Abstract
Femtosecond time-resolved infrared spectroscopy is used to study the formation of cyclobutane dimers in the all-thymine oligonucleotide (dT)18 by ultraviolet light at 272 nanometers. The appearance of marker bands in time-resolved spectra indicate that dimers are fully formed ∼1 picosecond after ultraviolet excitation. The ultrafast appearance of this mutagenic photolesion points to an approximately barrierless excited-state reaction for bases that are properly oriented at the instant of light absorption. The low quantum yield of this photoreaction is proposed to result from infrequent conformational states in the unexcited polymer, revealing a strong link between conformation prior to light absorption and photodamage.
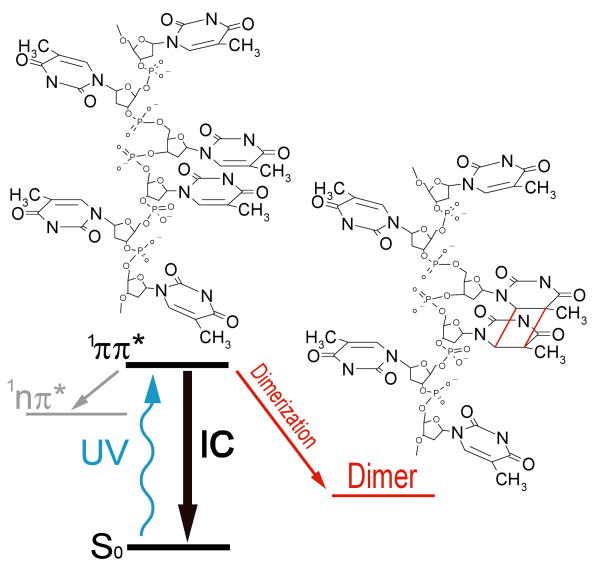
The most abundant lesion in UV-irradiated DNA is the cyclobutane pyrimidine dimer (CPD) formed between adjacent thymine bases (Fig. 1) (1). This mutagenic photoproduct disrupts the normal cellular processing of DNA, and leads to a complex web of biological responses including apoptosis, immune suppression, and carcinogenesis (2-4). Organisms possess elaborate repair pathways to counter this constant threat to genomic integrity. Aside from their biological significance, CPDs are of interest as structural reporters. Thymine dimer yields are not the same at all TT doublets in a given DNA sequence, but depend in poorly understood ways on the identity of the flanking bases and on local conformation (1). By exposing DNA to UV light and then measuring the relative photoproduct yields with single nucleotide resolution it has been possible in favorable cases to obtain structural information (5-7). In order for this methodology to realize its full potential, molecular-level understanding of the dimerization mechanism is essential. We report here a dynamical study of thymine dimerization, which provides insight into the coupling between structure and DNA photodamage.
Figure 1.
Simplified schematic of the photodynamics of the DNA oligomer (dT)18. UV excitation populates a singlet ππ* state. This state decays overwhelmingly via internal conversion (IC) to the S0 ground state. To a smaller extent population branches to a singlet nπ* state. Intersystem crossing to a triplet state has been detected in thymine but not in polymeric DNA. Finally the ππ* state can decay to a dimer photoproduct with the indicated structure contingent on the conformation at the time of excitation.
CPD formation is a [2+2] photocycloaddition reaction in which the carbon-carbon double bonds of proximal pyrimidine bases react to form a cyclobutane ring. In the analogous reaction between two ethylene molecules, electronic excitation and the proper orientation of the reacting double bonds are needed for reaction to occur (8). Unlike the case of free ethylene molecules, pyrimidine bases in DNA are tethered to the sugar-phosphate backbone and this restricts the achievable orientations. Some conformations are simply impossible due to backbone constraints. Thus, in UV-irradiated oligo- and polynucleotides a single CPD isomer (the cis-syn isomer shown in Fig. 1) is formed, while two thymine molecules diffusing freely in aqueous solution yield all six stereoisomers (1). Because DNA is moderately flexible, a vast number of conformations exist. Some of these have the bases positioned favorably for reaction, while others do not. Importantly, DNA is highly dynamic and motions such as the stacking and unstacking of bases, base pair breathing and opening, torsional oscillations, and helix bending will incessantly bring a given bipyrimidine doublet into and out of favorable geometries for dimerization. The impact of these motions on the reaction kinetics depends on how their rates compare to the rate of reaction by favorably oriented bases (9). Direct kinetic measurements of dimerization can elucidate the potentially complex interactions between conformational dynamics and photodamage, and is one of the main motivations of this work.
In an excited-state reaction, motion along the reaction coordinate occurs in competition with energy wasting steps like fluorescence and internal conversion to the electronic ground state. In the last few years it has become possible to directly observe the dynamics of excited electronic states in DNA model compounds by femtosecond spectroscopy (10, 11). The very high rate of nonradiative decay by the singlet ππ* (1ππ*) states of single nucleobases has been proposed to greatly restrict photodamage (10). However, recent work has revealed the presence of additional, rather long-lived singlet states in DNA (11) and single bases (12). In oligodeoxynucleotides, lifetimes of less than 1 ps to more than 100 ps have been observed, depending on base stacking and base sequence (11). Additionally, at least 10% of all singlet excitations in single pyrimidine bases like thymidine 5′-monophosphate (TMP) decay to singlet nπ* (1nπ*) excited states with lifetimes in excess of 10 ps (11). Kinetic measurements can determine which of these diverse excited states is the dimer precursor.
Past efforts to observe dimerization kinetics have been unsuccessful. It was shown by flash photolysis that photodimers are formed in (dT)20 in < 200 ns, the time resolution of the laser system used (13). Femtosecond transient electronic spectroscopy (11) has not provided direct evidence for dimer formation because CPDs do not absorb significantly at wavelengths longer than about 270 nm. Due to its chemical bond specificity, vibrational spectroscopy can often unambiguously identify transient species and stable photoproducts (14). We therefore recorded time-resolved infrared spectra of a DNA model compound excited by a femtosecond UV pump pulse (15). The system studied is the single-stranded all-thymine oligodeoxynucleotide (dT)18 in order to maximize the number of dimers formed with each laser pulse. In this DNA model system, every absorbed photon excites a residue capable of dimerization. Quantum yields in the closely related systems poly(dT) (0.033) (16) and (dT)20 (0.028) (13) are among the highest reported for any DNA compound. In contrast, the dimerization quantum yield is over thirty times lower in double-stranded, genomic DNA (17). This large reduction is due to the low frequency of TT doublets and absorption by non-thymine bases in mixed-sequence DNA. After presenting our results for (dT)18, we will discuss the implications for double-stranded nucleic acids.
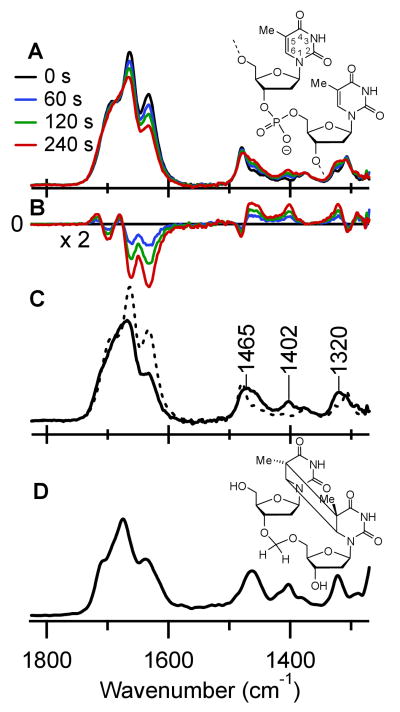
Steady-state IR absorption spectra of (dT)18 in D2O were recorded before and after UV-irradiation at 266 nm in order to locate IR marker bands indicative of dimerization. In the spectrum prior to UV-irradiation (black curve in Fig. 2A), three strong bands are observed at 1632, 1664, and 1693 cm-1. These bands, which arise from double-bond stretches associated with the two carbonyl groups and the C5=C6 double bond (18), bleach strongly after several minutes of UV exposure (Fig. 2A). Difference spectra were calculated by subtracting the steady-state IR spectrum from each spectrum of the UV-irradiated oligomer (Fig. 2B). Negative bleaching signals are apparent in the double-bond stretching region above 1600 cm-1. In addition, positive peaks between 1300 and 1500 cm-1 grow in with increasing exposure time. The IR absorption spectrum of the photoproduct (solid curve, Fig. 2C) was obtained from the difference spectra in Fig. 2B by target analysis (19), assuming that a single photoproduct is formed. In fact, a pyrimidine (6-4) pyrimidone photoadduct is also generated at TT doublets, but can be neglected because its quantum yield is fifty times lower in poly(dT) (20). The absorption spectrum (Fig. 2D) of a previously described model compound of the cis-syn thymine dimer (21) is in excellent agreement with the solid trace in Fig. 2C, showing that this is the only significant photoproduct under these conditions. Bands in the dimer spectrum substantially overlap those of unirradiated (dT)18 above 1500 cm-1. In contrast, a trio of marker bands is evident at 1320, 1402, and 1465 cm-1 (Fig. 2C), and these bands became the focus of the time-resolved experiments.
Figure 2.
(A) IR absorption spectra of (dT)18 (structure shown at the right) in D2O after exposure to UV laser pulses at 266 nm for the times indicated. (B) Difference IR spectra from data in part A. (C) IR absorption spectrum of the photoproduct obtained from data in part B by target analysis (solid curve), showing three distinctive marker bands between 1300 and 1500 cm-1. The IR absorption spectrum of (dT)18 prior to UV irradiation is shown for comparison (dashed curve). (D) Steady-state IR absorption spectrum of the cis-syn dimer model compound in D2O (structure shown at the right).
Broadband IR transient absorption signals were recorded between 1300 and 1550 cm-1 after excitation of (dT)18 by a femtosecond pump pulse at 272 nm (15). For comparison, measurements were carried out on TMP, which cannot dimerize on the time scales of interest here due to the slow rate of diffusional encounter by two TMP molecules. Transient spectra measured for both solutes are compared side-by-side in Fig. 3. Negative absorbance changes (bleaches) are colored blue, while positive signals are red. The bleaches monitor re-population of the starting material, while positive signals arise from vibrational bands of excited states or photoproducts. At first glance, the transient IR spectra of TMP and (dT)18 are very similar. The quantum yield for dimerization in (dT)18 is just 2 – 3% and most excitations in both systems decay nonradiatively on similar time scales (11).
Figure 3.
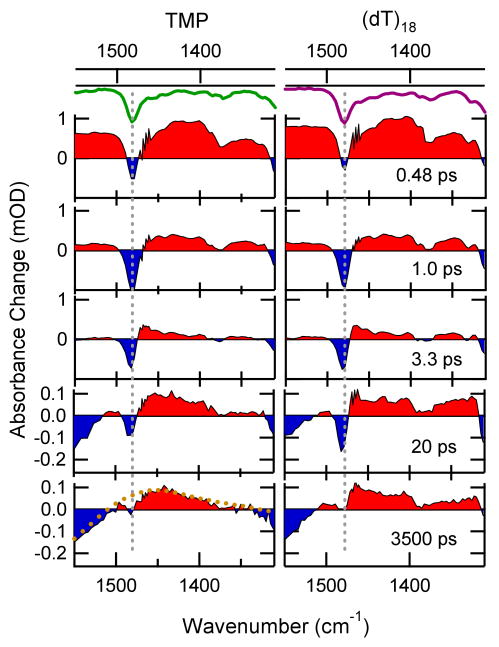
Transient IR difference spectra at the indicated times after 272 nm excitation of TMP and (dT)18 in D2O solution in the photodimer marker band region (typical errors are on the order of 10 μOD). Positive bands are shaded red, while negative signals are shaded blue. The green curves at top show the inverted steady-state IR spectra of each solute. Strong bands are indicated by the dashed gray lines. The dashed curve in the 3500 ps spectrum of TMP represents the steady-state difference IR spectrum obtained by raising the temperature of neat D2O (see supporting online material). After ultrafast internal conversion of excited molecules the transient spectra are dominated by cooling dynamics of the hot ground states on a time scale of some picoseconds. Transient spectra at later delay times show the broad signature of the heated solvent. The residual bleach seen for TMP at 3 ns is assigned mainly to intersystem crossing to a triplet state (estimated quantum yield ≤ (0.02) (6)). For (dT)18 one can see additional absorption due to the presence of thymine dimers.
We first describe the dynamical events revealed by the time-resolved spectra in Fig. 3 that are common to both solutes. Initially, UV excitation populates the lowest energy 1ππ* state, resulting in bleaches at frequencies corresponding to ground-state vibrations (dashed gray lines in Fig. 3). These bleaches have maximum amplitude near time zero as seen in the spectra recorded 0.48 ps after the pump pulse. Positive signals are seen at this time at all frequencies where bleaching is not observed. These broad bands decay with a lifetime close to that of the 1ππ* state (540 fs for thymidine (10)) and are no longer present in the 3.3 ps spectra. The short lifetime of this state is limited by internal conversion, which moves population nonradiatively from the 1ππ* state to the vibrationally excited electronic ground state. The photon energy is thus converted into sudden vibrational heating. This produces positive bands on the red edge of the negative bleach signals, resulting in distinctive sigmoidal line shapes (22) like the one seen near 1480 cm-1 in the 3.3 ps spectra. These features disappear by vibrational energy transfer to the solvent (vibrational cooling) with a time constant of 2 – 4 ps (11, 14) and are no longer visible at 20 ps.
The bleach near the maximum of each vibrational mode recovers in multiexponential fashion with similar kinetics as transient absorption signals recorded previously at UV wavelengths (11). The decay is 85 – 90% complete within 10 ps, while the remainder of the bleach recovers with time constants that vary between 100 and 1000 ps due to decay of 1nπ* population (12). A broad positive band near 1350 cm-1 decays on a 100 ps time scale and is tentatively assigned to this 1nπ* state. The spectra at 3 ns (Fig. 3) are dominated by a broad sigmoidal line shape, extending from 1300 to 1800 cm-1. This distinctive signature arises from a temperature jump effect, which is described in the supporting online text in more detail. The hot water contribution to the transient spectrum appears within a few picoseconds, but then remains constant in our time window due to slow heat transport out of the laser focus (23).
Now we address the subtle, but significant differences between the time-resolved IR spectra in Fig. 3. Greater modulation in the 20 ps and 3.5 ns spectra for (dT)18 is due to absorption in the oligomer at each of the three marker band frequencies identified in Fig. 2C. The difference is readily seen in a comparison of the transient spectra recorded for the two samples at 3 ns delay time in the top panel of Fig. 4A. The water heating signal is approximately the same for both samples due to the similar extent of ultrafast nonradiative decay. This signal can therefore be removed by subtracting the transient spectrum for TMP from the one recorded at the same delay time for (dT)18. Difference spectra constructed in this manner are shown by the blue curves in Fig. 4A and as a contour plot between 1 and 25 ps in Fig. 4B. The subtraction procedure is discussed at length in supporting online text.
Figure 4.
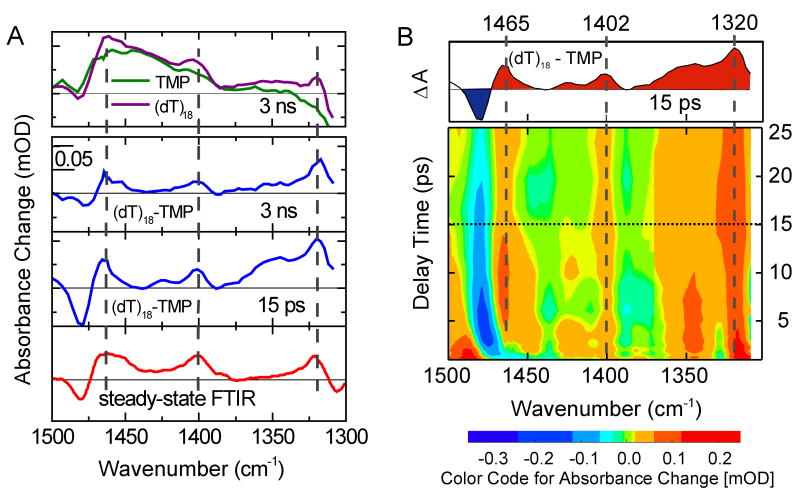
Difference spectra formed by subtracting the transient spectra of TMP from those of (dT)18. (A) The top panel shows transient spectra of TMP and (dT)18 at 3 ns. Their difference is plotted below as the blue curve together with a difference spectrum at 15 ps. The red curve at bottom represents the stationary IR difference spectrum of (dT)18 (dashed curve in Fig. 2C) and the dimer photoproduct (solid curve in Fig. 2C). It displays the absorption difference due to dimer formation. Dashed gray lines indicate the position of the cyclobutane dimer marker bands. (B) Contour plot of the difference spectrum. Red (blue) colors represent strong positive (negative) differences. A time slice showing the difference spectrum at 15 ps (dashed line) is shown in the top panel. Positive signals due to dimer formation are visible from ∼1 ps onwards for the bands at 1402 and 1320 cm-1, as indicated by the dashed gray lines. Because vibrational cooling dynamics differ for (dT)18 and TMP, incomplete subtraction in the spectral region around the 1480 cm-1 ground-state band obscures the 1465 cm-1 photodimer band at early times.
The red trace in Fig. 4A is the difference spectrum calculated by subtracting the ground-state absorption spectrum of (dT)18 from the dimer spectrum of Fig. 2C. This represents the expected absorption changes induced by dimer formation. The transient difference spectra at 15 ps and 3 ns show positive peaks at each of the dimer marker band frequencies and contributions from ground-state bleaching. The excellent agreement with the stationary spectrum shows unequivocally that thymine dimers are present ∼15 ps after excitation.
The dynamics of the marker bands at earlier times can be seen in a contour plot of the transient difference spectrum between 1 and 25 ps (Fig. 4B). The positive marker bands at 1402 and 1320 cm-1 are clearly visible over the entire time range shown. The marker band at 1465 cm-1 is visible down to 4 ps, but is obscured by vibrational cooling of hot thymine molecules at earlier delay times. Since TMP and (dT)18 exhibit different cooling dynamics (11), the vibrational cooling signatures do not fully cancel and show up in the difference plot in the vicinity of intense ground state bands. Thus, cooling dynamics by the hot 1480 cm-1 band (see Fig.3) covers the 1465 cm-1 marker band at early delay times. Cooling is also seen at other wavenumbers during the first few picoseconds, e.g. around 1350 cm-1. For delay times < 1 ps the signals are dominated by ultrafast relaxation of the electronically excited state, which obscures direct observation of dimer formation at the shortest times. Nevertheless, the observation of the dimer marker bands 1 ps after light absorption indicates that the reaction occurs on a femtosecond time scale.
The dimer yield can be estimated from the average amplitude of the marker bands in Fig. 4 of ∼30 μOD. This is 3% of the initial bleach of 1 mOD seen 1 ps after photoexcitation at 1480 cm-1. This band has a comparable cross section as the three marker bands, so the reported dimerization yield of 2 – 3 % (13, 24) should produce a signal of 20 – 30 μOD, as observed. The dimer yield at ∼1 ps thus equals the value from steady-state experiments within experimental uncertainty, demonstrating that dimerization is an ultrafast photoreaction. The remarkable speed of this bond-forming reaction is noteworthy, but not unprecedented. Ultrafast reaction rates are seen for some bimolecular reactions when the reactants are suitably preoriented (25). Also, the closely related intramolecular [2+2] photocycloaddition reaction of norbornadiene occurs in the gas phase in under 100 fs (26).
The ultrafast time scale of thymine dimerization suggests that an essentially barrierless path connects the initial 1ππ* state with the end product. This suggests that a conical intersection lies along this path as in computational studies of other pericyclic photoreactions (8). Significantly, dimerization in (dT)18 occurs more rapidly than many motions that could bring poorly oriented bases into a more favorable conformation for reaction. For example, base stacking and unstacking in thymine oligomers require tens of picoseconds according to a molecular dynamics study (27). Dimerization thus only occurs for thymine residues already in a reactive conformation at the instant of excitation (28, 29). Excited states of unfavorably oriented thymines are quenched before a change of conformation can occur. The extent of dimerization under steady-state irradiation thus depends on the fraction of time that a given doublet spends in reactive vs. nonreactive conformations. Control of CPD formation by ground-state structure is fully consistent with the rapid saturation of CPD formation in poly(U) and poly(dT) in a rigid glass at 77 K compared to room-temperature aqueous solution (30). This occurs because there are a finite number of reactive conformations in the low-temperature polymer, but the polymer in room-temperature solution is able to thermally fluctuate, allowing new reactive conformations to appear as exposure continues.
Because the rate of reaction by favorably aligned thymines is much faster than the rate of conformational change, the quantum yield is equal to the fraction of reactive conformations multiplied by the probability that a reactive conformation dimerizes upon excitation (9). The latter quantity is unknown, but is likely to approach unity based on the high quantum yields of dimerization in molecular crystals of some pyrimidine bases (31) and in dimers split in rigid matrices (32). With this assumption the quantum yield for dimerization is simply the fraction of favorably oriented conformations. The low yields for (dT)n thus reveal that only a few percent of TT doublets are favorably positioned for reaction at the time of excitation. This is consistent with the disordered structure of this rather flexible oligomer (33).
Excited-state modeling is needed to fully characterize the reactive conformations but some geometrical requirements are readily anticipated. Base stacking, which has been discussed in the past as a necessary criterion (30), reduces the distance between C5=C6 bonds compared to an unstacked geometry. However, the dimer geometry suggests that a low value of the dihedral angle between the reacting double bonds may also be important. The conformational changes observed near the site of a CPD like partial helix unwinding and bending (34) are likely to be the same ones needed to make a conformation favorable for reaction (7).
We fully expect thymine dimerization to be ultrafast in double-stranded DNA based on the speed of reaction in single-stranded (dT)18. Base pairing could affect the rates of nonreactive decay steps like internal conversion by the precursor excited state, but we consider this unlikely as recent time-resolved measurements show no effects due to base pairing on the dynamics of excited states in AT-containing oligodeoxynucleotides (11). We conclude that dimerization occurs with equal speed for bipyrimidine doublets in single- and double-stranded contexts provided that the thymine-thymine geometry is similar. Base pairing, on the other hand, will greatly influence quantum yields by altering the distribution of conformations. The structures of flexible all-thymine oligomers (27, 33) and double-stranded mixed-sequence DNA differ significantly, yet the quantum yields calculated per photon absorbed by a dimerizable thymine (supporting online text) are the same to within a factor of about two in room-temperature aqueous solution (17, 35). This means that a small percentage of TT doublets react in double-stranded DNA even though virtually all are well stacked. We propose that the winding of base pairs around the helix axis (the average twist angle is 36° in B-DNA (36)) keeps the C5=C6 double bonds too far apart. In contrast, although base stacking in single-stranded thymine oligomers is rare, the more flexible backbone does not prevent these rare stacks from adopting conformations suitable for dimerization.
A comparison of literature dimer yields in nucleic acids with A-type and B-type double helical structures supports the hypothesis that dimerization in double-stranded DNA occurs from uncommon conformations. The rate of dimer formation is decreased by up to a factor of two when double-stranded DNA is switched from B-type to A-type conformation (37). Even larger protective effects have been observed at TT steps in hairpins with A-type structure (38). The same base pairing is found in both structural classes, and the only difference is the distribution of accessible conformations. This establishes that conformation controls reactivity in duplex DNA just as in single-stranded (dT)18. The average twist angle between successive base pairs differs in A-DNA by only a few degrees compared to B-DNA, suggesting that ideal geometries in both helices are nonreactive. Instead, dimerization is proposed to take place at TT steps that deviate from the average duplex structure. Thus, the smaller amount of conformational variation in A-type vs. B-type structures (36) explains the greater resistance of A-DNA to CPD formation.
In summary, the model we have derived from our results implies that static thymine-thymine conformation (7, 29) and not conformational motions after photoexcitation (6, 39) determine the outcome of reaction. Flexibility does not help an excitation at a bipyrimidine doublet attain a better conformation within its lifetime, but a more flexible backbone can increase the fraction of reactive conformations present at the time of light absorption. With knowledge of the conformational criteria that make reaction inevitable, molecular dynamics simulation can be used to identify damage hotspots.
Supplementary Material
References and Notes
- 1.Cadet J, Vigny P. In: Bioorganic Photochemistry. Morrison H, editor. Wiley; New York: 1990. [Google Scholar]
- 2.Taylor JS. Acc Chem Res. 1994;27:76. [Google Scholar]
- 3.Vink AA, Roza L. J Photochem Photobiol, B. 2001;65:101. doi: 10.1016/s1011-1344(01)00245-7. [DOI] [PubMed] [Google Scholar]
- 4.Melnikova VO, Ananthaswamy HN. Mutation Research. 2005;571:91. doi: 10.1016/j.mrfmmm.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Becker MM, Wang JC. Nature. 1984;309:682. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- 6.Gale JM, Nissen KA, Smerdon MJ. Proc Natl Acad Sci U S A. 1987;84:6644. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pehrson JR. Proc Natl Acad Sci U S A. 1989;86:9149. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardi F, De S, Olivucci M, Robb MA. J Am Chem Soc. 1990;112:1737. [Google Scholar]
- 9.Wagner PJ. Acc Chem Res. 1983;16:461. [Google Scholar]
- 10.Crespo-Hernández CE, Cohen B, Hare PM, Kohler B. Chem Rev. 2004;104:1977. doi: 10.1021/cr0206770. [DOI] [PubMed] [Google Scholar]
- 11.Crespo-Hernández CE, Cohen B, Kohler B. Nature. 2005;436:1141. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 12.Hare PM, Crespo-Hernández CE, Kohler B. J Phys Chem B. 2006;110:18641. doi: 10.1021/jp064714t. [DOI] [PubMed] [Google Scholar]
- 13.Marguet S, Markovitsi D. J Am Chem Soc. 2005;127:5780. doi: 10.1021/ja050648h. [DOI] [PubMed] [Google Scholar]
- 14.Kuimova MK, et al. Proc Natl Acad Sci U S A. 2006;103:2150. doi: 10.1073/pnas.0506860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Information on materials and methods is available on Science Online.
- 16.Tramer Z, Wierzochowski KL, Shugar D. Acta Biochim Pol. 1969;16:83. [PubMed] [Google Scholar]
- 17.Douki T. J Photochem Photobiol, B. 2006;82:45. doi: 10.1016/j.jphotobiol.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhang SI, Michaelian KH, Loppnow GR. J Phys Chem A. 1998;102:461. [Google Scholar]
- 19.van Stokkum IHM, Larsen DS, van Grondelle R. Biochim Biophys Acta-Bioenergetics. 2004;1657:82. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell DL, Clarkson JM. Photochem Photobiol. 1984;40:735. doi: 10.1111/j.1751-1097.1984.tb04645.x. [DOI] [PubMed] [Google Scholar]
- 21.Butenandt J, Eker APM, Carell T. Chem--Eur J. 1998;4:642. [Google Scholar]
- 22.Schrader T, et al. Chem Phys Lett. 2004;392:358. [Google Scholar]
- 23.Lian TQ, Locke B, Kholodenko Y, Hochstrasser RM. J Phys Chem. 1994;98:11648. [Google Scholar]
- 24.Deering RA, Setlow RB. Biochim Biophys Acta. 1963;68:526. doi: 10.1016/0006-3002(63)90181-1. [DOI] [PubMed] [Google Scholar]
- 25.Scherer NF, Sipes C, Bernstein RB, Zewail AH. J Chem Phys. 1990;92:5239. [Google Scholar]
- 26.Fuss W, Pushpa KK, Schmid WE, Trushin SA. Photochem Photobiol Sci. 2002;1:60. doi: 10.1039/b107442c. [DOI] [PubMed] [Google Scholar]
- 27.Martinez JM, Elmroth SKC, Kloo L. J Am Chem Soc. 2001;123:12279. doi: 10.1021/ja0108786. [DOI] [PubMed] [Google Scholar]
- 28.Rahn RO. Science. 1966;154:503. doi: 10.1126/science.154.3748.503. [DOI] [PubMed] [Google Scholar]
- 29.Becker MM, Wang Z. J Mol Bio. 1989;210:429. doi: 10.1016/0022-2836(89)90120-4. [DOI] [PubMed] [Google Scholar]
- 30.Rahn RO, Hosszu JL. Photochem Photobiol. 1968;7:637. doi: 10.1111/j.1751-1097.1969.tb07230.x. [DOI] [PubMed] [Google Scholar]
- 31.Lisewski R, Wierzchowski KL. Photochem Photobiol. 1970;11:327. doi: 10.1111/j.1751-1097.1970.tb06006.x. [DOI] [PubMed] [Google Scholar]
- 32.Lamola AA, Eisinger J. Proc Natl Acad Sci U S A. 1968;59:46. doi: 10.1073/pnas.59.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills JB, Vacano E, Hagerman PJ. J Mol Biol. 1999;285:245. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 34.Park H, et al. Proc Natl Acad Sci U S A. 2002;99:15965. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosszu JL, Rahn RO. Biochem Biophys Res Commun. 1967;29:327. doi: 10.1016/0006-291x(67)90457-3. [DOI] [PubMed] [Google Scholar]
- 36.Saenger W. Principles of nucleic acid structure. Springer-Verlag; Berlin: 1984. [Google Scholar]
- 37.Becker MM, Zhou W. J Biol Chem. 1989;264:4163. [PubMed] [Google Scholar]
- 38.Kundu LM, Linne U, Marahiel M, Carell T. Chem--Eur J. 2004;10:5697. doi: 10.1002/chem.200305731. [DOI] [PubMed] [Google Scholar]
- 39.Lyamichev V. Nucleic Acids Res. 1991;19:4491. doi: 10.1093/nar/19.16.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B.K. thanks the National Institutes of Health (grant GM064563) and the Alexander von Humboldt Foundation for support.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.