Abstract
Pax6 genes encode evolutionarily highly conserved transcription factors that are required for eye and brain development. Despite the characterization of mutations in Pax6 homologs in a range of organisms, and despite functional studies, it remains unclear what the relative importance is of the various parts of the Pax6 protein. To address this, we have studied the Drosophila Pax6 homolog eyeless. Specifically, we have generated new eyeless alleles, each with single missense mutations in one of the four domains of the protein. We show that these alleles result in abnormal eye and brain development while maintaining the OK107 eyeless GAL4 activity from which they were derived. We performed in vivo functional rescue experiments by expressing in an eyeless-specific pattern Eyeless proteins in which either the paired domain, the homeodomain, or the C-terminal domain was deleted. Rescue of the eye and brain phenotypes was only observed when full-length Eyeless was expressed, while all deletion constructs failed to rescue. These data, along with the phenotypes observed in the four newly characterized eyeless alleles, demonstrate the requirement for an intact Eyeless protein for normal Drosophila eye and brain development. They also suggest that some endogenous functions may be obscured in ectopic expression experiments.
Keywords: Eyeless, Pax6, Transcription Factor, Paired Domain, Homeodomain, Drosophila, eye, brain, development
Introduction
Pax6/eyeless is involved in the development of the eye and the brain of both invertebrates and vertebrates. In Drosophila melanogaster, the Pax6 homolog eyeless is initially expressed throughout the undifferentiated eye disc, where it persists until the cells differentiate with the progression of the morphogenetic furrow (Halder et al., 1998; Quiring et al., 1994). eyeless is required for normal development of the Drosophila eye, as mutations in the eyeless gene result in irregular facets as well as in a reduction of eye size (Lindsley and Zimm, 1992). Misexpression of eyeless in antennal, leg, or wing imaginal discs is able to initiate the formation of ectopic eyes (Halder et al., 1995), establishing eyeless as a key regulator of D. melanogaster eye development. In the vertebrate eye, Pax6 is expressed in the optic vesicle/optic cup, lens and cornea (Walther and Gruss, 1991), and is required for normal development of each of these eye structures (Ton et al., 1991; Hill et al., 1991; Grindley et al., 1995; Quinn et al., 1996). Pax6 also has a role in retinogenesis, where it is required to maintain the retinogenic potential of retinal progenitor cells (Marquardt et al., 2001). Loss of Pax6 function results in aniridia or Peter's anomaly in humans, and the small eye phenotype in the mouse and rat. Ectopic expression of Pax6 in Xenopus laevis results in the formation of differentiated ectopic eyes (Chow et al., 1999), highlighting the conserved role of Pax6 during eye formation.
In the Drosophila embryonic brain, eyeless is expressed in all three neuromeres of the brain (Kammermeier et al., 2001) while in later stages of brain development, eyeless is primarily expressed in the protocerebrum in e.g. the optic lobes, the mushroom bodies, and the pars intercerebralis. Loss of eyeless activity leads to defects in these structures (Callaerts et al., 2001; Clements et al., 2008; Kurusu et al., 2000; Noveen et al., 2000). Vertebrate Pax6 is involved in the development of the forebrain (Mastick et al., 1997; Mastick and Andrews, 2001; Warren and Price, 1997), midbrain (Matsunaga et al., 2000), and hindbrain (Osumi et al., 1997; Stoykova et al., 2000; Takahashi and Osumi, 2002), where it has roles in the patterning of neuromeres, neuronal differentiation, and axon guidance.
The Pax6/Eyeless protein contains two DNA-binding domains, the 128-amino acid (aa)-long paired domain containing two helix-turn-helix (HTH) motifs (Bopp et al., 1986; Treisman et al., 1991) and a 60-aa-long paired-like homeodomain (Frigerio et al., 1986; Ton et al., 1991; Walther and Gruss, 1991). In addition, Pax6/Eyeless contains a proline, serine, and threonine-rich domain at its carboxy-terminus and a glycine-rich region of variable length that links the amino-terminal paired domain with the more carboxy-terminal homeodomain. A comparison of Pax6 genes from many species revealed a high degree of structural conservation, particularly within the two DNA-binding domains (Callaerts et al., 1997; Callaerts et al., 2006). The high degree of sequence conservation observed in Pax6 is suggestive of a high degree of conservation of macromolecular interactions as well, despite the vast differences in both structure and development of brain and eye in diverse phyla. Indeed, not only are all Pax6 genes studied so far expressed in the brain/nerve ring and the eye/photoreceptor, several of the target genes are conserved as well. These include the lens crystallins, the sine oculis/Six gene family, the eyes absent/Eya gene family and the dachshund/Dach gene family (Callaerts et al., 1997; Wawersik and Maas, 2000; see Gehring and Ikeo,1999; van Heyningen and Williamson, 2002; Callaerts et al., 2006; Kozmik, 2008 for further reviews on Pax6).
The high degree of sequence conservation argues for conserved functions for the DNA binding paired and homeodomains, as well as the transactivating C-terminal domain. Pax6 has effectively three DNA binding HTH domains: the amino-terminal (PAI) and carboxy-terminal (RED) subdomains of the paired domain and the homeodomain. By using multiple combinations of DNA binding domains, Pax proteins potentially regulate a multitude of different functions (Jun and Desplan, 1996). This hypothesis is supported by data in which a missense mutation in the paired domain can either abolish or increase the DNA binding and transcription dependent on homeodomain binding sites (Singh et al., 2000). Furthermore, point mutations in the transactivation domain can reduce or abolish the DNA binding activity of the paired domain or the homeodomain (Singh et al., 2001). The activity of the transactivation domain requires the entire domain (152-aa in human) for maximal activity (Tang et al., 1998). These data suggest a requirement of the paired domain, homeodomain, and transactivation domain for Pax6 function. However, given the high degree of conservation and its known DNA binding roles, surprisingly few homeodomain mutations have been found in comparison to the many paired domain and transactivation domain mutations, suggesting that the homeodomain may not be necessary for Pax6 function. Indeed, in Drosophila, the eyeless phenotype in the eye was rescued in 79% of cases by an Eyeless protein lacking the homeodomain, which was also able to induce the formation of ectopic eyes (Punzo et al., 2001). In contrast, the characterization of missense mutations affecting only the homeodomain of Pax6 (Azuma et al., 2003; Favor et al., 2001; Morrison et al., 2002; Redeker et al., 2008; Thaung et al., 2002) suggest that the homeodomain is required for normal function.
Here we describe a requirement for the paired domain, the linker region, the homeodomain, and the C-terminal domain of the D. melanogaster Pax6 homolog eyeless during eye and brain development. Furthermore, we demonstrate that the C-terminal domain of Eyeless functions as a transactivation domain as in Pax6. We describe four new eyeless alleles created by mutagenesis of the eyeless Gal4 driver OK107. This Gal4 driver expresses the yeast transcriptional activator Gal4 in an eyeless pattern in the Drosophila eye and brain (Adachi et al., 2003), allowing the targeted expression of any gene or construct that is preceded by the Gal4 upstream activating sequence (UAS) in cells that normally express eyeless (Brand and Perrimon, 1993). The four new alleles of eyeless also maintained the Gal4 activity of OK107, and thus allowed us to attempt rescue of the eye and brain phenotypes of homozygous mutant animals by driving the expression of Eyeless deletion constructs in an eyeless expression pattern. We describe, for the first time, complete rescue of the eyeless phenotype with full-length Eyeless protein. On the other hand, deletion of the paired domain, the homeodomain, or the C-terminal domain of the Eyeless protein resulted in the failure of these proteins to rescue either the eye or the brain phenotype of homozygous eyeless mutants. The new alleles are each caused by a single missense mutation in one of the four major domains of the Eyeless protein, resulting in abnormal eye and brain development. These data, along with the rescue experiments, strongly suggest that an Eyeless protein with each domain intact is necessary for normal Eyeless function. In addition, in one of the alleles, eyOK107/6, Eyeless protein failed to localize to the nucleus, revealing a nuclear localization signal (NLS) in the recognition helix of the homeodomain.
Materials and Methods
Mutagenesis screen
In order to generate eyeless alleles on the OK107 chromosome, we mutagenized 1500 homozygous OK107 males with 35 mM EMS as described (Lewis and Bacher, 1968; see Supplemental Figure 1 for mutagenesis scheme). These males were then crossed en masse to ciD females in order to obtain OK107*/ciD males (asterisk indicates mutagenized chromosome). 3314 males of this genotype were crossed individually to eyD1Da/cilacZ females. The progeny of this cross consisted of four possible genotypes (OK107*/eyD1Da, OK107*/cilacZ, eyD1Da/ciD, and cilacZ/ciD, which is lethal). Flies without the ciD phenotype were then screened for the eyeless phenotype (eyes reduced or lost), identifying multiple independent putative alleles. Of these, non-ciD flies without an eyeless phenotype (presumed genotype: OK107*/cilacZ) were crossed to eyD1Da/ciD flies in order to (1) verify the phenotype and (2) establish the eyOK107/X/ciD stocks (the name eyOK107/X refers to the entire series of alleles generated in this screen).
Analysis of phenotypes
The eye was examined by comparing the size of each mutant eye to the eye of a wildtype (Oregon-R) animal. Mutant eyes were then assigned to one of the following categories: 75 – 100% (eye relatively normal in size), 50 – 75%, 25 – 50%, 1 – 25% (eye present but extremely small, usually less than 50 ommatidia), or no eye (eye completely absent). Brain phenotypes were gauged on the basis of the morphology of the mushroom bodies and the central complex, both of which are dependent on eyeless for normal development. This was determined by whole-mount staining of adult brains with the monoclonal antibody 1D4. Brain defects were assigned to one of the following categories: no defects (structure phenotypically wildtype), mild (a few, subtle defects observed), severe (significant disorganization of neuropil structure), or very severe (structure completely or nearly completely absent).
Fly stocks
Flies were maintained at 25°C on standard agar-cornmeal-molasses medium. Fly stocks or alleles used were OK107 (Connolly et al., 1996), eyD1Da and eyJD (Callaerts et al., 2001), eyEH (Benassayag et al., 2003), ey2 (available from the Bloomington Stock Center), Pabp2CC00380 (Buszczak et al., 2007) and cilacZ (from Thomas Kornberg). Fly stocks used for rescue of the eyeless phenotype were UAS-ey (Halder et al., 1995) and UAS-eyΔPD, UAS-eyΔHD, and UAS-eyΔCTD (Punzo et al., 2001).
Immunohistochemistry
Whole-mount immunostaining with antibodies against Eyeless, Fasciclin II or GFP was performed on D. melanogaster adult brains or larval brain-imaginal disc complexes. These were dissected in PBS, fixed for 15 minutes in 4% formaldehyde in PBS, followed by three 10 minute washes in PBS and preincubation in PAXD solution (PBS containing 5% BSA, 0.3% Triton X-100, and 0.3% sodium deoxycholate) for at least 10 minutes. Incubations in primary and secondary antibodies were performed overnight at 4°C and followed by extensive washing in PAXD solution at room temperature. The primary antibodies were a rat polyclonal antibody against the Eyeless linker region (1:500 dilution), a rabbit polyclonal antibody against GFP (1:500 dilution; Invitrogen, Eugene, OR) and the monoclonal antibodies 1D4 against FasII (1:20 dilution; Developmental Studies Hybridoma Bank) and A11H6 against Eyeless (1:10 dilution; Clements et al., 2007). The secondary antibodies were FITC- or Cy3-conjugated (1:200 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA). Brains or dissected imaginal discs were mounted in Vectashield mounting medium for fluorescence (Vector Laboratories, Burlingame, CA) and stored at 4°C. Observations were made on an Olympus AX70 microscope equipped for epifluorescence and images were collected using a Magnafire digital camera. Some images were generated using a Leica TCS SP2 laser confocal microscope.
Adult head preparations
Adult fly heads were dissected and placed in 70% ethanol and stored at 4°C for several hours to several days, then mounted in Hoyer's medium and incubated at 50°C to make the cuticle more transparent (Sullivan et al., 2000).
Molecular characterization of eyeless alleles
The characterization of the eyOK107/X alleles was performed as described by Callaerts et al. (2001) with the following modifications. Exons of eyeless were PCR-amplified with either Pwo polymerase or Tgo polymerase (Roche Molecular Biochemicals, Indianapolis, IN) and subcloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) for sequencing. Sequencing was done as a service by Seqwright (Houston, TX).
Analysis of transactivation by the Eyeless C-terminal domain in cell culture
Plasmids
The possible role of the C-terminal domain (CTD) of Eyeless in transactivation was studied by transient cotransfection experiments in Drosophila Schneider-2 culture cells. Hybrid cDNAs encoding fusion proteins of the GAL4 DNA-binding domain and the wildtype or eyD1Da mutant version of the CTD of Eyeless were cloned under a metallothionein promoter in the pRmHa3 plasmid (Wisniewski et al., 1996). A hybrid protein of the GAL4 DNA-binding domain with the viral VP16 transactivating domain was used as a positive control (Wisniewski et al., 1996). The reporter plasmids consist of the Upstream Activating Sequences to which GAL4 can bind, a minimal promoter and the firefly luciferase reporter gene taken from the pGL2 luciferase vector (Promega™). The efficiency of the cotransfection experiments was normalized by cotransfecting the Renilla luciferase reporter gene under the control of a promotor that is constitutively active in S2 cells (actin promotor).
Transfection
The procedure consisted of the following steps: (1) Schneider S2 cells were cultured in Schneider's medium containing 10% Fetal Bovine Serum and grown to late log phase. (2) Confluent cells were harvested and plated in 60 mm petridishes at 5×106 cells/ml. (3) 10 μg of each plasmid was mixed with FuGENE transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN) and incubated with the cells for 30 minutes. (4) The supernatant was removed, new medium added, and the cells were allowed to recover for 48 hours. (5) Cells were harvested with cell scrapers and the luciferase assays were performed according to the manufacturer's protocol (Promega, Madison, WI). Each transfection assay was done five times and statistical significance was determined using Duncan's test.
Reporter gene activity measurements
Firefly and Renilla luciferase activity was determined with the Dual-Luciferase Reporter Assay System (Promega) and measured with a Packard Top Count NXT luminometer.
Results
Isolation of new eyeless alleles by EMS mutagenesis of the OK107 Gal4 enhancer trap
Eyeless encodes a transcription factor with two DNA binding domains, the paired domain and the homeodomain, and a PST-rich carboxy-terminal domain (Quiring et al., 1994). We sought to determine which domains were required for normal development of the eye and the brain, and which, if any, were dispensable. Ideally, these experiments would be done using the endogenous regulatory sequences of eyeless. The Gal4 driver line OK107 (Connolly et al., 1996), an enhancer trap line with a P-element insertion 6.5 kb upstream of the first exon of eyeless, drives Gal4 expression in an eyeless-specific pattern in the eye and brain (Adachi et al., 2003), thus making it an ideal driver for our studies. However, while this Gal4 line does exhibit developmental defects in the mushroom body lobes in approximately 50% of homozygous animals (results not shown) the eye and remaining brain structures are phenotypically wildtype, as the defects are likely caused by a disruption of a mushroom body enhancer near the P-element insertion site (Adachi et al. 2003). Since OK107 Gal4 does not have any eye defects, nor does it result in brain defects when in trans to other eyeless alleles, we mutagenized OK107 in order to generate mutations affecting the Eyeless protein itself while maintaining the Gal4 activity of the driver. We isolated four new eyeless alleles (eyOK107/4, eyOK107/6, eyOK107/10, and eyOK107/16: collectively referred to here as the eyOK107/X alleles) by their failure to complement the known allele eyD1Da. Once identified, a complementation analysis was carried out with the alleles eyD1Da, eyJD, and ey2 to assess the severity of the new alleles (Table 1). The eyOK107/X alleles failed to complement, and significant eye and brain defects were observed in almost all transheterozygous allelic combinations. Phenotypes were most severe in eyOK107/4 and eyOK107/16, while the phenotypes observed in eyOK107/10 flies were the mildest of the eyOK107/X alleles. Eye and brain development was not significantly affected in either eyOK107/4/ey2, eyOK107/6/ey2, and eyOK107/10/ey2 animals, similar to previous observations for eyJD/ey2 and eyD1Da/ey2 (Callaerts et al., 2001). However, the eyOK107/16/ey2 combination exhibited a moderate eye phenotype. All allelic combinations not involving ey2 resulted in significant eye and brain defects of differing magnitude. All eyOK107/X alleles displayed severe mushroom body and central complex phenotypes in all combinations (except with ey2). The observed central complex defect, but not the mushroom body defect, was markedly less severe in combinations involving the eyOK107/10 allele.
Table 1.
Genetic complementation analysis. “Total eyes” shows the total number of eyes that were scored to obtain the percentages displayed. The numbers in the different categories refer to the number of eyes with the indicated eye size, ranging from 0% wild type eye size (i.e. no eye) to 1-25%, 25-50%, 50-75%, and 75-100%. Brain defects were ranked according to degree of disruption and range from no defects (−) over mild (+), severe (++) to very severe (+++). The severity was determined by analyzing the frequency of the lobes missing or lobes reduced phenotype for mushoom bodies (MB), and the level of disorganization of the central complex (CC). The genotypes are listed by subscripts. For example: [OK107/4]×[JD] refers to the transheterozygous progeny of the genotype eyOK107/4/eyJD.
| Genotype | Total eyes | % Wild type eye size | MB Defects | CC Defects | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1-25 | 25-50 | 50-75 | 75-100 | ||||
| [OK107/4] × [JD] | 10 | 3 | 1 | 3 | 2 | 1 | +++ | +++ |
| [OK107/4] × [D1Da] | 46 | 7 | 4 | 11 | 14 | 10 | +++ | ++ |
| [OK107/4] × [OK107/10] | 76 | 0 | 3 | 0 | 8 | 65 | +++ | + |
| [OK107/4] × [2] | 100 | 0 | 0 | 0 | 0 | 100 | − | − |
| [OK107/10] × [JD] | 54 | 2 | 0 | 2 | 1 | 49 | ++ | − |
| [OK107/10] × [D1Da] | 68 | 0 | 0 | 10 | 49 | 9 | +++ | + |
| [OK107/10] × [2] | 132 | 0 | 0 | 0 | 1 | 131 | − | − |
| [OK107/6] × [JD] | 48 | 6 | 2 | 9 | 18 | 13 | +++ | ++ |
| [OK107/6] × [D1Da] | 70 | 2 | 0 | 7 | 54 | 7 | +++ | + |
| [OK107/6] × [OK107/4] | 52 | 0 | 4 | 15 | 30 | 3 | +++ | ++ |
| [OK107/6] × [OK107/10] | 104 | 0 | 0 | 0 | 9 | 95 | ++ | + |
| [OK107/6] × [2] | 110 | 0 | 0 | 0 | 0 | 110 | − | − |
| [OK107/16] × [JD] | 46 | 0 | 0 | 14 | 25 | 7 | ++ | +++ |
| [OK107/16] × [D1Da] | 28 | 2 | 7 | 14 | 5 | 0 | +++ | ++ |
| [OK107/16] × [OK107/4] | 22 | 2 | 2 | 13 | 5 | 0 | ++ | +++ |
| [OK107/16] × [OK107/6] | 90 | 1 | 6 | 23 | 54 | 6 | ++ | ++ |
| [OK107/16] × [OK107/10] | 34 | 0 | 3 | 8 | 18 | 5 | ++ | + |
| [OK107/16] × [2] | 114 | 0 | 0 | 7 | 42 | 65 | − | − |
The new eyOK107/X alleles encode missense mutations in the Eyeless protein
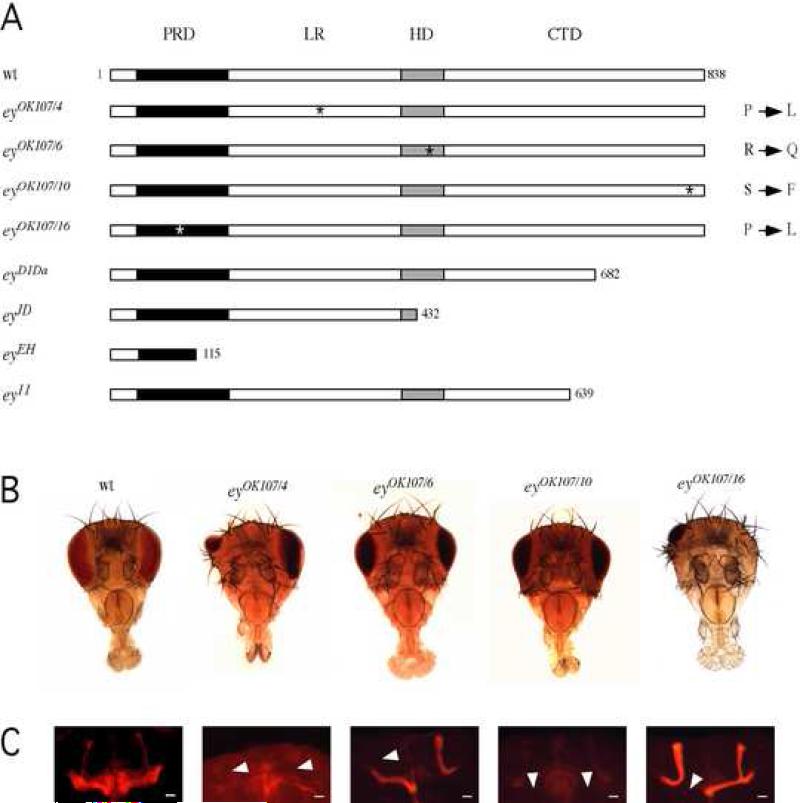
eyeless encodes a transcription factor comprised of an amino-terminal paired domain, a glycine-rich linker region, a paired-like homeodomain, and a carboxy-terminal domain. To date, all characterized eyeless alleles encode either truncated proteins (eyD1Da, eyJD, ey11, eyEH; Benassayag et al., 2003; Callaerts et al., 2001), are protein null (eyJ5.71 and presumably eyC7.20; Kammermeier et al., 2001; Punzo et al., 2001), or affect eyeless expression by disruption of intronic enhancer elements (ey2 and eyR; Quiring et al., 1994). In contrast, the four new eyeless alleles represent single amino acid substitutions in each of the four major domains of Eyeless protein (Fig. 1A). The allele eyOK107/4 is caused by a transition in which cytosine 993 in exon 5 is replaced with thymine, resulting in the P302L substitution in the linker region (nucleotide and amino acid numbering system as used by Quiring et al., 1994). The allele eyOK107/6 encodes a missense mutation in position 12 of the recognition helix of the homeodomain (R463Q) as a result of a guanine to adenine transition in nucleotide position 1476 of exon 7. The allele eyOK107/10 affects the carboxy-terminal domain of the protein causing the mutation S831F, as encoded by a transition mutation in nucleotide 2580 in exon 9 in which cytosine is replaced with thymine. The fourth allele, eyOK107/16, is also a cytosine to thymine transition, affecting nucleotide 390 of exon 4 and results in the missense mutation P101L in the carboxy-terminal tail of the paired domain. None of the eyOK107/X mutations affect protein synthesis or stability of the Eyeless protein (Fig. S2).
Figure 1.
(A) Schematic representation of the predicted protein products encoded by wild type eyeless, the four eyOK107/X alleles (stars indicate position of the amino acid substitution; this paper), eyD1Da and eyJD (Callaerts et al., 2001), and eyEH and ey11 (Benassayag et al., 2003). The Eyeless protein consists of the paired domain (PRD, black shading), the linker region (LR), the homeodomain (HD, grey shading), and the C-terminal domain (CTD). (B) Head and eye defects in homozygous eyOK107/X mutants. (C) Anti-Fasciclin II antibody staining reveals α, β and γ lobes of the mushroom bodies. One of the frequently observed phenotypes is the loss of lobes (marked with arrowheads in the different panels) as was previously documented in Callaerts et al. (2001). Size bars are 25 μm.
All four eyOK107/X alleles cause abnormal development of the eye and brain (Figs. 1B and 1C). The eyes in the mutant are significantly reduced, although there is significant variability between mutant animals as well as between the two eyes of the same animal, as is typical for eyeless alleles. In the brain, organization of major neuropils such as the mushroom bodies, the central complex, and the pars intercerebralis is severely disrupted. Interestingly, one allele, eyOK107/6, is homozygous viable despite its apparent brain defects, a phenomenon atypical of eyeless alleles.
In summary, these data confirm the previously established importance of the paired domain and the C-terminal domain, while revealing the functional importance of both the linker region and the homeodomain in eye and brain development.
All domains of the Eyeless protein are essential for normal development
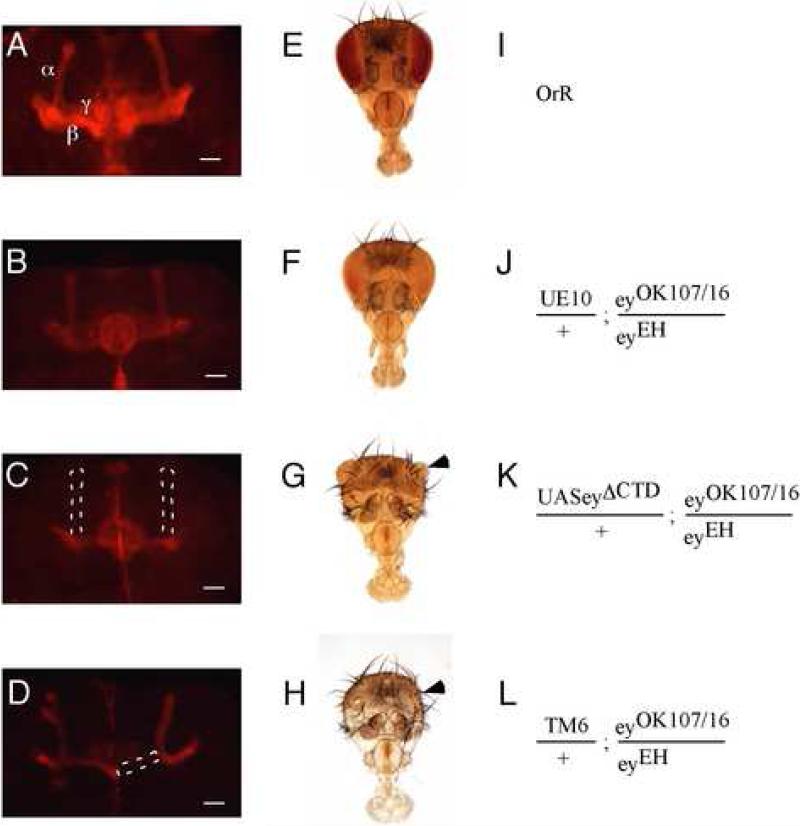
The newly isolated alleles allowed us to study the requirement for the various domains of Eyeless by driving the expression of eyeless cDNA deletion constructs in a homozygous eyeless mutant background using the Gal4 system (Brand and Perrimon, 1993). The ability of the deletion constructs to rescue the eye and/or brain phenotypes typical of eyeless mutants would therefore be expected to reveal the relative contribution of these domains for development of these structures. Specifically, we expressed deletion constructs encoding proteins in which either the paired domain (eyΔPD), the homeodomain (eyΔHD), or the C-terminal domain (eyΔCTD) was deleted from the protein (Punzo et al., 2001) in a transheterozygous eyeless mutant background using the allele eyOK107/16. The constructs ey (full-length), eyΔPD and eyΔHD were previously shown to give comparable expression levels (Punzo et al., 2001). We confirmed this by Western blotting and quantitation and extended the observations to ey CTD, with the deletion constructs maximally expressed at about twofold the level of the full-length (results not shown). While we found that truncated Eyeless proteins did provide some degree of rescue when compared to homozygous mutants, only expression of the intact Eyeless protein resulted in complete rescue of the eyeless phenotype (Table 2 and Fig. 2). In the eye, deletion of the paired domain or the carboxy-terminal domain only provided a negligible amount of rescue in which no more than 10% of eyes are larger than 50% of wildtype size. In contrast, deletion of the homeodomain led to a greater degree of rescue, in which the majority of eyes were larger than 50% of wildtype size, although this value was still significantly lower than that observed for the complete Eyeless protein. In the brain, rescue of both the mushroom body and central complex phenotypes was attained only with the full-length Eyeless UAS construct, with the exception of the homeodomain deletion construct, which rescued the brain phenotype in an eyOK107/16/eyJD mutant background, but not in an eyOK107/16/eyEH background. All other Eyeless deletion constructs did not provide any significant degree of rescue.
Table 2.
Phenotypic rescue experiments using the UAS-GAL4 system to express variants of the Eyeless protein. The ranking of phenotypes was done as in Table 1. “Total eyes” shows the total number of eyes that were scored to obtain the percentages displayed. The numbers in the different categories refer to the number of eyes with the indicated eye size, ranging from 0% wild type eye size (i.e. no eye) to 1-25%, 25-50%, 50-75%, and 75-100%. Brain defects were ranked according to degree of disruption and range from no defects (−) over mild (+), severe (++) to very severe (+++). The severity was determined by analyzing the frequency of the lobes missing or lobes reduced phenotype for mushoom bodies (MB), and the level of disorganization of the central complex (CC). As in Table 1, the genotypes of the eyeless alleles are listed by subscripts. Other abbreviations: UE10: UAS-Eyeless Full Length; UASΔPD: UAS-Eyeless minus paired domain; UASΔHD UAS-Eyeless minus homeodomain; UASΔCTD: UAS-Eyeless minus C-terminal domain.
| Genotype | Total eyes | % Wild type eye size | MB Defects | CC Defects | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1-25 | 25-50 | 50-75 | 75-100 | ||||
| [EH]/[OK107/16] | 92 | 29 | 15 | 20 | 16 | 10 | +++ | ++ |
| UE10;[EH]/[OK107/16] | 62 | 0 | 0 | 0 | 0 | 62 | +/− | +/− |
| UAS-ΔPD;[EH]/[OK107/16] | 62 | 8 | 18 | 25 | 10 | 1 | +++ | +++ |
| UAS-ΔHD; [EH]/[OK107/16] | 46 | 0 | 4 | 9 | 14 | 19 | ++ | ++ |
| UAS-ΔCTD; [EH]/[OK107/16] | 68 | 4 | 19 | 22 | 22 | 1 | +++ | ++ |
| [JD]/[OK107/16] | 90 | 2 | 10 | 14 | 37 | 27 | +++ | ++ |
| UE10;[JD]/[OK107/16] | 96 | 0 | 0 | 0 | 3 | 97 | +/− | − |
| UAS-ΔPD;[JD]/[OK107/16] | 50 | 0 | 0 | 1 | 3 | 46 | +++ | ++ |
| UAS-ΔHD;[JD]/[OK107/16] | 64 | 0 | 3 | 5 | 15 | 41 | +/− | − |
| UAS-ΔCTD;[JD]/[OK107/16] | 44 | 1 | 6 | 10 | 24 | 3 | +++ | ++ |
Figure 2.
The eye, head, and brain phenotypes observed in homozygous eyeless flies can be rescued only by full-length Eyeless (UE10 = UAS full-length Eyeless) when driven by the Gal4 activity present in the eyOK107/16 mutant. Eyeless UAS constructs in which one domain of the Eyeless protein has been removed do not result in a phenotypic rescue in the same mutant background (see Table 2 for complete results). (A-D) Anti-Fasciclin II antibody staining reveals α, β and γ lobes of the mushroom bodies. Note the lack of lobes in C and D (marked as an outline where the lobe is expected to be) and the phenotypic rescue with presence of normal mushroom lobes in B. Size bars are 25 μm. (E-H) Head and eye phenotypes in wild type control (E) and in the eyOK107/16 mutant in the presence of different UAS-Eyeless rescue constructs. Note that expression of full-length Eyeless completely rescues the eye and head phenotypes (F) whereas an Eyeless transgene encoding an Eyeless protein without the C-terminal domain (G) is not different from the siblings carrying the balancer chromosome (H) and yields flies with head and eye (arrowhead) defects and no phenotypic rescue. (I-L) Genotypes of the animals displayed in the corresponding panels on the same row (e.g. I represents the genotype of the animals shown in A and E).
The eyOK107/6 allele reveals a nuclear localization signal
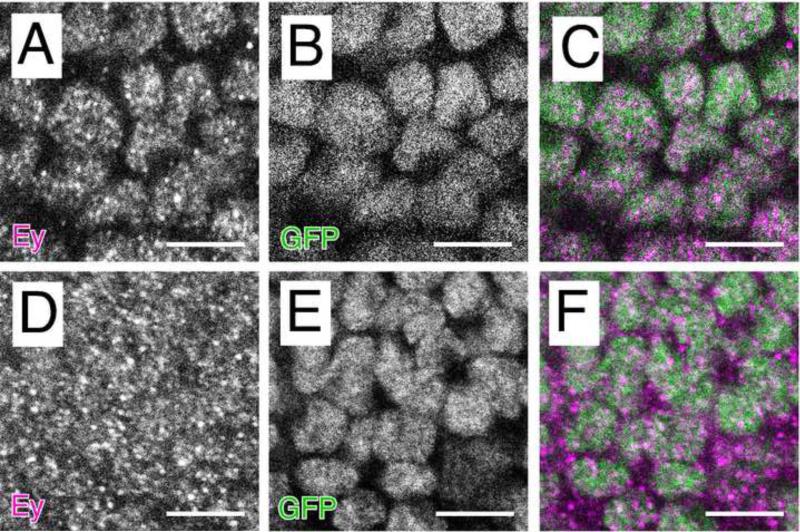
Consistent with its role as a transcription factor, three potential nuclear localization signals have been identified in Pax6 (Carrière et al., 1995; Glaser et al., 1995), although the requirement of any one particular NLS is unclear. The first of these is located within the paired domain, as isoforms lacking this region of the protein are localized predominantly in the cytoplasm (Carrière et al., 1995). The second putative NLS is found at the amino-terminus of the homeodomain, and is similar to the SV40 T-antigen NLS (Glaser et al., 1995). Although a missense mutation affecting this NLS has been identified in a patient with aniridia (Hanson et al., 1993), there is some debate whether this mutation affects nuclear localization (Carrière et al., 1995; Glaser et al., 1995). Lastly, there is a third putative NLS at the carboxy-terminus of the homeodomain (Glaser et al., 1995). All three putative NLSs are also conserved in Eyeless. The identification of the missense mutation (Arg to Gln) in the eyOK107/6 allele demonstrates the functionality of the carboxy-terminal NLS. In this mutant, Eyeless protein is abundant in the cytoplasm as well as the nucleus, contrary to the exclusive nuclear localization of wildtype Eyeless protein (Fig. 3 and Fig. S2). This demonstrates a functional requirement of this NLS for proper localization of Eyeless protein to the nucleus. In addition, the observation that some protein is localized to the nucleus in eyOK107/6 mutants suggests the existence of at least one other functional NLS within the Eyeless protein.
Figure 3.
Abnormal subcellular localization of protein encoded by the allele eyOK107/6. (A-C) Eyeless localizes to the nucleus in wild type eye discs. Single channel image of wildtype eye discs in a heterozygous Pabp2CC00380 background, which ubiquitously expresses a nuclear-localized Pabp2-GFP fusion protein, stained with Eyeless (A) or GFP (B). (C) Overlay of Eyeless (magenta) and GFP (green) reveals nuclear colocalization of Pabp2-GFP and Eyeless. (D-F) Eyeless is primarily cytoplasmic in eyOK107/6 mutant eye discs. Single channel image stained with Eyeless (D) or GFP (E). (F) Overlay of Eyeless (magenta) and GFP (green) reveals limited overlap between Pabp2-GFP and Eyeless and thus cytoplasmic localization of mutant Eyeless protein. Size bars are 5 μm.
The carboxy-terminus of Eyeless is required for transactivation
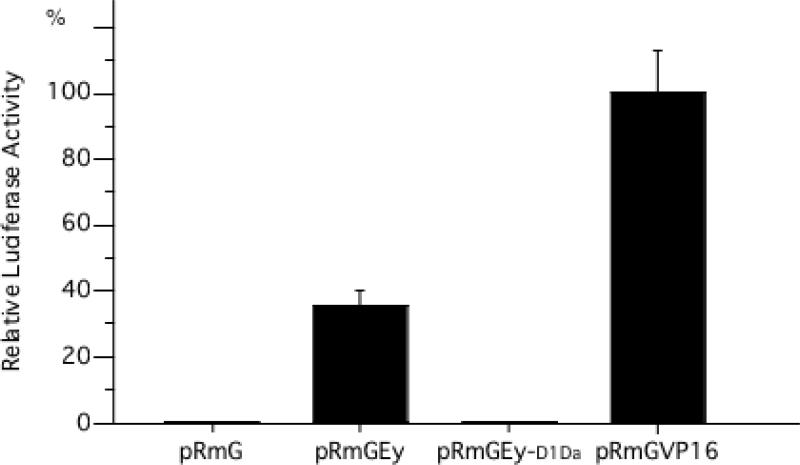
Given that the PST-rich C-terminal domains of mammalian and sea urchin Pax6 function as transactivation domains (Czerny and Busslinger, 1995; Glaser et al., 1994), and that truncations in the Eyeless C-terminal domain are associated with mutant phenotypes (Benassayag et al., 2003; Callaerts et al., 2001), we sought to determine if the PST-rich C-terminal domain of Eyeless also demonstrated transactivation activity. Transient transfection experiments with plasmids encoding fusion proteins of the GAL4 DNA-binding domain with variants of the Eyeless C-terminal domain, and luciferase reporter genes, revealed that the Eyeless C-terminal domain potently induces transcription of reporter genes to approximately 35% of GAL4-VP16-induced transcription levels (Fig. 4). Relative transcription was reduced to negative control levels (GAL4-DNA binding domain-only), when the mutation associated with the eyD1Da allele was introduced in the C-terminal domain. In the eyD1Da allele, a premature stop codon results in the loss of the most C-terminal 30% of the PST-rich domain (Callaerts et al., 2001), suggesting that the PST-rich C-terminus and in particular its most C-terminal portion is a key determinant for transactivation by Eyeless.
Figure 4.
The C-terminal domain of Eyeless functions as a transcriptional activator. Luciferase activity relative to pRmGVP16 for four protein fusions with the GAL4 DNA-binding domain. pRmG: empty vector negative control encoding the GAL4 DNA-binding domain only. pRmGEy: Eyeless C-terminal domain fusion. pRmGEy-D1Da: Eyeless C-terminal domain fusion with the eyD1Da mutation. pRmGVP16: Fusion with the strong transactivation domain of Herpes simplex VP16 as positive control. pRmGEy is significantly different from pRmGEy-D1Da and pRmG (p=0.0001, Duncan test).
Discussion
Since the original identification of Pax6 and its association with defective eye development in mouse, human and Drosophila (Walther & Gruss, 1991; Hill et al., 1991; Ton et al., 1991; Quiring et al., 1994), complementary approaches have been used to gain insight into the mechanistic basis of Pax6 function in eye and brain development. The identification of mutations in human PAX6 (current number of 494 entries in LOVD (https://lsdb.hgu.mrc.ac.uk/home.php), of which 299 are unique) underscores the importance of all parts of the PAX6 protein. Contrary to this, it has been shown that truncated versions of the Drosophila Pax6 homolog Eyeless are able to induce ectopic eyes and also display a considerable capacity to rescue eyeless mutant phenotypes (Punzo et al., 2001). We addressed this apparent discrepancy by a mutational analysis of eyeless and in vivo rescue experiments using variants of eyeless encoding proteins that lack distinct protein domains. Our results reveal that only an intact protein provides all properties for full function when rescue experiments are conducted using endogenous regulatory sequences of the eyeless gene. Furthermore, we provide novel insight into the in vivo role of each domain. Lastly, our data suggest that endogenous functions of genes may be obscured in ectopic or overexpression experiments.
In vivo structure-function analysis reveals new features of the Eyeless protein
Paired Domain
The paired domain is a 128-aa DNA-binding motif characteristic of the Pax family of proteins. The paired domain consists of two HTH motifs, the N-terminal PAI domain and the C-terminal RED domain (Jun and Desplan, 1996), which are joined by a 16-aa extended linker. Unlike the D. melanogaster Paired paired domain (Xu et al., 1995), both the PAI and the RED domains of the Pax6 paired domain make major groove contacts with DNA, although the contacts via the N-terminal subdomain are more numerous and of greater affinity (Xu et al., 1999). However, the Pax6a isoform, which has a fourteen amino acid insertion into the N-terminal domain, binds solely through the C-terminal RED domain (Epstein et al., 1994; Kozmik et al., 1997; Walther and Gruss, 1991). Furthermore, cooperative interactions of the paired domain with other DNA-binding domains such as the homeodomain and the Ets domain have been demonstrated (Fitzsimmons et al., 1996; Jun and Desplan, 1996; Sheng et al., 1997; Underhill et al., 1995; Underhill and Gros, 1997). Lastly, single missense mutations affecting the paired domain can lead to the loss or strengthening of homeodomain binding or it can influence the transactivation that normally follows DNA-binding (Fortin et al., 1997; Singh et al., 2000). Together, these data show that DNA-binding by the paired domain is context-dependent, and that it influences the activity of other parts of the protein, i.e. DNA binding by the homeodomain or transactivation.
We describe here the isolation of an allele of eyeless (eyOK107/16) that results in the substitution of proline 65 (Pro65) with a leucine in the paired domain linker region. The crystal structure of the Pax6 paired domain complexed to DNA reveals that the extended linker that connects the PAI and RED subdomains makes numerous contacts with bases and the sugar phosphate backbone. Pro65 makes van der Waals contacts with the phosphate backbone of the minor groove of the DNA and also interacts with Arg16 in the β2 turn unit of the N-terminal domain (Xu et al., 1999). The proline that is substituted in the eyOK107/16 allele is conserved in all Pax proteins (Walther et al., 1991), suggesting that it is crucial for normal paired domain linker function. Indeed, a patient with bilateral morning glory disc anomaly was identified in which a missense mutation in the homologous amino acid (P68S) was found (Azuma et al., 2003). It is likely that the P65L substitution of eyOK107/16 results in a major conformational alteration as a result of the bulkier Leu sidechain, and consequently affects the ability of the paired domain linker to interact with the minor groove of its target by disrupting the van der Waals contacts with the phosphate backbone or by destabilizing β-turn docking with the DNA. In accordance with this prediction, human Pax6 protein in which the P68S mutation was introduced showed a severe impairment in its ability to activate reporter gene expression in a chloramphenicol acetyltransferase assay (Azuma et al., 2003). A direct role for the paired domain linker region in target site selection and DNA binding has also been shown in Pax3 (Vogan et al., 1996; Vogan and Gros, 1997).
Linker
The linker is a glycine-rich region of variable length that joins the paired domain and the homeodomain. Invertebrate Pax6 linkers are usually longer than those found in vertebrate species, such as the D. melanogaster Eyeless linker that is three times as long as the 78-aa mouse and human Pax6 linker, for example (Callaerts et al., 1997; Callaerts et al., 2006). The linker of Pax6 does not contain the octapeptide that is found in the majority of Pax family members. However, there is a highly conserved undecapeptide which is usually found near the C-terminus of the paired domain, as well as a string of twelve conserved amino acids that precedes the N-terminus of the homeodomain (Callaerts et al., 1997; Callaerts et al., 2006). Although the conservation of these regions implies a structural or functional role, the exact function of the linker remains elusive. We have identified a missense mutation in the linker region of the eyeless gene in the eyOK107/4 allele, which results in severe eye defects, as well as extreme mushroom body and central complex defects. The only other missense mutation in the Pax6 linker was described by Azuma et al. (1998), and resulted in bilateral total aniridia in a 35-year-old woman and her 6-year-old son, demonstrating a functional requirement for the linker in humans. Combined, these data demonstrate that the Pax6 linker region is functionally important. Thus far, the only documented role for the linker region concerns Pax3, in which the linker is important for the interaction of the paired domain and homeodomain (Fortin et al., 1998).
Homeodomain
Pax6/Eyeless contains a 60-aa-long paired-like homeodomain. The homeodomain is a DNA-binding domain, consisting of an N-terminal arm and three -helices, which has a critical role in the development of higher eukaryotes (Scott et al., 1989). Helix 2 and helix 3 of the homeodomain form a helix-turn-helix motif, in which the third helix (the recognition helix) fits into the major groove of the DNA, while Helix 2 holds it in position by laying across it (Laughon and Scott, 1984). Two amino acids of the recognition helix contact DNA directly, whereas the remaining amino acids of Helix 3 interact with the DNA indirectly via contact with water molecules that in turn interact with the nucleotides of the major groove (Billeter et al., 1993; Wilson et al., 1995). The amino acid at position 50 of the homeodomain is crucial in determining DNA binding specificity, since substitution of the amino acid at this position results in an alteration of binding site recognition (Treisman et al., 1989). Arg-2 and Arg-5 of the N-terminal arm of the homeodomain interact directly with the core of the recognition motif, the tetranucleotide TAAT, suggesting that this arm has an important role in sequence recognition as well (Wilson et al., 1995). It has been shown for the protein encoded by the D. melanogaster gene paired (Wilson et al., 1993) and for mouse and sea urchin Pax6 (Czerny and Busslinger, 1995) that the paired class homeodomains bind DNA as cooperative dimers on so-called P3 sites. The latter are palindromic sequences consisting of two inverted TAAT half sites separated by three nucleotides (Wilson et al., 1993). This binding occurs via the N-terminal arm of each homeodomain interacting with Helix 2 of the other (Wilson et al., 1995).
The allele eyOK107/6, which results in the substitution of an arginine for a glutamine at position 12 of the recognition helix, for the first time reveals several important features of the Eyeless homeodomain. Arginine 12 forms salt bridges with the DNA backbone of the major groove (Billeter et al., 1993), and is conserved in paired class homeodomains, and in most other classes of homeodomains (Treisman et al., 1992; Wilson et al., 1995). It is likely that loss of this salt bridge destabilizes the interaction between the recognition helix and the major groove, resulting in reduced affinity for the recognition site.
Interestingly, the arginine – glutamine substitution found in eyOK107/6 also disrupts the function of a required nuclear localization signal, resulting in Eyeless protein that is found in both the nucleus and cytoplasm. Unlike vertebrates to Pax6, Drosophila heterozygous for an eyeless mutation do not generally exhibit any obvious phenotypes. Therefore it seems probable that the developmental defects observed in eyOK107/6 mutants are due to an impaired ability of the homeodomain to bind DNA, and not due to an inadequate level of nuclear Eyeless protein.
C-Terminal Transactivation Domain
We have demonstrated previously the importance of the C-terminus of Eyeless, as the alleles eyD1Da and ey11, which encode proteins with truncated C-termini, display severe developmental defects (Benassayag et al., 2003; Callaerts et al., 2001). The location of a transactivation domain in the PST-rich C-terminus of Pax6 in human (Glaser et al., 1994), mouse and sea urchin (Czerny and Busslinger, 1995) was previously demonstrated. By means of transient transfection experiments we show that the Eyeless C-terminal domain is a transactivation domain. The loss of transactivation following the introduction of the eyD1Da mutation in the C-terminal domain identifies the C-terminal 156 amino acids as critical for this function.
Finally, we have characterized a new allele of eyeless, eyOK107/10, which results in the substitution of a serine for a phenylalanine in the C-terminus of the Eyeless protein located eight amino acids from the carboxyterminal end of the protein. Like the other eyOK107/X alleles, eyOK107/10 exhibits both eye and brain developmental defects, although the phenotypes are milder than those caused by the other eyOK107/X alleles. The eyOK107/10 allele appears to be very sensitive to genetic background, as an observable eye phenotype in homozygous mutant animals is lost after only a few generations. This suggests that the functional effects of this mutation are easily overcome by other gene products in the D. melanogaster proteome. Since this mutation is located in the C-terminal domain of Eyeless, it is conceivable that the substitution has an effect on transactivation. However, by analogy to a missense mutation described for the human PAX6 C-terminal domain, it is also possible that the substitution in Eyeless negatively modulates DNA-binding by the homeodomain (Singh et al., 2001)
Supplementary Material
Supplementary Material
Supplemental Figure 1. Mutagenesis scheme for the eyOK107/X mutants
Males of the Gal4 line OK107 were mutagenized en masse by EMS feeding, and crossed to eyD1Da/ciD females, generating males of the genotype OK107*/ciD (asterisk indicates mutagenized OK107 chromosome). These males were then crossed individually to eyD1Da/cilacZ females, and their non-ciD progeny were screened for an eyeless phenotype. Of those found, their non-ciD siblings without eyeless phenotypes (OK107*/ cilacZ) were crossed to eyD1Da/ciD females in order to establish stocks and confirm the observed eyeless phenotype.
Supplemental Figure 2. Eyeless protein expression is not affected in the eyOK107/X mutants
Neither Eyeless protein synthesis nor stability is affected by the missense mutations in the eyOK107/X mutants. Here Eyeless expression in the Kenyon cell cluster of the mushroom body is shown for each of the four eyOK107/X alleles (homozygous for mutant allele) as well as a wildtype control (Oregon-R).
Acknowledgements
We thank Claudio Punzo and Walter Gehring for providing transgenic flies, Sean Sweeney for drawing our attention to the OK107 GAL4 enhancer trap line and for providing the stock, David Sherry for help with confocal microscopy, and the members of the Callaerts lab for technical assistance and scientific discussion. We also thank Grady Saunders for the use of the luminometer and Sanjaya Singh for initial help with the transactivation assays. This work was supported by grants from the March of Dimes Foundation (Basil O'Connor Award), NIH RO1-MH59763-02, the Research Foundation – Flanders (FWO contract number G.0285.05) and VIB. HGdC acknowledges the support of the American Heart Association (ref. no. 0051591Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, Kang YY, Eggert T, Walldorf U, Furukubo-Tokunaga K, et al. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech. Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hotta Y, Tanaka H, Yamada M. Missense mutations in the PAX6 gene in Aniridia. Invest. Ophthalmol. Vis. Sci. 1998;39:2524–2528. [PubMed] [Google Scholar]
- Azuma N, Yamaguchi Y, Handa H, Tadokoro K, Asaka A, Kawase E, Yamada M. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am. J. Hum. Genet. 2003;72:1565–1570. doi: 10.1086/375555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassayag C, Plaza S, Callaerts P, Clements J, Romeo Y, Gehring WJ, Cribbs DL. Evidence for a direct functional antagonism of the selector genes proboscipedia and eyeless in Drosophila head development. Development. 2003;130:575–586. doi: 10.1242/dev.00226. [DOI] [PubMed] [Google Scholar]
- Billeter M, Qian YQ, Otting G, Müller M, Gehring W, Wüthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J. Mol. Biol. 1993;234:1084–1097. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. The Carnegie Protein Trap Library: A versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J. Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Clements J, Francis C, Hens K. Pax6 and eye development in Arthropoda. Arthropod Struct. Dev. 2006;35:379–391. doi: 10.1016/j.asd.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Carrière C, Plaza S, Caboche J, Dozier C, Bailly M, Martin P, Saule S. Nuclear localization signals, DNA binding, and transactivation properties of quail Pax-6 (Pax-QNR) isoforms. Cell Growth Differ. 1995;6:1531–1540. [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Clements J, Hens K, Francis C, Schellens A, Callaerts P. Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc. Natl. Acad. Sci. USA. 2008;105:16183–16188. doi: 10.1073/pnas.0708330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJH, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol. Cell. Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Favor J, Peters H, Hermann T, Schmahl W, Chatterjee B, Neuhäuser-Klaus A, Sandulache R. Molecular characterization of Pax62Neu through Pax610Neu: An extension of the Pax6 allelic series and the identification of two possible hypomorph alleles in the mouse Mus musculus. Genetics. 2001;159:1689–1700. doi: 10.1093/genetics/159.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- Fortin AS, Underhill DA, Gros P. Reciprocal effect of Waardenburg syndrome mutations on DNA binding by the Pax-3 paired domain and homeodomain. Hum. Mol. Genet. 1997;6:1781–1790. doi: 10.1093/hmg/6.11.1781. [DOI] [PubMed] [Google Scholar]
- Fortin AS, Underhill DA, Gros P. Helix 2 of the paired domain plays a key role in the regulation of DNA-binding by the Pax-3 homeodomain. Nucleic Acids Res. 1998;26:4574–4581. doi: 10.1093/nar/26.20.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Cai J, Epstein JA, Jepeal L, Maas RL. PAX6 gene mutations in aniridia. In: Wiggs JL, editor. Molecular Genetics of Ocular Disease. Wiley-Liss, Inc.; New York: 1995. pp. 51–82. [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Hanson IM, Seawright A, Hardman K, Hodgson S, Zaletayev D, Fekete G, van Heyningen V. PAX6 mutations in aniridia. Hum. Mol. Genet. 1993;2:915–920. doi: 10.1093/hmg/2.7.915. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Kammermeier L, Leemans R, Hirth F, Flister S, Wenger U, Walldorf U, Gehring WJ, Reichert H. Differential expression and function of the Drosophila Pax6 genes eyeless and twin of eyeless in embryonic central nervous system development. Mech. Dev. 2001;103:71–78. doi: 10.1016/s0925-4773(01)00328-8. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z. The role of Pax genes in eye evolution. Brain Res. Bull. 2008;75:335–339. doi: 10.1016/j.brainresbull.2007.10.046. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and dachshund genes. Proc. Natl. Acad. Sci. USA. 2000;97:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A, Scott MP. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984;310:25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lewis EB, Bacher F. Method of feeding ethyl methane sulphonate (EMS) to Drosophila males. Dros. Inf. Serv. 1968;43:193–194. [Google Scholar]
- Lindsley D, Zimm G. The Genome of Drosophila melanogaster. Academic Press; New York: 1992. [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Davis NM, Andrews GL, Easter SS., Jr. Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol. Cell. Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H. Pax6 defines the di-mesencephalic boundary by repressing En1 and Pax2. Development. 2000;127:2357–2365. doi: 10.1242/dev.127.11.2357. [DOI] [PubMed] [Google Scholar]
- Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J. Med. Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;127:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S, Eto K. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development. 1997;124:2961–2972. doi: 10.1242/dev.124.15.2961. [DOI] [PubMed] [Google Scholar]
- Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001;15:1716–1723. doi: 10.1101/gad.196401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JC, West JD, Hill RE. Multiple functions for Pax6 in mouse eye and nasal development. Genes Dev. 1996;10:435–446. doi: 10.1101/gad.10.4.435. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Redeker EJW, de Visser ASH, Bergen AAB, Mannens MMAM. Multiplex ligation-dependant probe amplification (MLPA) enhances the molecular diagnosis of aniridia and related disorders. Mol. Vis. 2008;14:836–840. [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Tamkun JW, Hartzell GW. The structure and function of the homeodomain. Biochim. Biophys. Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Sheng G, Harris E, Bertuccioli C, Desplan C. Modular organization of Pax/Homeodomain proteins in transcriptional regulation. Biol. Chem. 1997;378:863–872. doi: 10.1515/bchm.1997.378.8.863. [DOI] [PubMed] [Google Scholar]
- Singh S, Stellrecht CM, Tang HK, Saunders GF. Modulation of PAX6 homeodomain function by the paired domain. J. Biol. Chem. 2000;275:17306–17313. doi: 10.1074/jbc.M000359200. [DOI] [PubMed] [Google Scholar]
- Singh S, Chao LY, Mishra R, Davies J, Saunders GF. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum. Mol. Genet. 2001;10:911–918. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS, editors. Drosophila protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. [Google Scholar]
- Takahashi M, Osumi N. Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development. 2002;129:1327–1338. doi: 10.1242/dev.129.6.1327. [DOI] [PubMed] [Google Scholar]
- Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J. Biol. Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- Thaung C, West K, Clark BJ, McKie L, Morgan JE, Arnold K, Nolan PM, Peters J, Hunter AJ, Brown SDM, et al. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum. Mol. Genet. 2002;11:755–767. doi: 10.1093/hmg/11.7.755. [DOI] [PubMed] [Google Scholar]
- Ton CCT, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Treisman J, Gönczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Treisman J, Harris E, Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Treisman J, Harris E, Wilson D, Desplan C. The homeodomain: A new face for the helix-turn-helix? BioEssays. 1992;14:145–150. doi: 10.1002/bies.950140302. [DOI] [PubMed] [Google Scholar]
- Underhill DA, Vogan KJ, Gros P. Analysis of the mouse Splotch-delayed mutation indicates that the Pax-3 paired domain can influence homeodomain DNA-binding activity. Proc. Natl. Acad. Sci. USA. 1995;92:3692–3696. doi: 10.1073/pnas.92.9.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DA, Gros P. The paired-domain regulates DNA binding by the homeodomain within the intact Pax-3 protein. J. Biol. Chem. 1997;272:14175–14182. doi: 10.1074/jbc.272.22.14175. [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum. Mol. Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- Vogan KJ, Underhill DA, Gros P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol. Cell. Biol. 1996;16:6677–6686. doi: 10.1128/mcb.16.12.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan KJ, Gros P. The C-terminal subdomain makes an important contribution to the DNA binding activity of the Pax-3 paired domain. J. Biol. Chem. 1997;272:28289–28295. doi: 10.1074/jbc.272.45.28289. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Walther C, Guenet J-L, Simon D, Deutsch U, Jostes B, Goulding MD, Plachov D, Balling R, Gruss P. Pax: A murine multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- Warren N, Price DJ. Roles of Pax-6 in murine diencephalic development. Development. 1997;124:1573–1582. doi: 10.1242/dev.124.8.1573. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum. Mol. Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of Paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a Paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- Wisniewski J, Orosz A, Allada R, Wu C. The C-terminal region of Drosophila heat shock factor (HSF) contains a constitutively functional transactivation domain. Nucl. Acids Res. 1996;24:367–374. doi: 10.1093/nar/24.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplemental Figure 1. Mutagenesis scheme for the eyOK107/X mutants
Males of the Gal4 line OK107 were mutagenized en masse by EMS feeding, and crossed to eyD1Da/ciD females, generating males of the genotype OK107*/ciD (asterisk indicates mutagenized OK107 chromosome). These males were then crossed individually to eyD1Da/cilacZ females, and their non-ciD progeny were screened for an eyeless phenotype. Of those found, their non-ciD siblings without eyeless phenotypes (OK107*/ cilacZ) were crossed to eyD1Da/ciD females in order to establish stocks and confirm the observed eyeless phenotype.
Supplemental Figure 2. Eyeless protein expression is not affected in the eyOK107/X mutants
Neither Eyeless protein synthesis nor stability is affected by the missense mutations in the eyOK107/X mutants. Here Eyeless expression in the Kenyon cell cluster of the mushroom body is shown for each of the four eyOK107/X alleles (homozygous for mutant allele) as well as a wildtype control (Oregon-R).