Abstract
The p53 family consists of three transcription factors, p53, p63 and p73 that share domain architecture and sequence identity. The mTOR (mammalian target of rapamycin) kinase responds to growth factors and nutrient levels to regulate cellular growth and autophagy. Whereas p53 acts both upstream and downstream of mTOR, gene signature-based analyses have revealed that p73 is inhibited by mTOR activity. p53 can both activate and repress autophagy levels depending on cellular context. While less is known about p73, recent studies have shown that it induces cellular autophagy and multiple autophagy-associated genes downstream of mTOR. Chromatin immunoprecipitation analyses demonstrate that endogenous p73 binds the regulatory regions of genes such as ATG5, ATG7 and UVRAG. How p73 regulates the expression levels of these genes in response to different cellular stresses remains unknown. Because p53 family members play key roles in tumor suppression, development, aging and neurodegeneration, the context and manner by which these transcription factors regulate autophagy may have implications for a wide range of human diseases.
Keywords: p73, mTOR, p53, p63, chromatin immunoprecipitation, UVRAG, ATG5, ATG7, rapamycin, tumor suppressor
Recent discoveries linking p53 to core metabolic pathways, and to processes such as autophagy, mark a shift in our understanding of p53 as a tumor suppressor. Mammalian cells also contain two homologs of p53, called p63 and p73, that likely coordinate with p53 during select cellular responses to environmental and developmental cues.1 All three p53 family members are subject to a variety of posttranslations modifications and can exist as multiple protein isoforms, so there is the potential for substantial complexity in the p53 family transcriptional response.1–3 Indeed, each family member can regulate thousands of target genes.4–6
We hypothesized that the gene signature of a transcription factor could be viewed as a reflection of pathways that are upstream of the transcription factor.7 Genome-wide technologies were used to identify a p73 gene signature. This signature was compared to pathway-associated gene signatures in the Broad Institute Connectivity Map.7,8 Using this approach, mTOR was identified and validated as a negative regulator of p73.7 The identification of pathways upstream of p73 is of particular relevance because, unlike p53, p73 is not mutated in human tumors.9 Thus, mTOR inhibition might engage p73 and hence a p53-like cell death response in tumors that lack functional p53.
Further analysis of subcomponents of the p73 gene signature reveal autophagy-associated genes that are regulated by mTOR in a p73-dependent manner.7 In addition, a reported link between p73 and autophagy was confirmed, and a transactivation-competent isoform of p73 was identified as a positive regulator of autophagy.7,10 These recently published data suggest that there are both similarities and differences between p53 and p73 in relation to mTOR. While our data showed that mTOR negatively regulates p73, it has now been shown in multiple contexts that mTOR is a positive regulator of p53.11,12 Consistent with these observations, we observed a simultaneous increase in p73 and decrease in p53 and p63 protein levels in primary human mammary epithelial cells treated with rapamycin.7 Of note, at least in some basal cell types, p63 is expressed as an isoform that can inhibit p73.1,13 Thus, a coordinate upregulation of p73 and downregulation of p63 may be needed to fully activate p73 in response to metabolic stress.
How then do p53 family members engage metabolic stress responses such as autophagy? p53 itself plays opposing roles.11,14 Cytoplasmic p53 inhibits autophagy in multiple cell types and organisms,11 whereas nuclear p53 activates autophagy by upregulating genes such as DRAM.14 The cytoplasmic function of p53 represents a basal activity, whereas the nuclear function of p53 seems to require activating signals such as genotoxic stress.11,14 In addition, p53 can act upstream of mTOR, inhibiting mTOR by activating AMPK,15,16 and this may alter cellular autophagy levels as well.
Similar to p53, transactivation-competent p73 isoforms can activate autophagy, presumably through nuclear activity.10 However, the upstream signals that regulate p73 to do so, and the target genes that mediate autophagy downstream of p73 have been largely unknown. Unlike p53, p73’s ability to induce autophagy is DRAM-independent.10 In addition, it is not known whether p73 can translocate to the cytoplasm to regulate autophagy as p53 does.
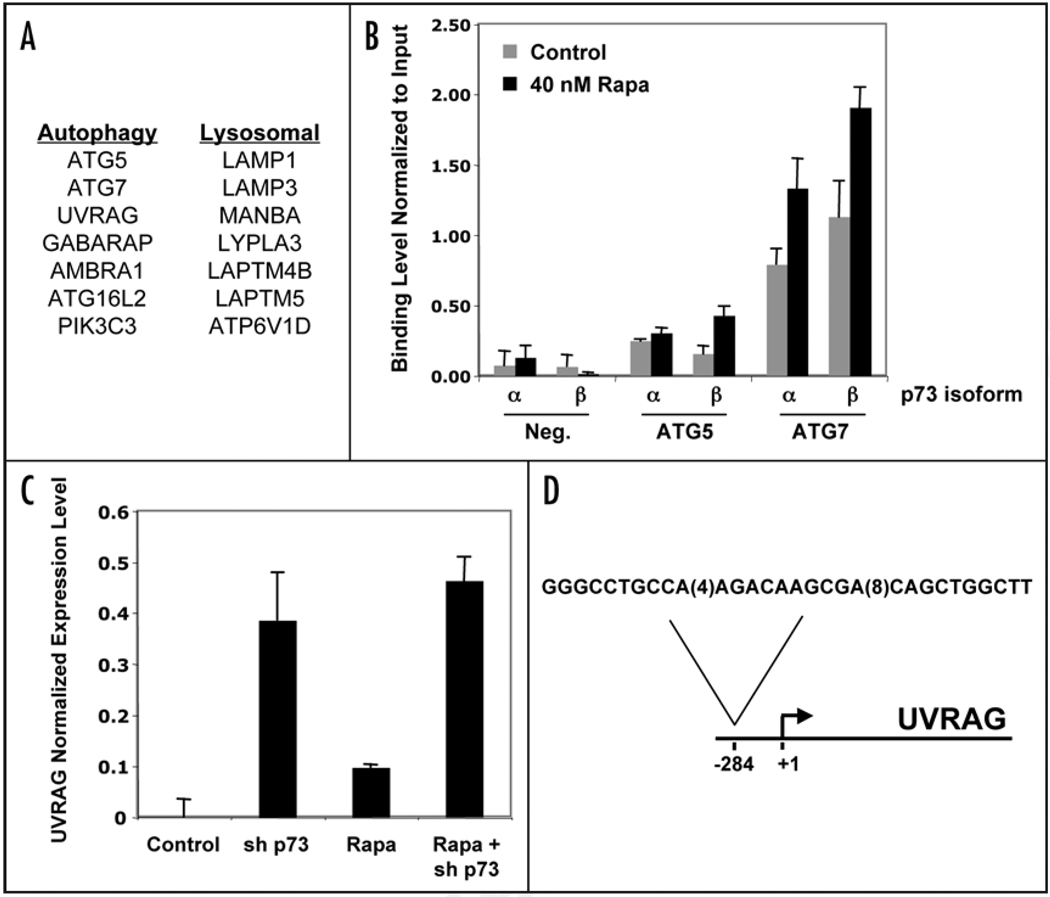
To explore the nuclear function of p73 further, we identified p73-regulated genes in a rhabdomyosarcoma cell line. Chromatin immunoprecipitation experiments demonstrated that p73 bound to genomic sites near many genes associated with autophagy or with lysosomal function (Fig. 1A). Both of the p73 isoforms expressed in the cell line used bound to sites near autophagy-associated genes such as ATG5 and ATG7, and in some cases the binding level was enhanced after treatment with rapamycin (Fig. 1B and data not shown). Thus, p73 regulates a network of genes, and the cumulative effect of this network may be to enhance and/or fine-tune the autophagic response.
Figure 1.
Analysis of autophagy-associated, p73-regulated genes. (A) Chromatin immunoprecipitation analysis was performed in Rh30 cells, and p73 binding was detected within 10 kb of the indicated autophagy-associated and lysosome-associated genes. (B) Chromatin immunoprecipitation coupled with real-time PCR was used to measure p73 binding at a negative control region, or at sites within 10 kb of the ATG5 and ATG7 genes, using antibodies specific for either the α or β isoform of p73. Cells were pre-treated with vehicle or 40 nM rapamycin in serum-free medium for 24 h before formaldehyde cross-linking and isolation of protein-DNA complexes. (C) Microarray analysis of duplicate samples was used to assess UVRAG RNA levels in Rh30 cells treated with vehicle or 40 nM rapamycin for 24 h and/or lentivirus expressing p73 RNAi. (D) Location of a p73-bound p53 response element in relation to the UVRAG transcriptional start site.
These data suggest a role for p73 that is similar to that of nuclear p53, to activate target genes that regulate autophagy. However, the role of p73 may not be entirely straightforward. In the case of at least one autophagy-associated gene (UVRAG), microarray analysis revealed that p73 knockdown increased UVRAG expression levels (Fig. 1C). In addition, chromatin immunoprecipitation showed that p73 bound to a region upstream of the UVRAG transcriptional start site that contains a p53 family response element (Fig. 1D). p73 may directly suppress UVRAG expression, perhaps through recruitment of transcriptional repressors such as histone deacetylases. In this case, p73 would be expected in inhibit autophagosome nucleation and maturation.17,18 Our recent studies have revealed an entire network of autophagy-associated genes that are regulated by p73, and the multiple mechanisms by which this pattern of genes may alter the autophagic process remains unknown.
Our observations that mTOR negatively regulates p73, and that p73 regulates many genes associated with autophagy, suggest both similarities and differences in the ways that p53 family members respond to metabolic stress and regulate autophagy. The role autophagy plays in the different in vivo functions of the p53 family remains to be determined. p53 is one of the most frequently mutated genes in human cancer. The roles of p63 and p73 in tumor suppression have long been controversial, although a recent isoform-specific p73 mouse model suggests that p73 is indeed a bona fide tumor suppressor.19 In addition, p63 and p73 play critical roles during development.1 For example, p63 plays an essential role in the development of the epidermis and mammary gland.20 Loss of p73 leads to hippocampal dysgenesis, gastrointestinal erosion, hemorrhage, and an increased propensity for neurodegenerative disease in mice.21,22 Furthermore, all three p53 family members are associated with aging or aging-related pathology.22–24 In fact, the effect of the p53 orthologue CEP-1 on the life span of C. elegans is mediated by autophagy.25 Thus, the discovery that p53 family members regulate autophagy may have implications for a wide range of human diseases.
Acknowledgements
We thank P. Houghton (St. Jude Children’s Research Hospital) for kindly providing Rh30 cells, and Deb Mays for technical assistance. Chromatin immunoprecipitation was performed using GenPathway, Inc., (San Diego, California) services. This work was supported by the National Institutes of Health Grants CA70856 and CA105436 (J.A. Pietenpol), ES00267 and CA68485 (core services), US Army Medical Research and Materiel Command Grant W81XWH-04-1-0304 (J.M. Rosenbluth), and GM07347 (MSTP training).
Footnotes
Previously published online as an Autophagy E-publication: http://www.landesbioscience.com/journals/autophagy/article/7293
Addendum to: Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol 2008; 28:5951-64; PMID: 18678646; DOI: 10.1128/MCB.00305-08.
References
- 1.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 2.Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: Rising from the shadow of p53. Drug Resist Updat. 2008 doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbluth JM, Pietenpol JA. The jury is in: p73 is a tumor suppressor after all. Genes & development. 2008;22:2591–2595. doi: 10.1101/gad.1727408. [DOI] [PubMed] [Google Scholar]
- 4.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nuc Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 9.Bell HS, Ryan KM. Targeting the p53 family for cancer therapy: ‘big brother’ joins the fight. Cell Cycle. 2007;6:1995–2000. doi: 10.4161/cc.6.16.4614. [DOI] [PubMed] [Google Scholar]
- 10.Crighton D, O’Prey J, Bell HS, Ryan KM. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ. 2007 doi: 10.1038/sj.cdd.4402108. [DOI] [PubMed] [Google Scholar]
- 11.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Inves. 2007;117:1370–1380. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK β1, TSC2 and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 17.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 19.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Devel. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 21.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 22.Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA, Cole CJ, Lerch JP, Spring S, Wang SH, Frankland PW, Henkelman RM, Josselyn SA, Sadikot AF, Miller FD, Kaplan DR. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer’s disease. Neuron. 2008;59:708–721. doi: 10.1016/j.neuron.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Matheu A, Maraver A, Serrano M. The Arf/p53 pathway in cancer and aging. Cancer Res. 2008;68:6031–6034. doi: 10.1158/0008-5472.CAN-07-6851. [DOI] [PubMed] [Google Scholar]
- 24.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Devel. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]