Scholar]
- Augoustides-Savvopoulou P, Luka ZS, Karyda S, Stabler SP, Allen RH, Patsiaoura K, et al. Glycine N-methyltransferase deficiency: a new patient with a novel mutation. J Inherit Metab Dis. 2003;26:745–759. doi: 10.1023/B:BOLI.0000009978.17777.33. [DOI] [PubMed] [Google Scholar]
- Balaghi M, Horne DW, Wagner C. Hepatic one-carbon metabolism in early folate deficiency in rats. Biochem J. 1993;291:145–149. doi: 10.1042/bj2910145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni GL, Richards HH, Chiang PK. Inhibitors of S-adenosylhomocysteine hydrolase and their role in the regulation of biological methylation. In: Usdin E, Borchardt RT, Creveling CR, editors. Transmethylation. Elsevier; North Holland, New York, N.Y.: 1978. pp. 155–164. [Google Scholar]
- Capdevila A, Wagner C. Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Anal Biochem. 1998;264:180–184. doi: 10.1006/abio.1998.2839. [DOI] [PubMed] [Google Scholar]
- Clarke S, Banfield K. S-adenosylmethionine-dependent methyltransferases. In: Carmel R, Jacobsen DW, editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge, U.K.: 2001. pp. 63–78. [Google Scholar]
- Cook RJ, Wagner C. Glycine N-methyltransferase is a folate binding protein of rat liver cytosol. Proce Nat Acad Sci USA. 1984;81:3631–3634. doi: 10.1073/pnas.81.12.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein JD, Martin JJ. Methionine metabolism in mammals. Adaptation to methionine excess. J Biol Chem. 1986;261:1582–1587. [PubMed] [Google Scholar]
- Fu Z, Hu Y, Konishi K, Takata Y, Ogawa H, Gomi T, et al. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]
- Luka Z, Cerone R, Phillips J, Mudd SH, Wagner C. Mutations in human glycine N-methyltransferase give insights into its role in methionine metabolism. Hum Genet. 2002;110:68–74. doi: 10.1007/s00439-001-0648-4. [DOI] [PubMed] [Google Scholar]
- Mato JM, Corrales A, Martin-Duce P, Ortiz P, Pajares MA, Cabrero C. Mechanisms and consequencs of the impaired trans-sulfuration pathway in liver disease: Part 1. Biochemical implications. Drugs. 1990;40(Suppl 3):58–64. doi: 10.2165/00003495-199000403-00006. [DOI] [PubMed] [Google Scholar]
- Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti GU, Caruso U, et al. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J Inherit Metab Dis. 2001;24:448–464. doi: 10.1023/a:1010577512912. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Gomi T, Fujioka M. Mammalian glycine N-methyltransferases. Comparative kinetic and structural properties of the enzymes from human, rat, rabbit and pig livers. Comp Biochem Physiol: Comp Biochem. 1993;106B:601–611. doi: 10.1016/0305-0491(93)90137-t. [DOI] [PubMed] [Google Scholar]
- Pakhomova S, Luka Z, Grohmann S, Wagner C, Newcomer ME. Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes. Proteins. 2004;57:331–337. doi: 10.1002/prot.20209. [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Newcomer ME, Wagner C. Crystal structure of apo-glycine N-methyltransferase (GNMT) Protein Sci. 1998;7:1326–1331. doi: 10.1002/pro.5560070608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook EF, Fritsch, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Suzuki N, Wagner C. Purification and characterization of a folate binding protein from rat liver cytosol. Arch Biochem Biophys. 1980;199:236–248. doi: 10.1016/0003-9861(80)90277-5. [DOI] [PubMed] [Google Scholar]
- Takusagawa F, Ogawa H, Fujioka M. Glycine N-methyltransferase, a tetrameric enzyme. In: Cheng X, Blumental RM, editors. S-adenosylmethionine-Dependent Methyltransferases: Structure and Functions. World Scientific Publishing; Singapore: 1999. pp. 93–122. [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl protooncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Wagner C, Briggs WT, Cook RJ. Inhibition of glycine N-methyltransferase activity by folate derivatives: implications for regulation of methyl group metabolism. Biochem Biophys Res Commun. 1985;127:746–752. doi: 10.1016/s0006-291x(85)80006-1. [DOI] [PubMed] [Google Scholar]
- Yeo EJ, Wagner C. Tissue distribution of glycine N-methyltransferase, a major folate-binding protein of liver. Proce Nat Acad Sci USA. 1994;91:210–214. doi: 10.1073/pnas.91.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]