Abstract
Productive HIV-1 replication is mainly controlled at the transcriptional level by HIV-1 Tat and at the post-transcriptional level by HIV-1 Rev. A number of host factors have been identified to be involved in these processes. Src-associated protein of 68 kDa in mitosis (Sam68) is a multi-functional RNA-binding protein and has been implicated in several important cellular processes. More evidence has accumulated to support an important regulatory function of Sam68 at two distinct steps of HIV-1 gene expression. Sam68 interacts with HIV-1 Rev protein and directly participates in nuclear exportation of HIV-1 unspliced or singly spliced RNA. In addition, Sam68 functions in the cytoplasmic processes of HIV-1 replication such as the translational regulation of HIV-1 RNA. Elucidation of the precise molecular function of Sam68 in HIV-1 gene expression is clearly warranted and is expected to unambiguously establish Sam68 as an important host factor for HIV-1 replication.
Keywords: Sam68, HIV-1, RNA nuclear export, RNA translation, gene expression
A cascade of sequential molecular interactions among DNA, RNA and proteins spatially and temporally control HIV-1 gene expression. This control mainly occurs at three levels. Following entry, uncoating, reverse transcription, nuclear import and integration, HIV-1 gene expression begins with basal transcription of the proviral DNA. The basal transcription is initiated by interaction of cellular transcription factors with their cognate DNA-binding sequences within the HIV-1 long-terminal repeat (LTR). NFκB recognizes and binds to the NFκB consensus DNA motif in the LTR; this DNA-protein interaction is one of the important events in this initial process.1 The next level of control involves the HIV-1 transcription trans-activator protein Tat.2 Tat recruits the eukaryotic positive transcription elongation factor b (p-TEFb), a cyclin dependent kinase comprised of Cdk9 and in humans cyclin T through a specific interaction between Tat and the viral TAR RNA stem-loop target sequence located at the 5' terminus of HIV-1 RNA.3,4 p-TEFb phosphorylates serine 2 at RNA polymerase C-terminal repeats, which in turn relieves RNA polymerase II from the negative elongation properties of DSIF and NELF and enters productive elongation resulting in transcription of full-length HIV-1 RNA. The last level of the regulation is mediated by HIV-1 Rev protein. In contrast to the basal and Tat-transactivated transcription, Rev functions post-transcriptionally. It interacts with a cis-acting RNA sequence, the Rev response element (RRE) present in all intron-containing singly spliced and unspliced HIV-1 RNA, and exports these RNA from the nucleus to the cytoplasm for synthesis of HIV-1 structural proteins and full-length HIV-1 genomic RNA, which both are needed for productive viral replication.5
A number of host factors have been identified to either positively or negatively regulate HIV-1 gene expression in these processes. Among those is Sam68, an Src-associated protein of 68 kDa in mitosis. Like other RNA-binding proteins, Sam68 is multifunctional. It regulates RNA metabolism and signal transduction and is implicated in a number of important cellular processes including cell cycle regulation, apoptosis, tumorigenesis, bone metabolism, and normal brain function.6 More evidence has accumulated to suggest an important role of Sam68 in HIV-1 replication. Here we intended to provide a brief update on our current understanding on Sam68 function in HIV-1 replication.
In 1999, Sam68 was first found being capable of substituting for and synergizing with HIV-1 Rev protein.7 Our studies have shown that a lower level of constitutive Sam68 is associated with the Rev functional defect in astrocytes and that overexpression of Sam68 overrides the Rev defect in these cells.8 The latter studies suggest that the synergistic effects between Sam68 and Rev are conditional and only occur in cells that express a lower level of constitutive Sam68. Thus, two competing pathways have been proposed for Sam68 role in nuclear export of HIV-1 viral RNA: CRM1-independent7 and CRM1-dependent.9 Initially, Sam68 was shown to bind RRE,7 assumingly through the UAAA sequence located within stem I of RRE. However, the relevance and specificity of this interaction remains controversial, since Sam68 can cooperate with RNA helicase A and TAP to facilitate exportation of intron-containing RNA in the absence of RRE, but in the presence of different retroviral RNA exportation elements such as CTE, RxRE, ERRE and RRE2;10–13 Meanwhile, Sam68 has been shown to be incapable of substituting for HIV-1 Rev function.9,10 A very recent report has shown that Sam68 is absolutely required for HIV-1 Rev function,14 which provides independent evidence to support our early similar findings.9 Thus, all these studies appear to argue against that Sam68 functions independently of the Rev/CRM1 pathway and support the notion that Rev/CRM1 requires Sam68 for nuclear export of HIV-1 RNA.
Despite the inconsistency regarding the role and the mechanisms of Sam68 in nuclear exportation of HIV RNA transcripts, various studies share two common biochemical features. First, Sam68 binding to RNA is required for transactivation of Rev function.10,15 Sam68 binds RNA through its single KH domain16–18 and a RG rich region that confers non-specific RNA binding activity to Sam68.15 Sam68 mutants defective for RNA binding activity (ΔKH or ΔRG) fail to functionally promote nuclear exportation of intron-containing RNA.10,15 Moreover, tyrosine phosphorylation of Sam68 by p59fyn and Sik/BRK inhibits its ability to bind RNA,10,19 and also abolishes its capacity to enhance RRE and CTE function.10,15 Second, Sam68 binds to Rev in vitro and in vivo.7,8 The early study has shown that Sam68 and Sam68Δ330–433, but not Sam68Δ1–329 GST fusion proteins interact with Rev protein in vitro.7 Our group has also shown that Sam68 forms a complex with Rev in vivo, and that the nuclear exportation signal (NES) of Rev and the region between aa residues 321 and 410 are directly involved in the protein-protein interaction.8 This suggests that the sequences spanning aa residues 321 to 329 play a critical role in Sam68 interaction with Rev and Rev function.
Compared to the limited studies in the direct role of Sam68 in HIV-1 replication, more studies have been focused on the Sam68 mutants that are deleted of the C-terminal nuclear localization signal (NLS). Several groups have shown that these cytoplasmic mutants potently inhibit HIV-1 replication.7,20,21 However, there is no consensus on the underlying mechanisms of this inhibition. The proposed mechanisms include impaired Rev localization and function and sequestration of RRE-containing RNAs from the translation machinery. In addition, Sam68 proteins with point mutations within the NLS localize in the cytoplasm and inhibit Rev/RRE transactivation,22,23 suggesting that cytoplasmic localization, but not the structural changes as a result of amino acid deletions, is a requirement for inhibition of HIV-1 replication. Recently, our group has provided the first direct evidence that Sam68 mutants inhibit HIV-1 replication by blocking Rev-dependent nuclear export of HIV-1 viral RNA.21 Moreover, the domain between aa residues 269 and 321 is required for inhibition of HIV-1 Rev function.
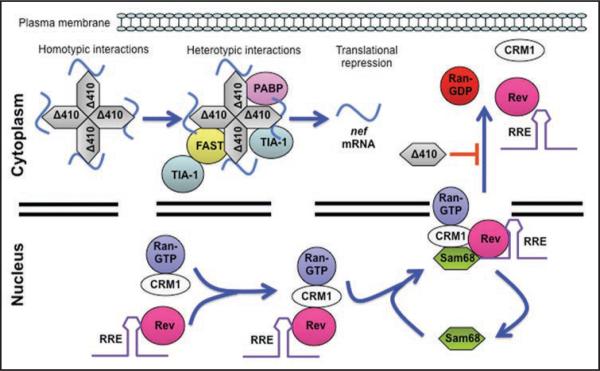
Using a series of Sam68 mutants, we have also found that Sam68 plays additional post-transcriptional roles in HIV-1 replication. We have shown that Sam68 cytoplasmic mutants potently suppress HIV-1 Nef expression, and that this is correlated with their ability to induce stress granules.24 The suppression is specific to Nef, and direct binding to nef mRNA 3'UTR confers the suppression specificity. Further characterization of the phenomenon led to three additional conclusions: first, deletion of the Sam68 NLS, resulting in cytoplasmic localization, is required for SG induction and Nef expression inhibition; second, RNA binding is mandatory since deletion of the KH domain abrogates the suppressive effect of NLS-deleted mutants (unpublished) and prevents Sam68 localization to SG; and third, Sam68 domain aa269–321 was indispensable but not sufficient for Nef suppression. The roles of the NLS and the KH domain on Sam68 function and localization have been extensively studied; however, the function of the domain aa269–321 is little known. This domain is located in the proline-rich region (P3), which overlaps the arginine-glycine (RG)-rich region.6 Some of the functions that have been attributed to these proline- and RG-rich regions include protein-protein interaction, intracellular localization and non-specific RNA binding.15,25,26 The RG-rich region is also a potential site for protein methylation.25 Nevertheless, whether any of these properties contribute to Nef suppression remains to be determined. Taken together, these studies indicate that Sam68 cytoplasmic mutants negatively regulates HIV-1 replication at two distinct steps of the virus life cycle: Rev-mediated nuclear export of incompletely spliced and unspliced HIV-1 RNA and translation of HIV-1 nef mRNA in the cytoplasm (Fig. 1).
Figure 1.
Sam68 function in HIV-1 gene expression. (A) Cytoplasmic regulation of HIV-1 gene expression. Overexpression of cytoplasmic Sam68-NLS mutants initiates nucleation of RNA-containing granules through homotypic interactions involving the RG-rich domain located between aa residues 269 and 321. Small primary aggregates are then formed and further cross-linked by direct or indirect heterotypic interactions between these mutants and SG core components such as TIA-1 (unpublished), FAST 30 or PABP-1,31 to generate larger secondary aggregates, followed by recruitment into mature SG. As a result, specific mRNA targets of these mutants, i.e., nef mRNA, are then brought into and enriched in the SG and made unavailable for translation. In addition, overexpression of these mutants inhibits Rev-mediated exportation of intron-containing RNA. (B) Nuclear regulation of HIV-1 gene expression. Rev, RRE-containing RNA, CRM1 and RanGTP form a complex in the nucleus in a Sam68-independent manner. Sam68 then associates with the complex via direct binding to Rev and transports the complex to the nuclear pore complex (NPC) and docks the complex onto NPC through CRM1 interaction with nucleoporins. Translocation of Rev, CRM1 and RRE-containing RNA into the cytoplasm leads to the release of Sam68 into the nucleus.
Meanwhile, another recent study has shown that Sam68 cytoplasmic mutants alter the binding of poly A-binding protein to unspliced HIV-1 RNA and selectively inhibit translation of some HIV-1 RNA.27 Moreover, two other studies have shown that Sam68 likely regulates HIV-1 replication at the translational level. Sam68 has been shown to enhance p24 expression from a gag-pol reporter plasmid containing a CTE.10 Sam68 overexpression increases the levels of cytoplasmic gag-pol RNA only by two fold, but protein expression by 45 fold. In addition, Sam68 has been shown to promote 3'-end processing (cleavage, polyadenylation) of HIV-1 unspliced mRNA, in the absence of a significant increase in the level of unspliced viral RNA in the cytoplasm.28 The latter effects appear to only occur in certain host cells and are not correlated with enhanced HIV-1 gene expression.29 Clearly, our understanding of Sam68 function in HIV-1 replication is just at the beginning, further elucidation of the underlying molecular mechanisms of Sam68 function in the Rev-mediated nuclear export pathway of HIV-1 RNA and in regulation of HIV-1 RNA translation is warranted.
Acknowledgements
This work was supported by the grants R01NS039804 and R01NS065785 (to J.J.H.) from the National Institutes of Health.
References
- 1.Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, et al. Absolute dependence on kappaB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–60. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadaie MR, Benter T, Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science. 1988;239:910–3. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Sharp PA. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–10. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 4.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–62. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 5.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–83. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 6.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Reddy TR, Xu W, Mau JK, Goodwin CD, Suhasini M, Tang H, et al. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–42. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Liu Y, Park IW, He JJ. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J Virol. 2002;76:4526–35. doi: 10.1128/JVI.76.9.4526-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Liu Y, Kim BO, He JJ. Direct participation of Sam68, the 68-kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol. 2002;76:8374–82. doi: 10.1128/JVI.76.16.8374-8382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coyle JH, Guzik BW, Bor YC, Jin L, Eisner-Smerage L, Taylor SJ, et al. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol Cell Biol. 2003;23:92–103. doi: 10.1128/MCB.23.1.92-103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JP, Reddy TR, Truong KT, Suhasini M, Wong-Staal F. Functional interaction of Sam68 and heterogeneous nuclear ribonucleoprotein K. Oncogene. 2002;21:7187–94. doi: 10.1038/sj.onc.1205759. [DOI] [PubMed] [Google Scholar]
- 12.Reddy TR, Xu WD, Wong-Staal F. General effect of Sam68 on Rev/Rex regulated expression of complex retroviruses. Oncogene. 2000;19:4071–4. doi: 10.1038/sj.onc.1203749. [DOI] [PubMed] [Google Scholar]
- 13.Reddy TR, Tang H, Xu W, Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19:3570–5. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- 14.Modem S, Badri KR, Holland TC, Reddy TR. Sam68 is absolutely required for Rev function and HIV-1 production. Nucleic Acids Res. 2005;33:873–9. doi: 10.1093/nar/gki231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Cote J, Carvajal HV, Richard S. Identification of Sam68 arginine glycine-rich sequences capable of conferring nonspecific RNA binding to the GSG domain. J Biol Chem. 2001;276:30803–11. doi: 10.1074/jbc.M102247200. [DOI] [PubMed] [Google Scholar]
- 16.Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–80. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 17.Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–71. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 18.Itoh M, Haga I, Li QH, Fujisawa J. Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucleic Acids Res. 2002;30:5452–64. doi: 10.1093/nar/gkf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LL, Richard S, Shaw AS. P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–3. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 20.Soros VB, Carvajal HV, Richard S, Cochrane AW. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J Virol. 2001;75:8203–15. doi: 10.1128/JVI.75.17.8203-8215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Liu Y, Henao J, Rugeles MT, Li J, Chen T, He JJ. Requirement of an additional Sam68 domain for inhibition of human immunodeficiency virus type 1 replication by Sam68 dominant negative mutants lacking the nuclear localization signal. Gene. 2005;363:67–76. doi: 10.1016/j.gene.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 22.Reddy TR. A single point mutation in the nuclear localization domain of Sam68 blocks the Rev/RRE-mediated transactivation. Oncogene. 2000;19:3110–4. doi: 10.1038/sj.onc.1203637. [DOI] [PubMed] [Google Scholar]
- 23.Lukong KE, Larocque D, Tyner AL, Richard S. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J Biol Chem. 2005;280:38639–47. doi: 10.1074/jbc.M505802200. [DOI] [PubMed] [Google Scholar]
- 24.Henao-Mejia J, Liu Y, Park IW, Zhang J, Sanford J, He JJ. Suppression of HIV-1 Nef Translation by Sam68 Mutant-Induced Stress Granules and nef mRNA Sequestration. Mol Cell. 2009;33:87–96. doi: 10.1016/j.molcel.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cote J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol Biol Cell. 2003;14:274–87. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–6. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 27.Marsh K, Soros V, Cochrane A. Selective translational repression of HIV-1 RNA by Sam68DeltaC occurs by altering PABP1 binding to unspliced viral RNA. Retrovirology. 2008;5:97. doi: 10.1186/1742-4690-5-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaren M, Asai K, Cochrane A. A novel function for Sam68: enhancement of HIV-1 RNA 3' end processing. RNA. 2004;10:1119–29. doi: 10.1261/rna.5263904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren M, Cochrane A. Mapping of determinants involved in the stimulation of HIV-1 expression by Sam68. Virology. 2009;385:93–104. doi: 10.1016/j.virol.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 30.Simarro M, Mauger D, Rhee K, Pujana MA, Kedersha NL, Yamasaki S, et al. Fasactivated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc Natl Acad Sci USA. 2007;104:11370–5. doi: 10.1073/pnas.0704964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–49. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]