Abstract
Genome-wide association studies (GWAS) have identified 38 larger genetic regions affecting classical blood lipid levels without adjusting for important environmental influences. We modeled diet and physical activity in a GWAS in order to identify novel loci affecting total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels. The Swedish (SE) EUROSPAN cohort (N SE = 656) was screened for candidate genes and the non-Swedish (NS) EUROSPAN cohorts (N NS = 3,282) were used for replication. In total, 3 SNPs were associated in the Swedish sample and were replicated in the non-Swedish cohorts. While SNP rs1532624 was a replication of the previously published association between CETP and HDL cholesterol, the other two were novel findings. For the latter SNPs, the p-value for association was substantially improved by inclusion of environmental covariates: SNP rs5400 (p SE,unadjusted = 3.6×10−5, p SE,adjusted = 2.2×10−6, p NS,unadjusted = 0.047) in the SLC2A2 (Glucose transporter type 2) and rs2000999 (p SE,unadjusted = 1.1×10−3, p SE,adjusted = 3.8×10−4, p NS,unadjusted = 0.035) in the HP gene (Haptoglobin-related protein precursor). Both showed evidence of association with total cholesterol. These results demonstrate that inclusion of important environmental factors in the analysis model can reveal new genetic susceptibility loci.
Author Summary
In this article we report a genome-wide association study on cholesterol levels in the human blood. We used a Swedish cohort to select genetic polymorphisms that showed the strongest association with cholesterol levels adjusted for diet and physical activity. We replicated several genetic loci in other European cohorts. This approach extends present genome-wide association studies on lipid levels, which did not take these lifestyle factors into account, to improve statistical results and discover novel genes. In our analysis, we could identify two genetic loci in the SLC2A2 (Glucose transporter type 2) and the HP (Haptoglobin-related protein precursor) gene whose effects on total cholesterol have not been reported yet. The results show that inclusion of important environmental factors in the analysis model can reveal new insights into genetic determinants of clinical parameters relevant for metabolic and cardiovascular disease.
Introduction
Genome-wide association studies (GWAS) have identified more than 38 larger genetic regions which influence blood levels of total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG) [1]–[3]. These studies modeled basic anthropometric confounders, such as sex and age, while leaving out important environmental influences, such as diet and activity. This strategy is statistically suboptimal since the unexplained variation in the phenotype can increase the measurement error and as a result require larger sample sizes to detect a significant effect. Manolio [4] argued strongly for modeling of environmental covariates in GWAS and recommended lipid levels as a paradigmatic phenotype for studying the genetic and environmental architecture of quantitative traits.
In order to explore the usefulness of including both environmental and genetic factors in the analysis model, we used lipid measurements from the EUROSPAN study, comprising 3,938 individuals for whom genome-wide SNP data (N SNP = 311,388) were available [5]. We measured daily intake of food and physical activity at work and at leisure and modeled the influence of those environmental covariates on serum lipid levels in a GWAS. First, data from the Northern Sweden Population Health Study (NSPHS) were used as a discovery cohort to screen for SNPs that displayed the lowest p-values when the model was adjusted for environmental covariates. We then used the other, non-Swedish EUROSPAN cohorts for replication of our strongest associations in a candidate gene association study (CGAS).
We chose a population living in northern Sweden for the selection of candidate loci because it shows strong natural heterogeneity in certain lifestyle factors (e.g. diet, activity), but homogeneity in other environmental aspects such as climate [6]. Whereas one group is living a modern, sedentary lifestyle found also in the southern part of Sweden and other western European countries, a subgroup of Swedes follows a traditional, semi-nomadic way of life based on reindeer herding. Reindeer herders typically show higher intake of game meat (reindeer, moose), which has a high protein and low fat content, and lower intake of non-game meat, fish, and dairy products among other, lesser differences. They also exert more physical activity at work to tend their reindeer herds, but less activity at leisure [7].
Results
Exploratory GWAS in NSPHS
We performed a GWAS with a lifestyle-adjusted model which included not only sex and age, but also daily intake of game meat, non-game meat, fish, milk products, physical activity at work and at leisure as covariates. We focused on the 0.05% of all SNPs with the lowest p-values in the diet- and activity-adjusted model (corresponding to about 150 SNPs per lipid). For total cholesterol, 88 of these were located in a gene and 14 in genes that have been associated with energy metabolism (http://www.ncbi.nlm.nih.gov/omim/). For LDL-C, 65 SNPs were located in a gene, of which 8 were functionally relevant. Several of the SNPs for LDL-C were identical with those affecting total cholesterol, as expected from the high correlation (r = 0.91) between both phenotypes. For HDL-C, SNP rs2292883, located in the MLPH gene (Melanophilin), showed a genome-wide significant p-value (p = 1.06×10−07). 69 SNPs for HDL-C were located in a gene and 14 of those genes were reported as having a metabolic effect. Finally, for triglycerides, 63 SNPs were located in a gene, but only 4 SNPs in genes with a functional annotation of interest (Table 1 and Table S1A, S1B, S1C, S1D).
Table 1. Candidate SNPs (n = 39) selected from the Swedish discovery cohort.
| SNP | p-value, unadjusteda | p-value, adjustedb | p-value ratioc | Gene Symbol | Product name (Product Symbol) |
| TC | |||||
| rs10513684 | 2.91E-05 | 1.08E-06 | 27 | SLC2A2 | Glucose transporter type 2 (GLUT-2) |
| rs1684885 | 3.47E-03 | 2.01E-04 | 17 | PRKCI | Protein kinase C iota type (nPKC-iota) |
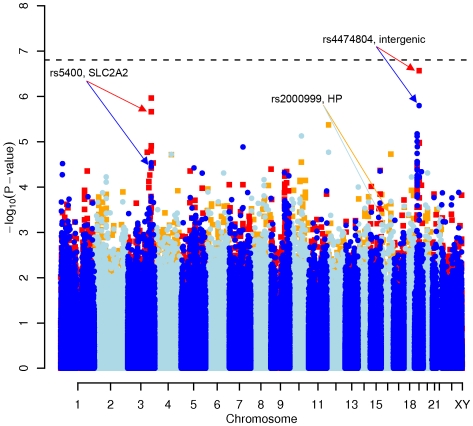
| rs5400 | 3.57E-05 | 2.18E-06 | 16 | SLC2A2 | Glucose transporter type 2 (GLUT-2) |
| rs47137 | 3.69E-03 | 2.63E-04 | 14 | SLC2A12 | Glucose transporter type 12 (GLUT-12) |
| rs669552 | 3.46E-03 | 2.87E-04 | 12 | FNDC3B | Factor for Adipocyte Differentiation 104 |
| rs2303324 | 1.49E-03 | 1.65E-04 | 9 | GALNT14 | Polypeptide GalNAc transferase 14 |
| rs12617790 | 2.26E-03 | 2.53E-04 | 9 | GALNT14 | Polypeptide GalNAc transferase 14 |
| rs10041333 | 3.04E-03 | 3.74E-04 | 8 | FABP6 | Gastrotropin (GT), alt. Fatty Acid-Binding Protein 6 |
| rs222014 | 2.93E-03 | 3.71E-04 | 8 | GC | Vitamin D-binding protein Precursor (DBP) |
| rs2000999 | 1.12E-03 | 3.84E-04 | 3 | HP | Haptoglobin-related protein Precursor |
| rs2070657 | 1.70E-04 | 1.43E-04 | 1 | APP | Alzheimer disease amyloid protein (ABPP) |
| rs2186830 | 2.66E-04 | 2.78E-04 | 1 | COLEC12 | Collectin-12 |
| rs2478571 | 3.07E-04 | 4.42E-04 | 1 | SLC39A12 | Zinc transporter (ZIP12) |
| LDL-C | |||||
| rs1684885 | 1.53E-04 | 1.61E-05 | 10 | PRKCI | Protein kinase C iota type (nPKC-iota) |
| rs1684881 | 2.80E-04 | 3.14E-05 | 9 | PRKCI | Protein kinase C iota type (nPKC-iota) |
| rs10513684 | 1.98E-04 | 2.61E-05 | 8 | SLC2A2 | Glucose transporter type 2 (GLUT-2) |
| rs5400 | 4.22E-04 | 6.10E-05 | 7 | SLC2A2 | Glucose transporter type 2(GLUT-2) |
| rs12617790 | 1.65E-03 | 2.96E-04 | 6 | GALNT14 | Polypeptide GalNAc transferase 14 |
| rs7583934 | 2.78E-04 | 1.84E-04 | 2 | LRP1B | Low-density lipoprotein receptor-related protein (LRP-DIT) |
| rs1864616 | 8.64E-05 | 1.36E-04 | 2 | TGFBR2 | Transforming growth factor-beta receptor type II (TGFR-2) |
| rs843319 | 6.74E-05 | 2.89E-04 | 4 | MBOAT1 | O-acyltransferase domain-containing protein 1 |
| HDL-C | |||||
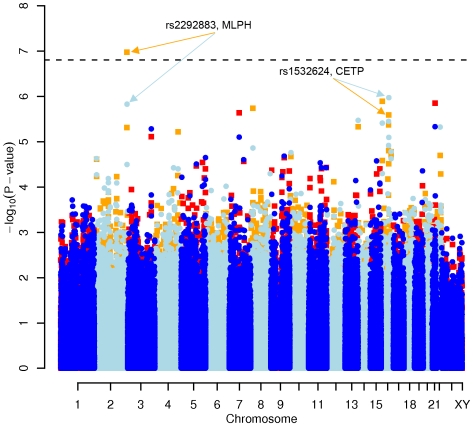
| rs2292883 | 1.48E-06 | 1.06E-07* | 14 | MLPH | Melanophilin |
| rs12712846 | 1.14E-03 | 3.02E-04 | 4 | MTA3 | Metastasis-associated protein |
| rs365578 | 8.39E-04 | 2.92E-04 | 3 | NDUFS4 | NADH dehydrogenase 8 iron-sulfur protein 4 (CI-AQDQ) |
| rs9866473 | 4.34E-04 | 2.55E-04 | 2 | CETP | Cholesteryl ester transfer protein |
| rs10519336 | 6.34E-04 | 3.82E-04 | 2 | MCC | Colorectal mutant cancer protein |
| rs2054247 | 3.73E-05 | 3.70E-05 | 1 | APLP2 | Amyloid-like protein 2 Precursor |
| rs11708205 | 2.67E-04 | 3.57E-04 | 1 | PLD1 | Phospholipase D1 |
| rs9863761 | 5.20E-06 | 7.74E-06 | 1 | CETP | Cholesteryl ester transfer protein |
| rs2124147 | 1.49E-04 | 2.23E-04 | 2 | CETP | Cholesteryl ester transfer protein |
| rs1567385 | 1.32E-04 | 1.99E-04 | 2 | MAP4K4 | Mitogen-activated protein kinase 4 (MEKKK4) |
| rs3776817 | 6.82E-05 | 1.31E-04 | 2 | ADAMTS2 | Procollagen I N-proteinase |
| rs1999088 | 7.19E-05 | 1.51E-04 | 2 | MBNL2 | Muscleblind-like protein 2 |
| rs1782644 | 1.39E-04 | 3.10E-04 | 2 | ZMIZ1 | Zinc finger MIZ domain-containing protein 1 |
| rs1532624 | 1.06E-06 | 2.55E-06 | 2 | CETP | Cholesteryl ester transfer protein |
| TG | |||||
| rs4304239 | 1.63E-03 | 2.40E-04 | 7 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 |
| rs11770192 | 1.82E-03 | 2.40E-04 | 8 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 |
| rs12540730 | 7.79E-04 | 2.43E-04 | 3 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 |
| rs3823763 | 9.58E-05 | 4.45E-05 | 2 | BBS9 | Parathyroid hormone-responsive B1 gene protein (PTHB1) |
All candidate SNPs show strongest associations (p-value, top 0.05% SNPs per lipid trait) and are located in a gene which has been reported to be relevant for energy metabolism. SNPs are sorted by p-value ratio (unadjusted∶unadjusted).
*: p≤1.6E-07 = genome-wide significant; All SNPs in genes with at least one replicated SNP are displayed in bold, replicated SNPs are formated in bold italics. a) unadjusted = covariates include sex and age; b) adjusted = covariates include sex, age, game meat, non-game meat, fish, milk products, physical activity at work and at leisure; c) p-ratio = max(p unadjusted∶p adjusted; p adjusted∶p unadjusted); p unadjusted∶p adjusted ratios are aligned left, p adjusted∶p unadjusted ratios are aligned right.
P-value changes
In order to evaluate the effect of including diet and activity covariates in the association analysis, we overlaid the p-values in the Manhattan plots from the NSPHS for the unadjusted and adjusted GWAS models (Figure 1, Figure 2, Figure 3, Figure 4). More refined GWAS results separating the effect of adjusting for either diet or physical activity are presented in Figure S1A, S1B, S1C, S1D; and Figure S2A, S2B, S2C, S2D. As expected, the p-values for a number of SNPs were sensitive to the inclusion of both diet and activity covariates in the model. We matched the 0.05% SNPs with the lowest p-values (top SNP list) between the unadjusted and the adjusted model. For TC, 83 (53%) SNPs were found in both top SNP lists. Those lists contained 102 (64%) identical SNPs for LDL-C and 103 (65%) for HDL-C. The analyses resulted in the same 74 (47%) top SNPs for TG levels (Table S1A, S1B, S1C, S1D). Finally, we compared the p-value changes of the resulting 39 candidate SNPs that are located in genes with a metabolic effect between the diet and activity-adjusted (full) model and the unadjusted (restricted) model resulting in an up to 27-fold p-value decrease (Table 1).
Figure 1. Manhattan plot of genome-wide effects on total cholesterol levels in the Swedish discovery cohort.
Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained food intake and physical activity as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
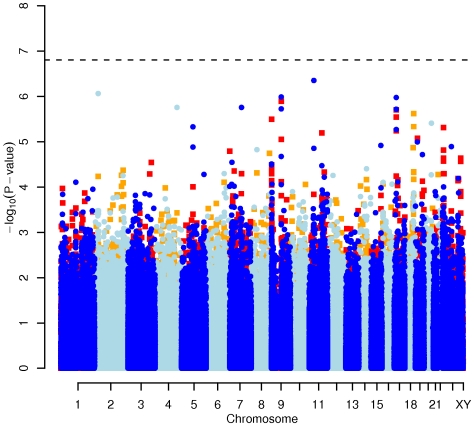
Figure 2. Manhattan plot of genome-wide effects on LDL cholesterol levels in the Swedish discovery cohort.
Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained food intake and physical activity as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
Figure 3. Manhattan plot of genome-wide effects on HDL cholesterol levels in the Swedish discovery cohort.
Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained food intake and physical activity as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
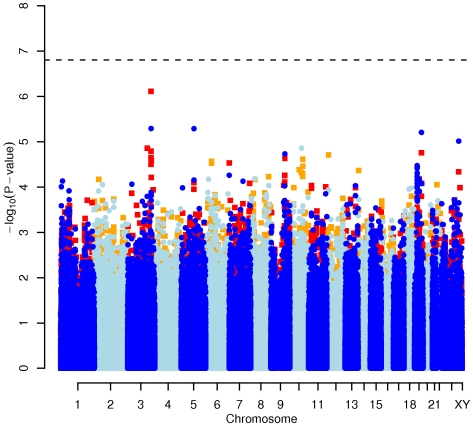
Figure 4. Manhattan plot of genome-wide effects on triglyceride levels in the Swedish discovery cohort.
Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained food intake and physical activity as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
Confirmatory CGAS in EUROSPAN
A food- and activity-adjusted candidate gene association study of the final 39 candidate SNPs in the Scottish (SC) sample (N = 714) was applied using similar lifestyle covariates (Table 2; Table S1E, S1F, S1G, S1H; Table S2). We replicated the effect of rs2000999 (p SC,unadj = 6.16×10−03, p SC,adj = 4.33×10−03) in the HP gene (Haptoglobin-related protein Precursor) on TC level and the effect of rs1532624 (p SC,unadj = 2.40×10−09, p SC,adj = 1.96×10−09) in CETP (Cholesteryl ester transfer protein) on HDL-C. In the Swedish cohort (SE), the unadjusted genetic effect of rs2000999 in the HP gene is equivalent to a moderately large difference in average TC level of 20.21 mg/dl between the homozyguous genotypes (MeanSE,unadj(TC|A/A)−MeanSE,unadj(TC|G/G) = 243.16−222.95, Effect SizeSE,unadj = 0.41, Effect SizeSE,adj = 0.44)(Effect Size (ES) = (M A/A−MB/B)/SD pooled). Equivalent effects were observed in the Scottish replication sample (M SC,unadj(TC|A/A)−M SC,unadj(TC|G/G) = 235.36 mg/dl−222.54 mg/dl = 12.82 mg/dl, ES SC,unadj = 0.29, ES SC,adj = 0.52). SNP rs1532624 in the CETP gene is associated with a large, unadjusted difference in HDL-C level of 9.99 mg/dl (M SE,unadj(HDL-C|A/A)−M SE,unadj(HDL-C|C/C) = 68.14 mg/dl−58.15 mg/dl, ES SE,unadj = 0.73, ES SE,adj = 0.48) in the discovery cohort and similar effects regarding direction and size in the replication cohort (M SC,unadj(HDL-C|A/A)−M SC,unadj(HDL-C|C/C) = 69.79 mg/dl−60.75 mg/dl = 9.04 mg/dl; ES SC, unadj = 0.59, ES SC, adj = 0.57).
Table 2. SNPs (n = 3) discovered in a Swedish and replicated in a non-Swedish EUROSPAN cohort.
| SNP | Gene | Trait | Cohort | p-value, unadjusteda | p-value, adjustedb | Mean Difference, unadjustedc | Effect Size, unadjustedd | Effect Size, adjustede |
| rs2000999 | HP | TC | Discovery, SE | 1.12E-03 | 3.84E-04 | 20.21 mg/dl | 0.41 | 0.44 |
| Replication, SC | 6.16E-03 | 4.33E-03 | 12.82 mg/dl | 0.29 | 0.52 | |||
| rs1532624 | CETP | HDL-C | Discovery, SE | 1.06E-06 | 2.55E-06 | 9.99 mg/dl | 0.73 | 0.48 |
| Replication, SC | 2.40E-09 | 1.96E-09 | 9.04 mg/dl | 0.59 | 0.57 | |||
| rs5400 | SLC2A2 | TC | Discovery, SE | 3.57E-05 | 2.18E-06 | 27.11 mg/dl | 0.57 | 0.66 |
| Replication, NS | 4.68E-02 | N.A. | 13.35 mg/dl | 0.30 | N.A. |
For all replicated SNPs p-values, mean differences, and effects sizes for unadjusted and adjusted lipid levels between homozygous genotypes are reported except for replication cohort NS. Discovery Cohort SE: Swedish EUROSPAN cohort (N SE = 656), Replication Cohort SC: Scottish EUROSPAN cohort (N Sc = 714), Replication Cohort NS: Non-Swedish EUROSPAN cohorts (Scotland, Croatia, Italy, Netherlands, N NS = 3,282), N.A.: not available; a) unadjusted: covariates include sex and age, b) adjusted: covariates include sex, age, game meat, non-game meat, fish, milk products, physical activity at work and at leisure, c) Genetic effect as mean difference of unadjusted lipid levels between homozygous genotypes: M(A/A)−M(B/B), d) Genetic effect as standardized effect size of unadjusted lipid levels: ES = (M A/A−M B/B)/SD pooled, e) Genetic effect as standardized effect size of adjusted lipid levels: ES = (M A/A−M B/B)/SD pooled.
We also performed an unadjusted candidate gene analysis of the 39 candidate SNPs in all non-Swedish (NS) EUROSPAN cohorts (Scotland, Croatia, The Netherlands, and Italy, N NS = 3,282) and aggregated the results in a meta-analysis (Table 2; Table S1I, S1J, S1K, S1L). We confirmed the effects of rs5400 (pNS = 4.68×10−02) in SLC2A2 on TC. We again found that rs2000999 (p NS,unadj = 3.54×10−2) in HP influences TC levels and rs1532624 (p NS,unadj = 2.87×10−20) in CETP (Cholesteryl ester transfer protein) affects HDL-C levels. The unadjusted genetic effect of rs5400 is equivalent to a moderately large difference in mean TC level of 27.11 mg/dl between homozyguous genotypes (M SE,unadj(TC|A/A)−M SE,unadj(TC|G/G) = 249.30 mg/dl−222.19 mg/dl, ES SE,unadj = 0.57, ES SE,adj = 0.66) in the Swedish Cohort and a small total effect in all non-Swedish samples (M NS,unadj(TC|A/A)−M NS,unadj(TC|G/G) = 236.69 mg/dl−223.34 mg/dl = 13.35 mg/dl, ES NS,unadj = 0.30).
No other associations, including LDL cholesterol or triglycerides levels, were replicated (all p>0.05). The genome-wide significant SNP rs2292883 in the Melanophilin (MLPH) gene found in the Swedish cohort was not confirmed.
Discussion
Environmental covariates may either act as moderators, mediators or even suppressors, thereby affecting the discovery of genetic susceptibility loci [8],[9]. Therefore, we conducted a GWAS, modeling genetic and important environmental effects, such as food intake and physical activity, on serum levels of classical lipids. To our knowledge, this is the first GWAS on blood lipid levels modeling environmental factors, in particular major food categories and physical activity, in international cohorts. Our analysis replicated one known locus in the CETP gene [1] and identified two other gene loci in the SLC2A2 and HP gene, respectively, involved in energy metabolism but not previously reported to be associated with cholesterol levels.
SLC2A2 encodes the facilitated glucose transporter member 2 (GLUT-2, Solute carrier family 2) and is predominantly expressed in the liver. Mice deficient in GLUT-2 are hyperglycemic and have elevated plasma levels of glucagon and free fatty acids [10]. Mutations in GLUT-2 cause the Fanconi-Bickel syndrome (FBS) characterized by hypercholesterolemia and hyperlipidemia [11],[12]. Cerf [13] argued that a high-fat diet causes a decreased expression of the GLUT-2 glucose receptor on β-cell islets. As a result, glucose stimulation of insulin exocytosis is impaired causing hyperglycemia, a clinical hallmark of type 2 diabetes. In addition, Kilpelainen et al. [14] found that physical activity moderates the genetic effect of SLC2A2 on type 2 diabetes. These studies suggest that these lifestyle factors could have masked genetic effects in previous, unadjusted GWAS. This is emphasized by the strong increase in statistical significance of the SLC2A2 polymorphisms after adjusting for diet and physical activity, indicating that the examined lifestyle factors modified the effect of this gene. Our supplemental results show that physical activity markedly moderated the genetic effect on total cholesterol.
The HP gene encodes the Haptoglobin-related Protein Precursor (Hp), which binds hemoglobin (Hb) to form a stable Hp-Hb complex and, thereby, prevents Hb-induced oxidative tissue damage. Asleh et al. [15] identified severe impairment in the ability of Hp to prevent oxidation caused by glycosylated Hb. Diabetes is also associated with an increase in the non-enzymatic glycosylation of serum proteins, so these authors suggested that there is a specific interaction between diabetes, cardiovascular disease and the Hp genotype. It results from the increased need of rapidly clearing glycosylated Hb-Hp complexes from the subendothelial space before they oxidatively modify low-density lipoprotein to form the atherogenic oxidized low-density lipoprotein. The p-value for association between the HP SNP rs2000999 and total serum cholesterol concentration decreased in the model adjusted for diet and physical activity, suggesting that the genetic effect is moderated by diet and physical activity. Our supporting material points out the moderating role of physical activity in particular.
We also observed a highly significant association between rs1532624 in CETP and HDL-C levels. The CETP protein catalyzes the transfer of insoluble cholesteryl esters among lipoprotein particles. Variation in CETP is known to affect the susceptibility to atherosclerosis and other cardiovascular diseases [16]. Adjustment for diet and physical activity in our model caused an increase of the p-value of this SNP. Our supporting results indicate that the genetic effect is mediated by diet or by physical activity in a similar way.
This study also has some limitations. First, we are aware that our candidate gene association approach covers only a very small fraction of all genomic loci, which is one of the potential reasons why some classical lipid-influencing genes, such as APOE, are not represented in our candidate SNP list. Therefore, our approach is not comprehensive and may have failed to identify other relevant lifestyle-sensitive genetic variants. Nonetheless, we decided to apply this approach to make the best out of the available lifestyle data. Second, our study provides only limited information on the role of individual lifestyle factors for a genetic variant. However, in this study we aimed at amplifying genetic effects by adjusting for a maximum amount of environmental variance in a single model and, therefore, we neglected some of these aspects here. Third, we did not model genetic covariates in known lipid-relevant genes which may also moderate the effect of other genetic predictors. This is due to the focus of this paper on gene-environment relationships.
In summary, we have demonstrated that modeling environmental factors, in particular major food categories and physical activity, can improve statistical power and lead to the discovery of novel susceptibility loci. Such models also provide an understanding of the complex interplay of genetic and environmental factors affecting human quantitative traits. Inclusion of environmental covariates represents a much needed next step in the quest to model the complete environmental and genetic architecture of complex traits.
Methods
Ethics statement
All EUROSPAN studies were approved by the appropriate research ethics committees according to the Declaration of Helsinki [17]. The Northern Swedish Population Health Study (NSPHS) was approved by the local ethics committee at the University of Uppsala (Regionala Etikprövningsnämnden, Uppsala). The Scottish ORCADES study was approved by the NHS Orkney Research Ethics Committee and the North of Scotland REC. The Croatian VIS study was approved by the ethics committee of the medical faculty in Zagreb and the Multi-Centre Research Ethics Committee for Scotland. The Dutch ERF study was approved by the Erasmus institutional medical ethics committee in Rotterdam, The Netherlands. The Italian MICROS study was approved by the ethical committee of the Autonomous Province of Bolzano, Italy.
Participants
The examined subjects stem from five different population-representative, pedigree-based cohorts from the EUROSPAN consortium (http://www.eurospan.org). All studies include a comprehensive collection of data on family structure, lifestyle, blood samples for clinical chemistry, RNA and DNA analyses, medical history, and current health status. All participants gave their written informed consent [18]. A brief description of each population is given below:
The Northern Swedish Population Health Study (NSPHS) represents a cross-sectional study conducted in the community of Karesuando in the subartic region of the County of Norrbotten, Sweden, in 2006 [5]. This parish has about 1500 eligible inhabitants of whom 740 participated in the study. The final sample consisted of 309 men and 347 women who were aged between 14 and 91 years. The inclusion of diet and activity covariates in the analytical model and according missing values reduced the effective sample size by less than 5%.
The Orkney Complex Disease Study (ORCADES) is a longitudinal study in the isolated Scottish archipelago of Orkney [19]. Participants from a subgroup of ten islands (N = 719) were used for the presented analysis. The sample comprised 334 men and 385 women aged between 18 and 100 years. The inclusion of diet and activity covariates in the analytical model and according missing values reduced the effective sample size by less than 5%.
The VIS study is a cross-sectional study in the villages of Vis and Komiza on the Dalmatian island of Vis, Croatia, and was conducted between 2003 and 2004 [20]–[22]. 795 participants who had both genotype and phenotypic data available were analysed. This cohort included 328 men and 467 women with an age between 18 and 93 years.
The Microisolates in South Tyrol Study (MICROS) is a cross-sectional study carried out in the villages of Stelvio, Vallelunga, and Martello, Venosta valley, South Tyrol, Italy, from 2001 to 2003 [23]. The 1,097 participants (475 males, 622 females, age between 18 and 88 years) presented in this study are those for whom both relevant genotype and phenotype data were available.
The Erasmus Rucphen Family Study (ERF) is a longitudinal study on a population living in the Rucphen region, the Netherlands, in the 19th century [24]. Fasting total cholesterol, HDL cholesterol and triglyceride levels were available. LDL cholesterol was estimated using the Friedewald formula [25]. The 918 individuals included in this study consisted of the first series of participants with 354 men and 564 women aged between 18 and 92 years.
Genotyping
DNA samples were genotyped according to the manufacturer's instructions on Illumina Infinium HumanHap300v2 or HumanCNV370v1 SNP bead microarrays. Both arrays have 311,388 SNP markers in common that are distributed across the human genome. Analysis of the raw data was done in the BeadStudio software with the recommended parameters for the Infinium assay and using the genotype cluster files provided by Illumina. Individuals with a call rate below 95% and SNPs with a call rate below 98%, deviating from Hard-Weinberg equilibrium (p HWE<1×10−6) or with a minor allele frequency of less than 1% were excluded from the analysis.
Lipids
Total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglycerides (TG) were quantified by enzymatic photometric assays using an ADVIA1650 clinical chemistry analyzer (Siemens Healthcare Diagnostics GmbH, Eschborn, Germany) at the Institute for Clinical Chemistry and Laboratory Medicine, Regensburg University Medical Center, Germany.
Diet
In the NSPHS cohort, we collected data with a food frequency questionnaire based on the Northern Sweden 84-item Food Frequency Questionnaire (NoS-84-FFQ) [26]. We included in the questionnaire several items on foods specific for the lifestyle in this geographic region, in particular on game consumption (reindeer, moose). The answer options consisted of an 11-point format: 0 = “Never”, 1 = “less than 1 time per month”, 2 = “1 to 3 times per month”, 3 = “1 time per week”, 4 = “2 to 4 times per week”, 5 = “5 to 6 times per week”, 6 = “1 time per day”, 7 = “2 to 3 times per day”, 8 = “4 to 5 times per day”, 9 = “6 to 8 times per day”, 10 = “9 to 10 times per day”. The questionnaire was applied in electronic format by a trained study nurse as an interviewer. For each food item we calculated daily intake in gram per day as a standardized unit of measurement and aggregated the items to food categories, such game meat, non-game meat, fish, and dairy products. We evaluated the construct validity (known-groups validity) of the added items on game consumption in the NoS-84-FFQ questionnaire. We compared reindeer herders (N = 94) versus non-reindeer herders (N = 505). We observed highly significant, large effect sizes in men (ES = 1.25, p = 9.7×10−04) and women (ES = 1.15, p = 2.9×10−05) in the expected direction corresponding with an approximately three times higher consumption of absolute overall game intake in reindeer herders compared to others. A similar approach was used for the measurement and analysis of dietary data collected with a food frequency questionnaire in the Scottish cohort (Table S2).
Physical activity
In the NSPHS cohort, we used two self-report scales to measure overall physical activity at work and at leisure. The Work Activity Scale (WAS, 6 items) addresses typical occupational physical activities: sitting, standing, walking, lifting, and general indicators of physical activity, i. e. sweating and tiredness after work. The Leisure Activity Scale (LAS, 4 items) asks for various typical freetime activities such walking, cycling, other sporting activities, and sweating as a general indicator of physical activity. Participants reported the frequency of each activity on a 5-point rating scale (1 = “never”, 2 = “seldom”, 3 = “sometimes”, 4 = “often”, and 5 = “always”). Both scales showed satisfying internal consistency with Cronbach's α(WAS) = 0.73 and Cronbach's α(LAS) = 0.70. A similar approach was used for the measurement and analysis of data on physical activity collected with a self-report questionnaire in the Scottish cohort (Table S2).
Statistical analysis
Model selection
Sex and age are chosen as standard moderators of medical outcomes. Food and physical activity covariates have been selected based on findings on natural variation in lifestyle factors in this (data not presented) and other [7] northern Swedish populations between a modern, sedentary and a traditional, semi-nomadic lifestyle based on reindeer herding. Mostly significant associations between diet and activity covariates and lipid levels were found in the examined Swedish EUROSPAN cohort in the following ranges: r = [−0.01;0.12] (p = [1.28×10−02;0.16]) for game meat, r = [−0.13;−0.05] (p = [8.63×10−04;0.74]) for non-game meat, r = [0.06;0.16] (p = [2.12×10−05;0.12]) for fish, r = [0.04;0.13] (p = [2.51×10−09;3.85×10−06]) for physical activity at work, and r = [−0.11;0.01] (p = [5.05×10−09;1.30×10−06]) for physical activity at leisure (Table S3). We finally selected sex, age, game meat, non-game meat, fish, dairy products, physical activity at work, and physical activity at leisure as covariates in our diet- and activity-adjusted model (“adjusted” model) in the Swedish EUROSPAN sample. Sex and age were used as covariates in the “unadjusted” model.
We tested whether the inclusion of those covariates in the explanatory model led to a statistical significant improvement of the goodness of model fit compared to a restricted model by applying a maximum likelihood ratio (MLR) test. We inferred a significant better model fit of the full model if the difference of the χ2 value between both models had an equal or lower probability than p = 0.05 (one-sided, upper tail) on a χ2 distribution with k degrees of freedom. The degrees of freedom k are equal to the difference of the number of parameters in each model. The difference of χ2 values between both models is calculated according to the following formula with MLE indicating the maximum likelihood estimates per model: χ2(rest−full) = −2 (log 10(MLE rest)−log 10(MLE full)). The comparison of the goodness of fit between the unadjusted and the diet- and activity-adjusted full model, using a MLR test, showed a statistically significant improvement for all four lipid traits (TC: χ2 diff = 59.69, df = 6, p = 5.21×10−11; LDL-C: χ2 diff = 39.45, df = 6, p = 5.85×10−07; HDL-C: χ2 diff = 29.57, df = 6, p = 4.75×10−05; TG: χ2 diff = 69.32, df = 6, p = 5.65×10−13). All included polygenic, anthropometric and lifestyle factors (with the effect of including only the polygenic, sex, and age effects in parentheses) explained 64.07% (58.02%) of the variation of TC, 59.47% (56.47%) of the variation of LDL-C, 83.73% (82.59%) of the variance of HDL-C and 58.68% (41.80%) of the variation of TG levels. Dietary measures accounted for 22% (TC), 40% (LDL-C), 74% (HDL-C), and 7% (TG), respectively, of the variance explained by lifestyle factors with physical activity being responsible for the rest. GWAS results for models adjusted for sex, age, and diet only (Figures S1A, S1B, S1C, S1D) or physical activity only (Figures S2A, S2B, S2C, S2D) are presented in the supporting figures.
The confounding effect of treatment with statins on total cholesterol level and LDL cholesterol level was adjusted for by imputing untreated lipid concentrations of medicated individuals using the npsubtreated() function of the R/GenABEL package which implements the algorithm of Tobin et al. [27]. Additionally, we conducted the same analysis in subsamples which did not receive any lipid-lowering treatment and found overall converging, but somewhat weaker results for rs2000999 (p SE,adj = 2.55×10−04; p SC,adj = 2.07×10−02, p NS,unadj = 5.93×10−02), rs1532624 (p SE,adj = 2.26×10−05; p SC,adj = 2.28×10−09, p NS,unadj = 2.37×10−19), and rs5400 (p SE,adj = 5.34×10−06; p SC,adj = 2.23×10−01, p NS,unadj = 8.04×10−02) (Table S4).
Genome-wide association analysis
First, deviations from normality for all quantitative traits (lipids, age, diet, and physical activity) were corrected by inverse-normal transformation without adjusting for covariates. Second, linear mixed effects models were fitted for the transformed outcomes (TC, LDL-C, HDL-C, TG) using the above mentioned covariates in the Swedish EUROSPAN sample and corresponding measures in the Scottish EUROSPAN sample (Table S2). The analysis was performed using the “polygenic” linear mixed effects model function polygenic() of the R/GenABEL package. Third, genome-wide association analysis was performed using a score test, a family-based association test [28], implemented in the mmscore() function of R/GenABEL. It uses the residuals and the variance-covariance matrix from the polygenic model and additional the SNP fixed effect coded under an additive model (0 = A/A, 1 = A/B, 2 = B/B). Fourth, genome-wide significance of a genetic loci was based on a local type I error of α = 0.05/311 388 SNPs = 1.6×10−7 according to a Bonferroni adjustment.
Candidate gene association analysis
The same statistical approach was used for association analysis of candidate loci with a local type I error of α = 0.05. No Bonferroni adjustment was applied to protect against α inflation since this method would be biased for the following reasons. The applied selection procedure for candidate loci makes the assumption of a global null hypothesis highly unlikely. Additionally, the phenotypes and some of the genotypes are highly correlated decreasing the number of independent tests. Instead all confirmatory tests are reported to allow the reader to evaluate the overall significance of the findings [29].
Relatedness
λ coefficients of lifestyle-adjusted genome-wide analysis varied in a low range between 1.00 and 1.04 in the Swedish cohort (see QQ-plots, Figures S3A, S3B, S3C, S3D, and Figure S4A, S4B, S4C, S4D) and between 1.00 and 1.01 in the Scottish cohort across all lipid traits. λ values for the unadjusted model used in the other three EUROSPAN cohorts did not exceed 1.01. These values indicate that our statistical model adequately handled relatedness in our pedigree-based samples since deflation of λ values is expected after correction for family structure.
Software and databases
We performed all analysis with the statistical analysis system R (V2.8.1) [30] mainly using the packages GenABEL (V1.4.2) [31] and biomarRt (V1.16.0) [32]. We accessed the following databases: Ensembl (http://www.ensembl.org) and Online Mendelian Inheritance in Men (http://www.ncbi.nlm.nih.gov/omim/).
Supporting Information
Manhattan plots of genome-wide effects on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained dietary measures (game meat, non-game meat, fish, milk products) as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
(0.31 MB DOC)
Manhattan plots of genome-wide effects on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained physical activity measures (job, leisure) as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
(0.31 MB DOC)
QQ-Plots for the unadjusted GWAS on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. The analysis model was only adjusted for sex and age, but not for diet and activity measures (black line = expected slope under no inflation, red line = slope fitted to observations).
(0.12 MB DOC)
QQ-Plots for the adjusted GWAS on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. The analysis model was adjusted for sex, age, diet and activity measures (black line = expected slope under no inflation, red line = slope fitted to observations).
(0.12 MB DOC)
GWAS results for all top candidate SNPs (0.05%) in the Swedish (SE) discovery cohort, the Scottish (SC), and all non-Swedish (NS) replication cohorts.
(0.41 MB XLS)
Comparison of the diet- and activity-adjusted analysis model in the Swedish and the Scottish cohort.
(0.04 MB DOC)
Pearson correlations, determination coefficients (explained variance), and p-values of the inverse-normal transformed lipid, dietary, and physical activity measures in the Swedish cohort.
(0.03 MB XLS)
GWAS results for all top SNPs (0.05%) in the Swedish (SE) discovery cohort, and for all candidate SNPs in the Scottish (SC), and in the non-Swedish (NS) replication cohorts including only individuals without lipid-lowering treatment.
(0.34 MB XLS)
Acknowledgments
We would like to thank the many colleagues who contributed to collection and phenotypic characterization of the samples, genotyping and analysis of the GWAS data, as well as lipid species analysis. We would also like to acknowledge those who agreed to participate in these studies.
NSPHS: We are grateful for the contribution of samples from the Medical Biobank in Umeå and for the contribution of the district nurse Svea Hennix. ORCADES: We would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney. VIS: We collectively thank a large number of individuals for their individual help in organizing, planning and carrying out the field work related to the project and data management: Professor Pavao Rudan and the staff of the Institute for Anthropological Research in Zagreb, Croatia (organization of the field work, anthropometric and physiological measurements, and DNA extraction); Professor Ariana Vorko-Jovic and the staff and medical students of the Andrija Štampar School of Public Health of the Faculty of Medicine, University of Zagreb, Croatia (questionnaires, genealogical reconstruction and data entry); Dr Branka Salzer from the biochemistry lab “Salzer”, Croatia (measurements of biochemical traits); local general practitioners and nurses (recruitment and communication with the study population); and the employees of several other Croatian institutions who participated in the field work, including but not limited to the University of Rijeka and Split, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia. SNP Genotyping of the Vis samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh. MICROS: We thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the research project. ERF: We are grateful to all patients and their relatives, general practitioners, and neurologists for their contributions and to P. Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection. UPPMAX: The computations were performed on UPPMAX (http://www.uppmax.uu.se) resources under Project p2008027. Further information about The European Special Populations Research Network (EUROSPAN) consortium is available at http//www.eurospan.org.
Footnotes
The authors have declared that no competing interests exist.
The European Special Populations Research Network (EUROSPAN) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947). High-throughput genome-wide association analysis of the data was supported by joint grant from Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). Lipidomic analysis was supported by the European Commission FP7 grant LipidomicNet (2007-202272). The NSPHS study was supported by grants from the Swedish Natural Sciences Research Council, the European Commission through EUROSPAN, the Foundation for Strategic Research (SSF), and the Linneaus Centre for Bioinformatics (LCB). The ORCADES study was supported by the Scottish Executive Health Department and the Royal Society. DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. The VIS study in the Croatian island of Vis was supported through the grants from the Medical Research Council UK to HC, AW, and IR and the Ministry of Science, Education, and Sport of the Republic of Croatia to IR (number 108-1080315-0302). The MICROS study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano and the South Tyrolean Sparkasse Foundation. The ERF study was supported by grants from the NWO, Erasmus MC, and the Centre for Medical Systems Biology (CMSB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabatti C, Service SK, Hartikainen A, Pouta A, Ripatti S, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 5.Johansson A, Marroni F, Hayward C, Franklin CS, Kirichenko AV, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18:373–380. doi: 10.1093/hmg/ddn350. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross AB, Johansson A, Ingman M, Gyllensten U. Lifestyle, genetics, and disease in Sami. Croat Med J. 2006;47:553–65. doi: 16909452. [PMC free article] [PubMed] [Google Scholar]
- 7.Ross A, Johansson Å, Vavruch-Nilsson V, Hassler S, Sjölander P, et al. Adherence to a traditional lifestyle affects food and nutrient intake among modern Swedish Sami. International Journal of Circumpolar Health. 2009;68:313–416. doi: 10.3402/ijch.v68i4.17371. [DOI] [PubMed] [Google Scholar]
- 8.Pearl J. Statistics and causal inference: A review. TEST. 2003;12:281–345. doi: 10.1007/BF02595718. [Google Scholar]
- 9.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–82. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 10.Guillam MT, Hümmler E, Schaerer E, Yeh JI, Birnbaum MJ, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet. 1997;17:327–330. doi: 10.1038/ng1197-327. doi: 10.1038/ng1197-327. [DOI] [PubMed] [Google Scholar]
- 11.Santer R, Schneppenheim R, Dombrowski A, Götze H, Steinmann B, et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17:324–326. doi: 10.1038/ng1197-324. doi: 10.1038/ng1197-324. [DOI] [PubMed] [Google Scholar]
- 12.Manz F, Bickel H, Brodehl J, Feist D, Gellissen K, et al. Fanconi-Bickel syndrome. Pediatr Nephrol. 1987;1:509–518. doi: 10.1007/BF00849262. [DOI] [PubMed] [Google Scholar]
- 13.Cerf ME. High fat diet modulation of glucose sensing in the beta-cell. Med Sci Monit. 2007;13:RA12–17. [PubMed] [Google Scholar]
- 14.Kilpelainen TO, Lakka TA, Laaksonen DE, Laukkanen O, Lindstrom J, et al. Physical activity modifies the effect of SNPs in the SLC2A2 (GLUT2) and ABCC8 (SUR1) genes on the risk of developing type 2 diabetes. Physiol Genomics. 2007;31:264–272. doi: 10.1152/physiolgenomics.00036.2007. doi: 10.1152/physiolgenomics.00036.2007. [DOI] [PubMed] [Google Scholar]
- 15.Asleh R, Marsh S, Shilkrut M, Binah O, Guetta J, et al. Genetically determined heterogeneity in hemoglobin scavenging and susceptibility to diabetic cardiovascular disease. Circ Res. 2003;92:1193–1200. doi: 10.1161/01.RES.0000076889.23082.F1. doi: 10.1161/01.RES.0000076889.23082.F1. [DOI] [PubMed] [Google Scholar]
- 16.Dullaart RPF, Sluiter WJ. Common variation in the CETP gene and the implications for cardiovascular disease and its treatment: an updated analysis. Pharmacogenomics. 2008;9:747–763. doi: 10.2217/14622416.9.6.747. doi: 10.2217/14622416.9.6.747. [DOI] [PubMed] [Google Scholar]
- 17.World Medical Association (WMA) World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects [Internet]. 2000. Available: http://www.wma.net/e/policy/pdf/17c.pdf. Accessed 30 Sep 2009.
- 18.Mascalzoni D, Janssens ACJ, Stewart A, Pramstaller P, Gyllensten U, et al. Comparison of participant information and informed consent forms of five European studies in genetic isolated populations. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.155. Available: http://www.ncbi.nlm.nih.gov/pubmed/19826451. Accessed 19 Oct 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McQuillan R, Leutenegger A, Abdel-Rahman R, Franklin CS, Pericic M, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barać L, Pericić M, Klarić IM, Rootsi S, Janićijević B, et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet. 2003;11:535–542. doi: 10.1038/sj.ejhg.5200992. doi: 10.1038/sj.ejhg.5200992. [DOI] [PubMed] [Google Scholar]
- 21.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Collegium Antropologicum. 1999;23:531–46. doi: 10646227. [PubMed] [Google Scholar]
- 22.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. doi: 5201589. [DOI] [PubMed] [Google Scholar]
- 23.Pattaro C, Marroni F, Riegler A, Mascalzoni D, Pichler I, et al. The genetic study of three population microisolates in South Tyrol (MICROS): study design and epidemiological perspectives. BMC Med Genet. 2007;8:29. doi: 10.1186/1471-2350-8-29. doi: 1471-2350-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aulchenko Y, Heutink P, Mackay I, Bertoli-Avella AM, Pullen J, et al. Linkage disequilibrium in young genetically isolated Dutch population. Eur J Hum Genet. 2004;12:527–34. doi: 10.1038/sj.ejhg.5201188. doi: 15054401. [DOI] [PubMed] [Google Scholar]
- 25.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 26.Johansson I, Hallmans G, Wikman A, Biessy C, Riboli E, et al. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002;5:487–96. doi: 10.1079/phn2001315. doi: 10.1079/PHNPHN2001315. [DOI] [PubMed] [Google Scholar]
- 27.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 28.Chen W, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proschan MA, Waclawiw MA. Practical guidelines for multiplicity adjustment in clinical trials. Control Clin Trials. 2000;21:527–539. doi: 10.1016/s0197-2456(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing 2006 [Google Scholar]
- 31.Aulchenko Y, Ripke S, Isaacs A, van Duijn C. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. doi: btm108. [DOI] [PubMed] [Google Scholar]
- 32.Smedley D, Haider S, Ballester B, Holland R, London D, et al. BioMart - biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan plots of genome-wide effects on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained dietary measures (game meat, non-game meat, fish, milk products) as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
(0.31 MB DOC)
Manhattan plots of genome-wide effects on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. Results for two GWAS analysis models are presented. The unadjusted model (dark blue and light blue circles) included only sex and age as covariates. The adjusted model (red and orange squares) additionally contained physical activity measures (job, leisure) as predictors. The dashed line indicates the local Bonferroni-adjusted α error = 1.6×10−7.
(0.31 MB DOC)
QQ-Plots for the unadjusted GWAS on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. The analysis model was only adjusted for sex and age, but not for diet and activity measures (black line = expected slope under no inflation, red line = slope fitted to observations).
(0.12 MB DOC)
QQ-Plots for the adjusted GWAS on total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels in the Swedish discovery cohort. The analysis model was adjusted for sex, age, diet and activity measures (black line = expected slope under no inflation, red line = slope fitted to observations).
(0.12 MB DOC)
GWAS results for all top candidate SNPs (0.05%) in the Swedish (SE) discovery cohort, the Scottish (SC), and all non-Swedish (NS) replication cohorts.
(0.41 MB XLS)
Comparison of the diet- and activity-adjusted analysis model in the Swedish and the Scottish cohort.
(0.04 MB DOC)
Pearson correlations, determination coefficients (explained variance), and p-values of the inverse-normal transformed lipid, dietary, and physical activity measures in the Swedish cohort.
(0.03 MB XLS)
GWAS results for all top SNPs (0.05%) in the Swedish (SE) discovery cohort, and for all candidate SNPs in the Scottish (SC), and in the non-Swedish (NS) replication cohorts including only individuals without lipid-lowering treatment.
(0.34 MB XLS)