Abstract
Mussel byssal threads contain unusual block copolymer-like proteins that combine collagen with flanking domains that resemble silk-fibroin (preCol-D) or elastin (preCol-P). These are distributed in complementary gradients along the length of the threads and as precursors in the mussel foot. We discuss a 76-kDa precursor, preCol-NG, from a cDNA library of the foot where it has no gradient but rather is distributed evenly along the distal to proximal axis. A pepsin-resistant fragment of preCol-NG has been confirmed in byssal threads. Like preCol-D and -P, this protein has a central collagenous domain, flanking domains, an acidic patch, and histidine-rich termini. The flanking domains of preCol-NG resemble the glycine-rich proteins of plant cell walls with tandem XGlyn repeats where X denotes alanine, leucine, or asparagine but not proline. Similarity with the (glycine–alanine) repeats and poly(alanine) runs of arthropod silks also exists. Based on available evidence, a model of preCol axial assembly is proposed in which preCol-NG functions as a mediator between preCol-D/-P molecules. This is consistent with the observed progression of mechanical properties in byssal threads.
Keywords: collagen/glycine-rich cell wall proteins/byssus/Mytilus/nongradient distribution
A primary goal in the study of biomolecular materials is a fundamental understanding of how chemical and physical structure determines mechanical behavior. Byssal threads of marine mussels have three attributes that make them an attractive model system in pursuit of this goal: a comparatively simple morphology, rapid, reproducible formation, and interesting biomechanical properties. Byssal threads are proteinaceous tethers used by marine mussels to attach themselves to hard surfaces in the sea (Fig. 1A). Based on their collagen content, displacement outside the confines of living tissue, and proximal fusion with the byssal retractor muscles, the threads often are described as extracorporeal tendons (1, 2).
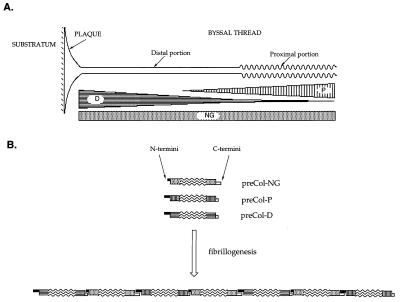
Figure 1.
A mussel byssal thread and the distribution of exotic collagens therein (approximate dimensions 300 × 0.1 mm). (A) The graded distribution of Col-D and -P along the axis of each byssal thread. The distribution of Col-NG is proposed based on RT-PCR data. (B) Model of axial assembly of byssal preCols is based on strict maintenance of N→C polarity and joining of long and short His-domains that are denoted by black (N-terminal) or white (C-terminal) half-laps. The collagen domains (wavy lines) are flanked by either amorphous fibroin (speckled), elastic (vertical stripes), or crystalline β-sheet fibroin (horizontal stripes) blocks in the three preCols. The joints in the assembled product may involve His-mediated intermolecular binding of metals such as zinc.
Ironically, biomechanical analyses of byssal threads have revealed properties that differ significantly from those of vertebrate tendons (3, 4). Distally, they are as strong as tendon but with 3–5 times more extensibility; proximally, they are weaker but 15–20 times more extensible. In terms of strain energy density, byssal threads are ≈6 times tougher than tendon, and the distal portion may actually approach the toughness of silk (3). Even more intriguing is the observation by Bell (as cited in ref. 5) that, given sufficient relaxation time, stretched threads can recover their initial length and stiffness after yield.
Recent biochemical analyses have sought a molecular explanation for the nontendon-like behavior of byssus. To wit, byssal collagens have departed from several hallmarks of the type I–III fibrillar collagens: There is no apparent quarter-stagger array during fibrillogenesis of byssal collagens (6); there is no procollagen precursor that requires activation to a tropocollagen (7, 8), and maturation is not based on lysyl oxidase-catalyzed cross-linking (9). Detailed biochemical analysis of byssal threads, however, has been largely thwarted because of extensive quinone-tanning (10). To date, the only effective strategy has been prolonged pepsinization at 4–10°C. Two pepsin-resistant collagenous fragments, Col-D and Col-P, have been isolated and characterized from the distal and proximal portions of byssal threads, respectively (11). These appear to be homotrimers having masses corresponding to less than half that of pepsinized type I collagen, Gly-X-Y repeat sequences, and a complementary graded distribution along the axis of each thread as shown in Fig. 1A. Few additional details have emerged from direct analyses of byssal threads.
The molecular characterization of byssal precursors from the foot of the mussel has been much more revealing. Byssal threads are formed, one at a time, by the mussel foot in a 5-min process that resembles reaction injection molding: The ventral groove of the foot is filled with a milky dispersion of secreted protein that is molded, condensed, and coated before release (2). Two major thread precursors secreted by the foot are preCol-D and preCol-P. PreCol-P is a molecular hybrid of collagen with flanking domains having elastin-like sequences; its expression predominates in the proximal portion of the foot (7). PreCol-D, which is expressed strongly at the distal end of the foot, has a collagenous domain flanked by sequences resembling spider dragline silk motifs (8). Because the patterns of preCol expression in the foot mirror the graded distributions of Col-P and Col-D in byssal threads, the gradients appear to be predetermined in the foot before secretion. Accordingly, preCol-D would be maximal at the distal end of each thread decreasing proximally, whereas preCol-P would have a gradual crescendo beginning at mid-length of each thread and reaching a maximum at the proximal end (11). Although the flanking domains might be used conveniently to explain the nontendon-like attributes of byssal threads, the assembly scheme for these unusual molecules in the byssus is not known.
Here, we describe another hybrid collagen from the foot and byssus of the common mussel Mytilus edulis. Like the other two, it possesses a domain structure that includes histidine-rich termini and a central collagenous domain. In contrast to preCol-P and -D, its flanking domains are dominated by sequences akin to plant cell wall proteins, and it is uniformly present in foot sections. For this reason, it has been named a nongradient (NG) byssal precursor collagen or preCol-NG. It has unusual sequence attributes that fit a role as mediator or adaptor in preCol assembly.
METHODS
Screening cDNA Library.
A cDNA library of the foot (ZAP expression vector, Stratagene, La Jolla, CA) of Mytilus edulis initially was screened with an oligonucleotide probe encoding the signal peptide and N terminus of preCol-D with the aim of finding whole transcripts of preCol-D (8). Positive phages were converted into plasmids (Pbk-CVM) in the presence of helper phage (Stratagene). The plasmids were isolated by alkaline lysis and purified by anion exchange chromatography by using miniprep kits (Qiagen, Chatsworth, CA). Pure plasmids were subjected to restriction digestion analysis to determine insert size. Nested deletions of the full length clones were made by using the Nested Deletion Kit (Pharmacia) and were sequenced with a dideoxy termination kit (Perkin—Elmer). Sequence analysis was performed with commercial software (sequencher 3.0, Gene Codes, Ann Arbor, MI).
Reverse Transcriptase (RT)-PCR of Foot Sections.
Total RNA (1 μg) extracted from each of five foot sections and nonfoot tissues (muscle, gill, mantle) was reverse transcribed into cDNA with random hexamers (Life Technologies, Gaithersburg, MD) with a first strand cDNA synthesis kit (Life Technologies). One-tenth of the first strand reaction was PCR-amplified by using gene specific primers for preCol-NG (sense: 5′-GATTTGGAGACTTCGCTGAT-3′ and antisense 5′-GGTCCACCTGGGCCGTTCAA-3′) and the EukA and EukB primers specific to eukaryotic rRNA as positive controls (12). The preCol-D and -P gene-specific primers served as gradient controls (8). The amplifications were carried out for 35 cycles with 30 s at 94°C for denaturation, 30 s at 55°C for annealing the preCol-NG, -P, and –D-related cDNAs and at 60°C for annealing the eukaryotic rRNA controls, and 1.5 min at 72°C for extension with a final 5-min extension. No reverse transcriptase was added to foot and nonfoot tissues as a control for the presence of DNA contamination. Omission of first strand templates was used as a control for primer–dimer formation. The positive control for reverse transcriptase activity was RNA supplied with the kit. The plasmid containing preCol-NG insert was used as a positive control for PCR. PCR products from the RT-PCR amplification of foot sections subsequently were analyzed by restriction enzyme digestion and dideoxy sequencing to confirm their identity.
Extraction of Col-NG.
To obtain direct evidence that preCol-NG or some part of it is present in byssus, 2–3 g (wet weight) byssal threads were harvested from mussels maintained at 12°C in aquaria with running sea water. These were divided into two lots of proximal and distal portions. Each lot was disrupted mechanically in 100 ml 5% (vol/vol) acetic acid/g wet weight as per Qin and Waite (11). Thereafter, all procedures were performed at ≈4°C. The resultant slurries were subjected to pepsin digestion (1:10 enzyme to wet weight) for 48 h with constant stirring. Insoluble materials were removed by centrifugation at 17,000 × g for 30 min. Collagens were precipitated from the supernatant by overnight dialysis against 0.1 M borate pH 8.0. Precipitates were harvested by centrifugation 31,000 × g for 60 min, and, after decanting the supernatant, pellets were redissolved in 5% acetic acid with 8 M urea.
Isolation and Partial Characterization of Col-NG.
Solutions containing acid-soluble pepsin-resistant byssal proteins were neutralized by stepwise addition of 1 M Tris base (Bio-Rad) and were reduced at room temperature for 30 min with 50 mM DTT. The samples were clarified by a 10-min spin at top speed in an Eppendorf microfuge and filtered through a 0.45-μm membrane. Fractionation was accomplished by C4 HPLC by using a linear gradient of aqueous acetonitrile (18–35%) with 0.1% trifluoroacetic acid over 60 min. The eluted fractions were monitored at 214 nm, and peak fractions were freeze-dried for electrophoresis. The discontinuous Tris–glycine method was used for routine PAGE in the presence of SDS (SDS/PAGE). Running conditions on 10% gels were 15 mA for 3 h at 20°C. In preparation for sequencing, the proteins separated by SDS/PAGE were electroblotted horizontally to Immobilon P (Millipore) by using a methanolic glycine-free Tris–borate transfer buffer with 0.1% SDS (11) and a constant current of 200 mA for 3 h. Transferred proteins were stained with Coomassie Blue G-250, and the 60- to 64-kDa bands (120–180 pmol) were excised and sequenced. The N terminus of Col-NG was analyzed by an automated sequencing protocol optimized for posttranslational modifications (ref. 13; Porton Instruments, Tarzana, CA).
RESULTS
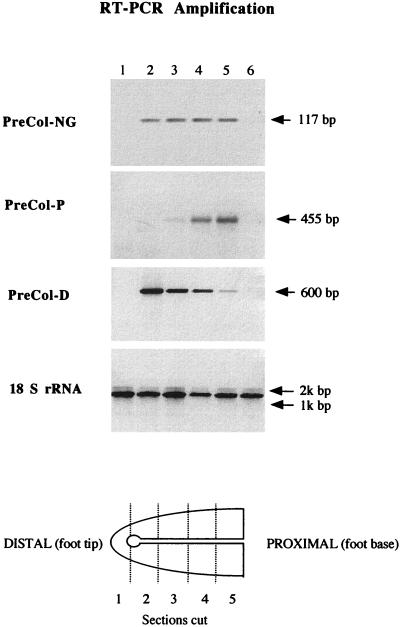
PreCol-NG has not been detected previously in mussel foot tissue or as a pepsin-resistant fragment from byssal threads. Its detection in a cDNA library resulted from strong hybridization to a probe designed from part of the signal peptide region of preCol-D. Sequence analysis of eight positive clones revealed that two were preCol-NG. Evidently, preCol-NG was picked up because of the high degree of identity (>50%) of its signal peptide sequence with that of preCol-D. The larger preCol-NG clone contains 2,796 bp and an ORF that encodes 904 aa (Fig. 2). The first 20 aa represent the signal peptide identified by its hydrophobicity (14). A further study of the tissue-specific expression of the latter using gene-specific RT-PCR revealed that it is foot-specific, but, unlike preCol-P and -D, its distribution in the foot exhibits no graded expression (Fig. 3). PreCol-NG is distributed uniformly in all foot sections except for the tip, where none of the byssal collagens are present.
Figure 2.
Deduced sequence of α-preCol-NG arranged to exhibit the block domains and align the characteristic repeat motifs. Cell wall (CW) and fibroin-like motifs are aligned at XGn and polyalanine clusters, respectively; the collagen domain is enclosed in a bold box and arrayed as a series of tripeptides. H, histidines in the His domains; A, polyalanines; •, imperfection. Underlining indicates sequenced peptide from byssus-derived protein; N-/C-terminal boxes denote Tyr-containing sequences common to all preCols. PreCol-NG cDNA sequence has been deposited with GenBank AF-043944.
Figure 3.
The tissue specificity and foot distribution of α-preCol-NG as determined by RT-PCR analysis by using primers that are gene-specific for preCol-NA (A), preCol-P (B), preCol-D (C), and a universal transcript, 18S rRNA (D; positive control). Total RNA (1 μg) was extracted from serial foot sections (lanes 1 to 5) and from nonfoot tissues (mantle, gill, adductor muscle) and reverse-transcribed and PCR amplified. The cartoon of the foot at the bottom shows the approximate location of segments prepared for RT-PCR. The hairpin loop in the middle represents the distal depression (bulb) and ventral groove (shaft) where byssal threads are assembled by injection molding of precursors.
Mature preCol-NG has a predicted pI of 8.0 and a molecular mass of 76 kDa based on sequence deduced from nucleic acid data. This is much less basic than the other two preCols (pIs 10.1 [D] and 11.6 [P]); the mass (76 kDa), however, is comparable. The deduced primary sequence of preCol-NG contains several structural motifs: a central collagenous domain that is flanked on both sides by domains that resemble Gly-rich cell wall proteins (Fig. 2). At the N and C termini are histidine-rich regions. The collagen domain accounts for 50% of the total mass of preCol-NG and, like fibrillar vertebrate collagens, consists of tandemly repeated Gly-X-Y triplets. In typical collagens, the amino acids in X and Y are unspecified; however, Pro is common as X, and trans 4-hydroxyproline is common as Y. Aromatic amino acids, Gly, and Cys are rare in X and Y. The collagenous domain of preCol-NG generally abides by these conventions although there are notable exceptions: First, the eleventh triplet is missing a Gly, i.e., GHS⋅PQ. A similar deletion exists in preCol-P at the same location (7). The significance of the imperfection on collagen structure is not clear at this stage; however, computer modeling of α chains having the Col-NG sequence indicates a bend of ≈35° at the deleted Gly (result not shown). A single Cys (374) is present in the first third of the collagenous domain. PreCol-D also has a single Cys near the middle of its collagen domain whereas preCol-P has none (7, 8). There are eight occurrences of Gly in the X position of the triplet and four in the Y. The abundance of Gly-Gly-Y triplets within the collagen domain is predicted to contribute to greater chain flexibility and decreased melting temperature for the collagen triple helix (15).
The collagen domain of preCol-NG frequently has Pro in the X and/or Y positions; those in Y are expected to be hydroxylated. Hyp-containing sequences have been detected directly in the collagen domains of Col-P (11) and preCol-D (8); in sequencing the N terminus of Col-NG, one residue of Hyp was detected (see below). The C-terminal end of the collagenous domain (res 658–709) is punctuated by an acidic-hydrophobic patch that has a significant homology with the first 10–12 residues of similar patches in preCol-D and -P.
Flanking the collagen domain on both sides are Gly- and Ala-rich domains. These resemble the flanking domains of preCol-D; however, the relative abundance of the polyalanine runs and XGlyn (n = 1, 2 or 3) repeats is reversed with respect to the N and C termini. There are two polyalanine runs on the N-terminal side in preCol-NG. Each is 12–20 residues long with one interruption, e.g., Ser, Asn, His or Arg per three Ala. XGlyn clusters are ≈50 residues in length and are found in both the N- and C- flanking domains of preCol-NG (Table 1). These are longer than those in preCol-P/-D and are rather similar in sequence to the Gly-rich proteins from plant cell walls (16), shell hinge ligaments (17), and a variety of trematode egg shell proteins (18) (Table 1). Gly-rich cell wall protein, ptGRP1, and the protein rubber, abductin, in particular, share the hydrophobic nature of residues in position X. The flanking domains also contain another β-sheet motif that is more typical of cocoon silks, i.e., GlyAlaGlyAla/Ser (19). These together with the polyalanines tend to form a “sandwich” around the XGlyn repeats in the flanking domains.
Table 1.
A comparison of the flanking domain sequences of preCol-NG with other glycine-rich proteins having repeats of the consensus X-[Gly]n in which X can be any residue but Gly or Pro
| [X-Glyn]m | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | X/n=1 | 2 | 3 | 4 | 5 | 6 | n > 6 | |
| preCol-NG | ||||||||
| N-side | L | 1 | 4 | 5 | 4 | 1 | 1 | 1 |
| F | - | 1 | 1 | - | - | - | - | |
| N | - | - | 1 | - | - | - | - | |
| V | - | - | 1 | - | - | - | - | |
| C-side | A | - | 13 | - | - | - | - | - |
| L | 1 | 4 | 2 | - | - | - | - | |
| N | - | 2 | 4 | - | - | - | - | |
| I | - | - | 1 | - | - | - | - | |
| F | 1 | 1 | - | - | - | - | - | |
| Cell wall protein ptGRP1 (24) | A | 4 | 1 | 13 | - | 3 | - | 1 |
| V | 5 | - | 8 | - | 1 | - | 1 | |
| F | 7 | 1 | 2 | - | - | - | 2 | |
| L | - | - | 2 | - | 5 | - | 1 | |
| H | - | - | 5 | - | - | - | - | |
| S | 5 | - | 1 | 1 | 1 | - | - | |
| I | 2 | - | 2 | - | - | - | - | |
| Abductin Ap4/7 (25) | M | 1 | 4 | 8 | - | - | - | - |
| F | - | 10 | - | - | - | - | - | |
| K | - | 2 | - | - | - | - | - | |
| I | - | 1 | - | - | - | - | - | |
| Egg shell protein pSMf61-46 (26) | Y | 4 | 2 | 9 | - | - | - | - |
| K | 3 | 4 | 1 | - | - | - | - | |
| S | 3 | 3 | - | - | - | - | - | |
| C | 2 | - | 1 | - | - | - | - | |
| N | 1 | 2 | - | - | - | - | - | |
Proteins and preCol-NG are listed according to the nature of X, the number of Gly residues (n), and the number of repeats (m).
Following the Gly-/Ala-rich domains are histidine-rich domains. The five His residues in the N-terminal region and 16 at the C terminus are dispersed within Gly- or Ala-rich sequences. Both termini also contain Lys- and Tyr-rich sequences. These resemble other byssal protein precursor sequences in which Tyr is usually posttranslationally modified to 3,4-dihydroxyphenylalanine (Dopa) (20); KPGY (26–30), for example, is close to the KPSY repeat sequence in the adhesive foot protein-1 of M. edulis (21). Indeed, N-terminal sequencing of preCol-D revealed that all four of its tyrosines were converted to Dopa (8).
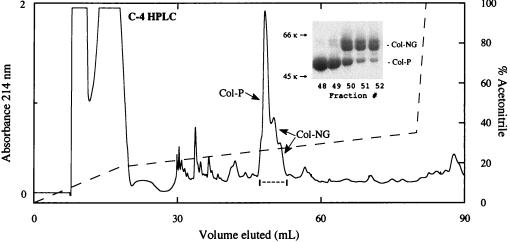
Detection of intact preCol-NG in byssal threads was not possible because of cross-linking. A pepsin-resistant fragment, Col-NG, however, can be prepared from foot and thread extracts. Although this normally comigrates with Col-D on SDS/PAGE, a clean separation of the two can be obtained by prior C4 HPLC of DTT-reduced samples. Col-NG derived from the pepsin-digested proximal portion of byssal threads elutes at 50 ml in a doublet after the large Col-P peak (Fig. 4). SDS/PAGE of the fractions reveals two peptides with apparent masses of 60–64 kDa. Both have unambiguous N-terminal sequences of ATGGGGFPLPGAP*GPQGP as determined by automated Edman degradation where P* denotes trans-4-hydroxyproline. Variable digestion at the C terminus may account for observed heterogeneity during SDS/PAGE and HPLC. The Edman sequence matches the cDNA-deduced amino acid sequence of preCol-NG between res 211–228 (Fig. 2).
Figure 4.
C-4 HPLC of pepsin-resistant collagens derived from the proximal portion of byssal threads. The collagens were reduced with DTT at room temperature before chromatography. HPLC fractions (brackets) were freeze-dried before running on SDS/PAGE (Inset). The two closely migrating bands labeled as “Col-NG” were subjected separately to automated Edman sequencing after electrotransfer to polyvinylidene difluoride membranes. Two standard markers (66 kDa and 45 kDa) are as shown. HPLC flow rate was 1 ml/min, and volume per collected fraction was 1 ml. Absorbance at 214 nm (solid line); acetonitrile gradient (dashed line).
DISCUSSION
Our results establish the existence of a cDNA that encodes preCol-NG, a third byssal collagen having a block copolymer structure. The uniform distribution of this collagen along the length of the foot is in marked contrast to the other precursors, preCol-P and –D, which have graded distributions. Although we have evidence confirming the presence of preCol-NG in the byssus, the pattern of its distribution and structural role there remain to be shown. So far, there has been a perfect correlation between the distribution of preCols in the foot and their distribution in the byssus (8, 11).
Gly-rich sequences [XGlyn]m dominate the flanking domains of preCol-NG (Table 1). Although the ordered structure of model synthetic poly(XGlyGly) in vitro is limited to type II β-turns when X = Ala, Val, Leu, or Ile, poly(Leu-Gly-Gly) had the least stable conformation of the four polymers (22). A new study of the XGlyGly repeats in spider dragline silk reports that they do not form β-sheets at all but rather 31 helices (23). Thus, the actual structure of XGlyGly repeats where X is often L in preCol-NG, ptGRP1, or spidroin remains uncertain.
There are some features of “silk fibroin-like” structure that preCol-NG shares with preCol-D (8). Both have the polyalanine runs typical of spider dragline silks (spidroin 1 or ADF-3) (24, 25); there are eight of these in preCol-D compared with three in preCol-NG. Moreover, preCol-NG has extended [GlyAlaGlyAla] clusters that are typical of the stiffer cocoon silks (18, 26). Together with the polyalanines, these tend to be arrayed in sandwich-like fashion around the extensive XGlyn repeats in both the N- and C-flanking domains of preCol-NG (Fig. 3). The effect of this blend of structural motifs on tensile properties is difficult to predict. In general, the polyalanine runs and GAGA clusters of silk fibroins form stiffer paracrystalline assemblies (27, 28) than XGlyn sequences, which are, by all accounts, less structured (29). However, silks are diverse materials. Although only a handful of primary sequences are available, at least five different categories of silk have been identified crystallographically with intersheet spacings ranging from 9 to 15 Å (26). These run the gamut of mechanical properties from ultrahigh tensile strength with low extensibility (<30%) to high tensile strength with high extensibility (300%) (30). The large number of XGlyn repeats in the flanking domains of preCol-NG suggests that these will make preCol-NG more extensible than preCol-D, but this remains to be demonstrated.
We have proposed that preCol-P, with its elastic domains, confers high extensibility and elastic recoil on the proximal thread (7), and preCol-D with silk-like domains contributes to the strength and extraordinary toughness of the distal portion of the thread (8). PreCol-NG may provide a molecular “thread of continuity” for preCol assembly between the proximal and distal ends of each thread, but the meaning of this to byssal function is unlikely to be revealed without additional information about the specific interrelationships of preCols during fibrillogenesis.
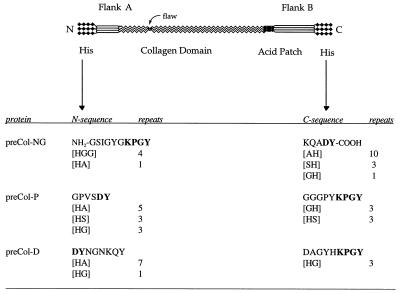
The three byssal precursors preCol-D, preCol-P, and preCol-NG are generally of homologous design. For example, all three consist of the same N→C block domain sequence: His-rich, flanking, collagen, acidic patch, flanking, and His-rich (Fig. 5). The acidic patch is an obvious deviation from the symmetrical array of domains around the central collagen but it is common to all three. More subtle asymmetries are apparent within the His-rich domains: (i) they are generally larger at one end of the protein than the other and (ii) they have distinctive Tyr-containing sequences. For example, preCol-NG has five residues of His and a signature sequence, LysProGlyTyr, on the N-terminal side, whereas on the C-terminal side, it has 16 His residues and the sequence AspTyr. N-terminal His residues are separated by Gly residues, whereas most at the C terminus are separated by Ala. The situation is reversed in preCol-P and -D (Fig. 5). PreCol-NG is thus the only precursor with a preponderance of His at the C terminus. The distribution of His and Tyr in the His-rich domains has important implications for the intra- and intermolecular stabilization of assembled preCols in the byssus. Dopa- and His-rich sequences frequently are involved in high affinity metal binding (31, 32), and byssal threads have elevated levels of Zn and other metals (33). Earlier, we (7, 8) proposed that intermolecular, His-mediated binding of Zn might serve to stabilize assembled preCols.
Figure 5.
Cartoon of the block domain structure of byssal preCols including the N- and C-terminal sequences and tally of the distribution of histidines and histidine-containing sequences from the N- and C-termini of three byssal collagens, preCol-NG, preCol-P and preCol-D.
The relative reversal in Tyr-containing sequences and His abundance in the terminal domains of preCol-NG suggests an unusual role for this collagen in the assembly of preCols. If we assume that byssal collagens, like the higher type I–III fibrillar collagens, are all unidirectional in polarity during assembly, i.e., all N termini point in one direction of the fiber axis and all C termini in the other, a role as “mediator” of axial assembly seems plausible for preCol-NG. The “LEGO”-like model in Fig. 1B suggests that by “dovetailing” the long and short histidine-rich domains of preCol-NG with the corresponding long and short domains in preCol-P and D, the axial assembly necessarily would have a preCol-NG at every other position in the chain, e.g., X-preCol-NG-X-preCol-NG-X. The identity of X would be determined by where in the byssus the fibers are: At the distal end, X would be preCol-D; beyond mid-length, preCol-D and preCol-P would occur with equal frequency; and at the proximal end, preCol-P would predominate as X. Although there are other possible models, this model is consistent with the graded distributions of preCol-P and -D, the relative stoichiometry of preCol-D and -NG, and the uniform distribution of preCol-NG. Obviously, additional evidence is needed for validation. Furthermore, lateral interactions of axial preCol microfibers and proper register of flanking domains will necessarily play a major role in determining biomechanical properties. The involvement of Cys residues in the lateral interactions of Col-D and Col-NG is possible. These will require additional scrutiny.
These results are important for two reasons: They suggest that (i) the proteins of natural fibers can combine collagen domains with a variety of structural motifs including elastin, cocoon and dragline silks, cell wall proteins, etc., and that (ii) collagen-containing macromolecules may undergo fibrillogenesis in ways that seem very bizarre.
Acknowledgments
We thank the laboratory of John McDonald for the generous use of his DNA sequenator, Kathryn Coyne for sharing her expression library of the foot of M. edulis, and George Luther for modeling an α-chain fragment of Col-NG, and we thank an anonymous reader for a number of useful criticisms This research was supported by grants from NIDR-Biomaterials and the Office of Naval Research.
ABBREVIATIONS
- NG
nongradient
- RT
reverse transcriptase
- Col-D
pepsin-resistant collagen from the distal portion of byssus
- Col-NG
pepsin-resistant NG collagen from the byssus
- Col-P
pepsin-resistant collagen from the proximal portion of byssus
- preCol-D
precursor of Col-D
- preCol-NG
precursor of Col-NG
- preCol-P
precursor of Col-P
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF043944).
References
- 1.Gathercole L J, Keller A. In: Structure of Fibrous Biopolymers. Atkins E D T, Keller A, editors. London: Butterworths; 1975. pp. 153–175. [Google Scholar]
- 2.Waite J H. Results Probl Cell Diff. 1992;19:27–54. doi: 10.1007/978-3-540-47207-0_2. [DOI] [PubMed] [Google Scholar]
- 3.Bell E C, Gosline J M. J Exp Biol. 1996;199:1005–1017. doi: 10.1242/jeb.199.4.1005. [DOI] [PubMed] [Google Scholar]
- 4.Smeathers J E, Vincent J F V. J Molluscan Stud. 1979;49:219–230. [Google Scholar]
- 5.Waite J H, Qin X X, Coyne K J. Matrix Biol. 1998;17:93–106. doi: 10.1016/s0945-053x(98)90023-3. [DOI] [PubMed] [Google Scholar]
- 6.Bairati A, Vitellaro Zuccarello L. Cell Tissue Res. 1976;166:219–234. doi: 10.1007/BF00227043. [DOI] [PubMed] [Google Scholar]
- 7.Coyne K J, Qin X X, Waite J H. Science. 1997;277:1830–1832. doi: 10.1126/science.277.5333.1830. [DOI] [PubMed] [Google Scholar]
- 8.Qin X X, Coyne K J, Waite J H. J Biol Chem. 1997;272:32623–32627. doi: 10.1074/jbc.272.51.32623. [DOI] [PubMed] [Google Scholar]
- 9.van Ness K P, Koob T J, Eyre D R. Comp Biochem Physiol. 1988;91B:531–534. doi: 10.1016/0305-0491(88)90017-x. [DOI] [PubMed] [Google Scholar]
- 10.Brown C H. Q J Micros Sci. 1952;93:487–502. [Google Scholar]
- 11.Qin X X, Waite J H. J Exp Biol. 1995;198:633–644. doi: 10.1242/jeb.198.3.633. [DOI] [PubMed] [Google Scholar]
- 12.Medlin L, Elwood H J, Stickel S, Sogin M L. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 13.Waite J H. Methods Enzymol. 1992;258:1–20. doi: 10.1016/0076-6879(95)58033-6. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–12. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Shah N K, Sharma M, Kirkpatrick A, Ramshaw J A M, Brodsky B. Biochemistry. 1997;36:5878–5883. doi: 10.1021/bi963146c. [DOI] [PubMed] [Google Scholar]
- 16.Condit C M, Meagher R B. Nature (London) 1986;323:178–181. [Google Scholar]
- 17.Cao Q, Wang Y, Bayley H. Curr Biol. 1997;7:R677–678. doi: 10.1016/s0960-9822(06)00353-8. [DOI] [PubMed] [Google Scholar]
- 18.Bobek L A, Rekosh D M, LoVerde P T. Mol Cell Biol. 1988;8:3008–3016. doi: 10.1128/mcb.8.8.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas F, Shaw J T B, Smith S G. Adv Protein Chem B. 1958;4:313–327. doi: 10.1016/s0065-3233(08)60599-9. [DOI] [PubMed] [Google Scholar]
- 20.Papov V V, Diamond T V, Biemann K, Waite J H. J Biol Chem. 1995;270:20183–20192. doi: 10.1074/jbc.270.34.20183. [DOI] [PubMed] [Google Scholar]
- 21.Filpula D R, Lee S M, Link R P, Strausberg S L, Strausberg R L. Biotech Prog. 1990;6:171–177. doi: 10.1021/bp00003a001. [DOI] [PubMed] [Google Scholar]
- 22.Tamburro A M, Guantieri V, Scopa A, Drabble J M. Chirality. 1991;3:318–323. doi: 10.1002/chir.530030417. [DOI] [PubMed] [Google Scholar]
- 23.Kümmerlen J, van Beek J D, Vollrath F, Meier B H. Macromolecules. 1996;29:2920–2928. [Google Scholar]
- 24.Lewis R V. Acc Chem Res. 1992;25:392–398. [Google Scholar]
- 25.Guerette P A, Ginzinger D G, Weber B H F, Gosline J M. Science. 1996;272:112–115. doi: 10.1126/science.272.5258.112. [DOI] [PubMed] [Google Scholar]
- 26.Gosline J M, DeMont M E, Denny M W. Endeavour. 1986;10:37–43. [Google Scholar]
- 27.Simmons A H, Michal C A, Jelinski L W. Science. 1996;271:84–87. doi: 10.1126/science.271.5245.84. [DOI] [PubMed] [Google Scholar]
- 28.Warwicker J O. J Mol Biol. 1960;2:350–362. doi: 10.1016/s0022-2836(60)80046-0. [DOI] [PubMed] [Google Scholar]
- 29.Thiel B L, Guess K B, Viney C. Biopolymers. 1997;41:703–719. doi: 10.1002/(SICI)1097-0282(199706)41:7<703::AID-BIP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Denny M W. J Exp Biol. 1976;65:483–505. [Google Scholar]
- 31.Taylor S W, Luther G W, Waite J H. Inorg Chem. 1994;33:5819–5824. [Google Scholar]
- 32.Lipscomb W N, Sträter N. Chem Rev. 1996;96:2375–2433. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 33.Coombs T L, Keller P A. Aquat Toxicol. 1980;1:291–300. [Google Scholar]