Abstract
Polydimethylsiloxane (PDMS) has become a staple of the microfluidics community by virtue of its simple fabrication process and material attributes, such as gas permeability, optical transparency, and flexibility. As microfluidic systems are put toward biological problems and increasingly utilized as cell culture platforms, the material properties of PDMS must be considered in a biological context. Two properties of PDMS were addressed in this study: the leaching of uncured oligomers from the polymer network into microchannel media, and the absorption of small, hydrophobic molecules (i.e. estrogen) from serum-containing media into the polymer bulk. Uncured PDMS oligomers were detectable via MALDI-MS in microchannel media both before and after Soxhlet extraction of PDMS devices in ethanol. Additionally, PDMS oligomers were identified in the plasma membranes of NMuMG cells cultured in PDMS microchannels for 24 hours. Cells cultured in extracted microchannels also contained a detectable amount of uncured PDMS. It was shown that MCF-7 cells seeded directly on PDMS inserts were responsive to hydrophilic prolactin but not hydrophobic estrogen, reflecting its specificity for absorbing small, hydrophobic molecules; and the presence of PDMS floating in wells significantly reduced cellular response to estrogen in a serum-dependent manner. Quantification of estrogen via ELISA revealed that microchannel estrogen partitioned rapidly into the surrounding PDMS to a ratio of approximately 9:1. Pretreatments such as blocking with serum or pre-absorbing estrogen for 24 hours did not affect estrogen loss from PDMS-based microchannels. These findings highlight the importance of careful consideration of culture system properties when determining an appropriate environment for biological experiments.
Introduction
Over the past decade, the silicon-based elastomer polydimethylsiloxane (PDMS) has found widespread use within the rapidly developing field of microfluidics. Its simple fabrication process and design adaptability have made it a popular polymer for applications ranging from surface micropatterning1 to the casting of 2D and 3D geometries2–4 from stiff master molds. PDMS is both optically transparent and gas permeable, and it spontaneously and reversibly adheres to commonly used substrates such as polystyrene and glass. These characteristics render it compatible with optical and fluorescence microscopy, sufficient for gas exchange over extended culture periods, and ideal for quickly assembled and disassembled microscale culture chambers.
In the last few years, PDMS-based microfluidic systems have been increasingly utilized in cell culture applications and are the subjects of recent reviews.5–7 Attempts have been made to characterize the effects of channel geometry and size, as well as cell type and seeding conditions, on cell behavior in PDMS-based culture devices;4,8–11 however, little attention has been paid to potential experimental artifacts introduced by the material and chemical properties of PDMS.12 While exhaustive validation may not be necessary for microfluidic sorting or separating systems in which cells are only transiently exposed to PDMS, the need to fully vet its impacts on cells becomes paramount as culture time increases and when proliferation and/or protein expression are desired endpoints. It has recently been observed that basal levels of glucose consumption, as well as metabolic and stress pathways, of mouse mammary fibroblasts are significantly different in PDMS-based microchannels from those seen in 96-well plates.13 The direct causes of these differences (whether specific to the microscale or the material) have yet to be determined. It is important to separate material-induced artifacts from biological effects resulting from microscale culture if these systems are to find widespread use.14
PDMS exists as a crosslinked polymer of hydrophobic dimethylsiloxane oligomers, which raises two concerns about its use in cell culture systems: firstly, that there are residual uncrosslinked oligomers that may leach from the bulk polymer into the culture medium; and secondly, that the porous, hydrophobic network may sequester small hydrophobic molecules (e.g. steroid hormones) from culture media.
Uncrosslinked oligomers
PDMS curing is a time- and temperature-dependent process that does not achieve 100% crosslinking. It has been demonstrated previously that after extensive curing, as much as 5% (w/w) of the PDMS bulk remains uncrosslinked and extractable with organic solvents.15 These freely diffusive oligomers are often referred to as low molecular weight (LMW) species16–18 and are held to be largely responsible for many of the elastomer's defining surface characteristics. Diffusion of LMWs to the polymer surface from the bulk plays a significant role in hydrophobic recovery after plasma treatment by helping to mask hydrophilic surface groups.16,18–20 Additionally, the collection of LMW residues on the surface results in a lubricating effect that facilitates both sealing to and removal from substrates. Several studies have attempted to address the LMW issue for the purpose of maintaining surface hydrophilicity following plasma treatments. Techniques previously used to neutralize uncured oligomers include removal by Soxhlet extraction,16 serial extractions in organic solvents,15 and extended oven baking to drive crosslinking to completion.17
Though the mobility of LMWs within the polymer bulk is known, there has been little exploration as to whether they escape the confines of the elastomer network to enter the aqueous culture environment of PDMS-based microfluidic channels. Removal of LMWs via extraction has been connected with extended survival time of neurons cultured at low density in microchannels,21 and LMWs have been shown to migrate into gelatin after extended contact with PDMS gel sheets,22 but the actual prevalence and consequence of PDMS oligomers in culture media are not well characterized.
Hydrophobic absorption
Due to its hydrophobic nature, PDMS has a high solubility in nonpolar organic solvents,15 resulting in swelling of the polymer. Interest in the utilization of microchannels for organic reactions has prompted the exploration of surface modification techniques such as coating with glass23 and metal oxides,24 as well as replacing PDMS altogether with fluorinated compounds,25 in an effort to reduce the absorption of organic solvents into the polymer bulk. From a cell culture perspective, this same need exists in the form of maintaining important hydrophobic serum-borne molecules available to cells in culture.
It has been demonstrated that the small, hydrophobic fluorophore nile red (318.37 Da) is strongly absorbed by PDMS.26 Toepke and Beebe calculated that the ability to maintain a quinine (324 Da) solution at a constant 2 μM would require the pre-absorption of the equivalent of 100 channel volumes of the quinine solution. While the exact number is specific to their microchannel dimensions, the principle that is illustrated is that few assumptions about media component concentrations are valid in the face of an actively participating polymer container. Chemically defined media constituents will selectively partition into the PDMS bulk in a solubility-dependent manner. For serum-borne steroids such as estradiol, progesterone, and aldosterone (272.39, 314.47, and 360.44 Da, respectively), which occur in serum in the low nanomolar range, absorption into the PDMS bulk could effect a significant alteration to the cellular microenvironment and have a corresponding impact on hormone-dependent cell behavior. Estrogen and progesterone are of paramount importance in mammary gland development, and hormone responsiveness has significant prognostic value in breast cancer. Studying the estrogen-dependent behavior of breast cancer cell lines therefore necessitates that a culture system does not interfere with medium estrogen levels or estrogen pathway components.
The leaching of LMWs and the absorption of hydrophobic molecules into PDMS could introduce material artifacts resulting from cytotoxicity or interference with cellular signaling pathways. Fundamentally, these issues preclude direct comparisons of cellular behavior between macroscale wells and microscale channels, limiting the validation of microfluidic systems as a powerful new development in the evolution of in vitro cell culture. In this paper we explore the extent to which PDMS oligomers escape the parent matrix and interact with cells in culture. In addition, we attempt to quantify the absorption of estrogen into PDMS and demonstrate the impact this has on the estrogen responsiveness of human breast cancer cell lines.
Results
Uncured PDMS oligomers are detectable in microchannel media and cell membranes
The solubility of crosslinked polymers is not measured by their dissolution in a solvent, but by the degree of swelling induced by a solvent.15 Swelling of the polymer network increases solvent availability to uncured oligomers within the bulk and facilitates the removal of those oligomers. Water is the solvent in which PDMS has the lowest solubility15 and is therefore the least likely to extract detectable amounts of uncrosslinked oligomers from the polymer bulk. However, as water is the universal biological solvent, any leaching of uncrosslinked PDMS into it is likely to have consequences for PDMS-based cell culture systems. To mimic 24 hours of cell culture without confounding signal contributions from serum or saline components, DIUF water was incubated for 24 hours in PDMS microchannels (Fig. 1) and subsequently analyzed via matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS, see experimental section). Uncured PDMS oligomers were visible as a regularly repeating signal extending from under 1500 m/z to over 7000 m/z (Fig. 2a). Given a mass of 74.15 Da for the dimethylsiloxane monomer, and assuming single charges on ionized oligomers, these bounds indicate that PDMS oligomers were present in a continuous range of fewer than 20 to more than 90 subunits. Detection of uncrosslinked PDMS in solution casts doubt on the biological inertness of the polymer because the potential interactions of PDMS oligomers with medium components, or with cells themselves, are unknown.
Fig. 1.
A PDMS-based microfluidic channel of the type used in this study.
Fig. 2.
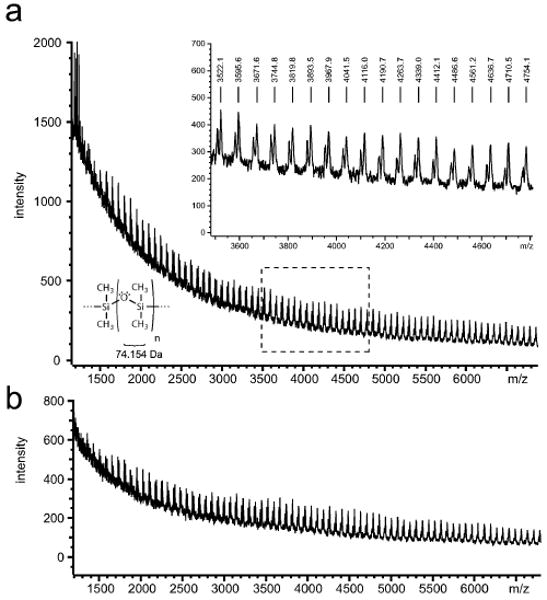
PDMS oligomers are detectable in microchannel media. (a) Uncured PDMS oligomers were detectable via MALDI-MS in DIUF water aspirated from microchannels after a 24-hour incubation. (b) Uncrosslinked oligomers were still detected in water aspirated from Soxhlet-extracted microchannels.
To reduce or eliminate the presence of uncured PDMS in microchannel media, PDMS microchannels underwent overnight Soxhlet extraction in ethanol. Though PDMS is not highly soluble in ethanol,15 ethanol was desirable as a readily available, volatile, and non-noxious organic solvent that would not require tedious stepwise removal from the polymer. PDMS devices that underwent overnight Soxhlet extraction consistently lost 4% of their total mass (Figure S1†); however, PDMS oligomers were still detectable via MALDI MS in the aspirate from extracted microchannels after 24 hours of incubation (Fig. 2b), indicating that complete extraction was not achieved.
The presence of PDMS oligomers in microchannel media is troublesome if they have any interaction with cells or media components. Due to the hydrophobicity of both PDMS oligomers and the lipid interior of cell membrane bilayers, it was expected that oligomers that came into contact with cells would be likely to remain there, and accumulated PDMS would therefore be detectable in cell membranes. Energy-dispersive X-ray spectroscopy (EDS) is a technique commonly employed in the analysis of minerals that produces spectral peaks indicating the atomic composition of the surface and near-surface of a sample. As silicon is not a typical component of cell membranes, and PDMS is nearly 40% silicon by weight, its presence there is an indicator of accumulation of uncrosslinked oligomers. EDS was utilized in conjunction with SEM (Fig. 3a and 3b) to assess the silicon content (Fig. 3c and 3d) of normal mouse mammary epithelial (NMuMG) cells that had been previously cultured for 24 hours in either extracted or non-extracted PDMS microchannels. To avoid background silicon signals arising from silicates within glass, the cells were cultured on polystyrene microscope slides; PDMS was removed after fixing and washing. Silicon was spread diffusely across cell populations cultured within non-extracted microchannels (Fig. 3c); the silicon peak at 1.7 keV was small but present (Fig. 3e). Cells from Soxhlet-extracted microchannels showed a qualitative reduction in both the spatial distribution (Fig. 3d) and spectral peak of silicon (Fig. 3f). That silicon was still detectable in the membranes of cells cultured in extracted microchannels was consistent with the finding that PDMS oligomers were still detectable in media from extracted microchannels (Fig. 2b). However, interpretation of these data are complicated by both the small peak sizes and by previous documentation that there is a minimum silicon signal associated with the X-ray detector itself.27
Fig. 3.
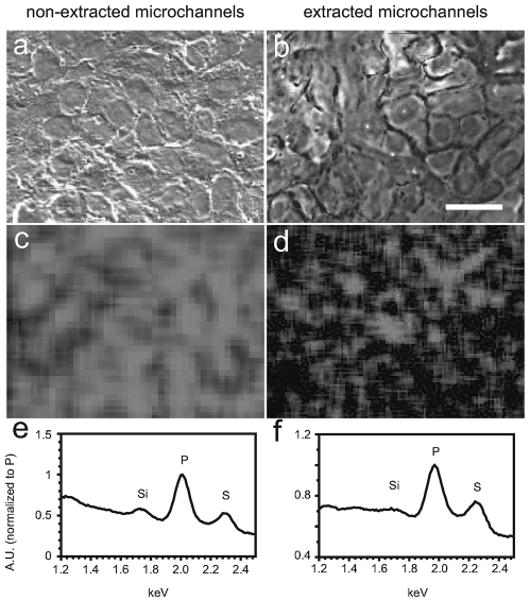
PDMS oligomers are detectable in cell membranes. SEM of NMuMG luminal cells cultured in non-extracted PDMS (a) or Soxhlet-extracted PDMS (b) microchannels, with corresponding EDX maps of silicon content over the same regions of interest (c, d). Silicon was profusely distributed in the plasma membranes of cells cultured in non-extracted PDMS microchannels for 24 h. The silicon content was substantially reduced but detectable in the membranes of cells cultured in extracted microchannels. Spectra showing silicon, phosphorus, and sulfur peaks for both cases represent full-image regions of interest (e, f). Scale bar = 20 μm.
Uncured PDMS leaching from the polymer bulk into culture medium complicates interpretation of data from microchannels. Free-floating low molecular weight oligomers may or may not interfere with normal cell behavior upon incorporation into the plasma membrane. The ability to isolate uncured PDMS oligomers as a side product of Soxhlet extraction theoretically provides a means of assessing the biological impact of the oligomers directly, though to date such experiments have not been performed.
PDMS absorbs culture media estrogen
In light of the hydrophobic nature of PDMS, it is expected that PDMS plays at least some role in the sequestration of hydrophobic molecules. The extent to which this directly affects steroid hormones remains unknown, as does the degree of consequence this has on cell signaling. Fluorophores of similar molecular weight and solubility to common steroid hormones have been shown to absorb readily into the polymer bulk,26 intimating that a similar behavior could be expected of the hormones.
It was expected that PDMS would sequester hormones to a degree inversely proportional to their water solubility. Estrogen (E2, 272 Da) was expected to absorb into the polymer while large hydrophilic molecules such as prolactin (PRL, 24 kDa) were not. To this end, MCF-7 human breast cancer cells28 were transiently transfected with a luciferase reporter driven by Activator Protein 1 (AP-1) enhancer elements. AP-1 activation is a downstream event in both estradiol and PRL signaling, which results in luciferase expression in this system.29 Under normal culture conditions in 12-well plates, both estrogen and PRL were able to elicit luciferase expression significantly above vehicle control (p < 0.05); however, when these cells were seeded on PDMS inserts inside the wells, expression of the AP-1-driven luciferase reporter was responsive to PRL (p < 0.001) but unchanged from control in response to estrogen (Fig. 4). This was consistent with expectations that PDMS selectively sequesters small hydrophobic molecules, such as steroid hormones, and significantly reduces their availability to cells.
Fig. 4.
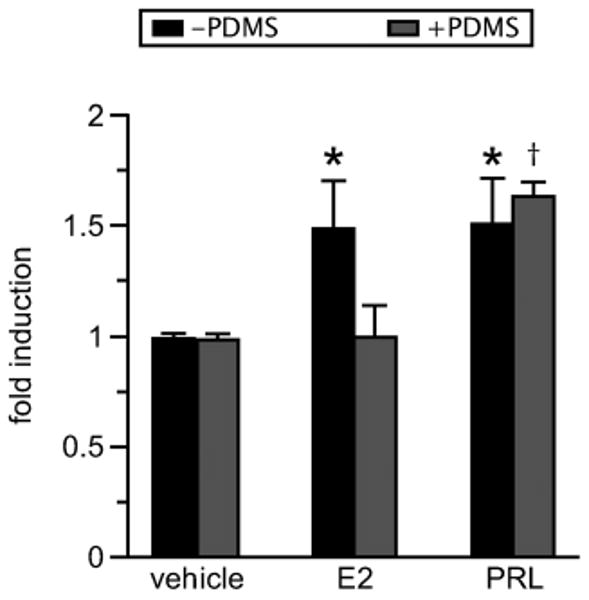
PDMS selectively inhibits estrogen signaling. MCF-7 cells were transiently transfected with a 4xAP-1-luciferase construct and seeded in 12-well plates with or without PDMS inserts. Cells in direct contact with PDMS (seeded on PDMS inserts) (+PDMS) displayed significant AP-1 activity in response to prolactin (PRL, 4 nM) but not estrogen (E2, 1 nM). Cells cultured in the absence of PDMS (−PDMS) displayed significant AP-1 activity in response to both estrogen and PRL (n = 3 experiments). *p < 0.05; †p < 0.001.
A sub-line of MCF-7 cells (designated MVLN by the creators30) was previously stably transfected with a construct containing the consensus estrogen response element upstream of the Herpes Simplex Virus-thymidine kinase promoter and luciferase gene. MCF-7 MVLN cells were cultured in opaque 96-well plates in order to provide a robust signal in a reliable plate reader format. The equilibrial interaction between PDMS and estrogen could be demonstrated by estrogen's absorption into and release from PDMS in a dose- and extraction-dependent manner, as evidenced by both MCF-7 MVLN luciferase induction and direct [E2] assessment via ELISA.
Just as PDMS strips serum-containing medium of its estrogen content, it also releases sequestered estrogen back into media that are otherwise devoid of it. Small, thin pieces of PDMS—either 10, 20, or 30 mg, both flat and cuboidal—that had been previously incubated for 24 hours in serum-free medium supplemented with 10−7 M estrogen were thoroughly rinsed and transferred to MCF-7 MVLN cells cultured in serum-free medium. The transferred PDMS induced luciferase expression almost two-fold over control (p < 0.005 each) in a manner independent of the shape or mass of the pieces, suggesting that the PDMS pieces had not become saturated with estrogen during the 24-hour incubation (Fig. 5a).
Fig. 5.
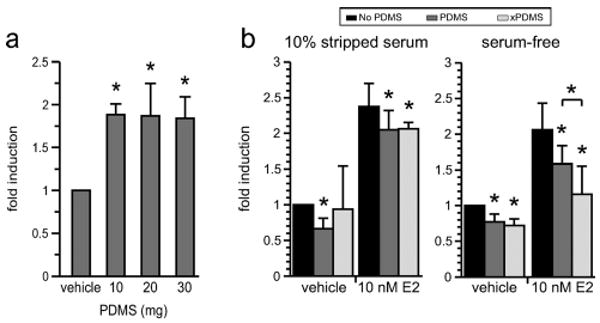
PDMS absorbs and releases estrogen. PDMS exhibits capacitance for small, hydrophobic molecules. (a) Pieces of PDMS of mass 10, 20, or 30 mg were incubated individually for 24 hours in 100 μl of serum-free media containing 10−7 M E2. When thoroughly rinsed and transferred to wells containing cells in stripped serum, estrogen was released from the PDMS in a manner independent of the mass, indicating that the PDMS was not saturated with estrogen (p < 0.005 each, n = 3 experiments). (b) MCF-7 MVLN cells stably expressing luciferase were cultured in 10% stripped serum or serum-free media in the presence of no PDMS (No PDMS), non-extracted PDMS (PDMS), or extracted PDMS (xPDMS) pieces. Left, Cells cultured in stripped serum exhibited no difference in luciferase induction between PDMS treatments, though both conditions caused a statistically significant drop in induction from no-PDMS controls. Right, Cells cultured in the absence of serum showed a clearer distinction between non-extracted and extracted PDMS in response to estrogen. Both PDMS conditions effected a statistically significant inhibition of estrogen signaling, and at 10−8 M estrogen were significantly different from each other (p < 0.05, n = 3 experiments).
MCF-7 MVLN cells were responsive to supplemented estrogen in a dose-dependent manner in both the presence and absence of floating PDMS. The presence of floating PDMS pieces, 10–14 mg each, decreased the luciferase signal resulting from estrogen addition over a range of concentrations (Figure S2†). Luciferase induction was generally indistinguishable between cell populations exposed to extracted and non-extracted PDMS, regardless of serum condition, though there was a significant difference in induction between PDMS treatments at the highest estrogen concentration in serum-free media (10−8 M, Fig. 5b). While extraction status did not significantly affect the outcome of PDMS exposure at most estrogen concentrations, a trend did exist within all serum-free experiments such that extracted PDMS inhibited estrogen signaling slightly more than non-extracted PDMS. This trend is visible in Figure S2†. Though it is not essential to the conclusions of the paper, it does corroborate our expectations that PDMS has a greater absorptive capacity upon extraction of uncrosslinked oligomers. That this trend is not visible in the corresponding 10% serum conditions is likely a result of the presence of water-soluble growth factors, which largely pass through charcoal stripping, that are capable of activating the estrogen receptor and partially masking the inhibition caused by PDMS.
While the luciferase signals from cell populations cultured in the presence of PDMS were significantly lower than the signals from those cultured without PDMS, the relative depletion of estrogen should be exacerbated in microfluidic channels in which fluid volumes are two orders of magnitude smaller and completely contained within PDMS. Estrogen depletion from microchannel media was assessable by pooling aspirated microchannel media and subjecting them to ELISA (Fig. 6). Phenol red-free DMEM supplemented with 1 nM estrogen was added to microchannels and collected immediately and after 1, 3, 6, 12, and 24 hours of incubation at 37 °C. No cells were cultured in the microchannels during incubation. The estrogen concentrations of collected media were then quantified via ELISA. The transfer of estrogen from microchannels to PDMS was rapid, with most loss occurring within the first three hours of incubation. The partitioning of estrogen into PDMS reached ∼90% within the observed 24 hours. To determine whether this initial absorption of estrogen achieved at least partial saturation of the PDMS, microchannel media were aspirated and replaced with new media at 24 hours. An identical time course was observed over the subsequent 24 hours, revealing the same pattern of estrogen partition into the PDMS. At 48 hours, microchannel estrogen concentrations were again approximately one order of magnitude lower than at filling, suggesting that PDMS saturation did not occur to any appreciable degree in the course of the experiment. That the measured concentration of estrogen in PDMS microchannels was the same at 24 and 48 hours, in spite of replenishment, reflects the high partition coefficient between saline water and PDMS, excepting that estrogen had not yet equilibrated within the entire PDMS bulk. An alternate hypothesis, that serum protein adsorption would serve to block the PDMS surface and mitigate estrogen absorption, also proved ineffective. Pretreatment of extracted microchannels for 24 hours with 100% stripped serum provided maximal serum protein presence while eliminating pre-absorption of estrogen into the polymer bulk; however, microchannels with this pretreatment showed no improvement in estrogen retention in the second 24 hours.
Fig. 6.
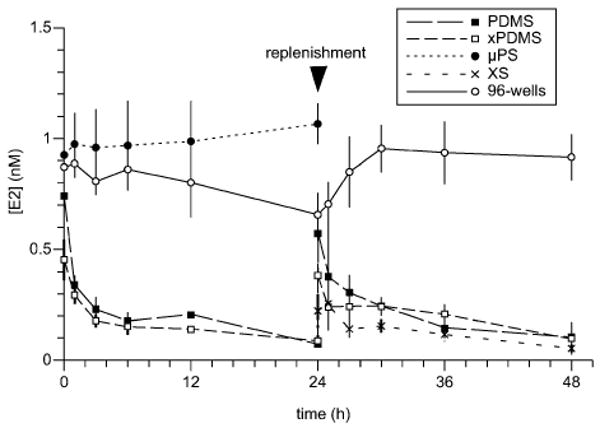
Time-dependent estrogen loss from microchannel media. Estrogen was lost to the surrounding PDMS in all PDMS-based microchannels, irrespective of pretreatment. Microchannels made from either non-extracted PDMS (PDMS) or Soxhlet-extracted PDMS (xPDMS) lost estrogen readily in the first 24 hours after filling with 1 nM estrogen-supplemented, phenol red-free DMEM. Other extracted channels were first incubated for 24 hours with estrogen-supplemented DMEM (xPDMS) or 10% stripped serum (XS) and sampled over the following 24 hours. Neither pretreatment was able to prevent estrogen absorption into the bulk during the second 24 hours. Polystyrene wells (96-wells) and tissue-culture treated polystyrene microchannels (μPS) did not lose estrogen progressively over time}.
The time-dependent reduction in estrogen concentration was not due to hormone degradation. Media from wells of a non-tissue culture treated 96-well plate were collected over the same time intervals and did not exhibit estrogen loss within 48 hours. Traditional well plates are constructed of dense polystyrene, and adsorption, not absorption, is the primary means of medium component reduction in wells. In addition, the low surface area-to-volume ratio (SAV) of wells reduces the impact of surface adsorption on medium component concentrations. Tissue culture-treated polystyrene microchannels (Bellbrook Labs) of similar SAV to PDMS microchannels were sampled over the course of 24 hours and likewise showed no reduction in medium estrogen concentration (Fig. 6). The apparent trend visible around 24 hours in the 96-well track is not real. It is most likely attributable to inconsistent mixing of well contents during collection that failed to homogenize the estrogen distribution. The same artifact is not expected to have occurred in microchannels because the entire contents of multiple microchannels were pooled together, as opposed to the fraction of the volume of a well.
Discussion
From an engineering standpoint, PDMS has been a tremendous asset to the microfluidics community. Its ease of use, as well as inherent optical and mechanical properties, have catered to a wide range of designs and applications and played well to the creativity of researchers. From a biological standpoint, several unknowns limit its acceptance as a culture environment. As a manually mixed, crosslinked polymer of hydrophobic precursors, the two most worrisome complications that PDMS poses for cell culture applications are that of incomplete crosslinking and absorption of small nonpolar molecules.
Incomplete crosslinking of PDMS would, in and of itself, be cause for little concern if no uncrosslinked oligomers made their way into solution. However, a wide range of oligomer lengths were detectable in microchannels via mass spectrometry. Furthermore, silicon atoms—implicating PDMS—appear to be localized to the plasma membranes of cells via EDS after 24 hours of culture in PDMS microchannels. To date, though, there has been no direct evidence indicting leaching PDMS in cell stress. Reduction of uncrosslinked oligomers from PDMS is straight-forward and consistent via Soxhlet extraction in ethanol. However, given the relatively low solubility of PDMS in ethanol on a scale of organic solvents,15 it is not likely that the extractions described in this paper ran to completion. Complete extraction would likely require the use of highly hydrophobic organic solvents in which PDMS is highly soluble, which would maximize PDMS swelling and solvent availability to uncrosslinked oligomers throughout the bulk. Many of these chemicals, however, are themselves extremely toxic and must be removed in a time-consuming step-wise manner. The degree of extraction achieved by ethanol was insufficient to reduce oligomers to undetectable levels, yet a lengthy stepwise extraction procedure involving toxic hydrocarbons and amines is unappealing from the standpoint of both safety and throughput.
It is not clear what precise effects, if any, leaching PDMS oligomers have on cell viability or behavior. Recently, mouse mammary fibroblasts (MMF) were found to have elevated basal levels of glucose consumption, as well as transiently high activation of ribosomal subunits and indications of increased endoplasmic reticulum stress, when cultured in PDMS microchannels. p16(Ink4a)−/− MMFs exhibited cell cycle defects in PDMS microchannels not seen in wells.13 The source of these alterations has not been elucidated, but speculation that plasma membrane-incorporated LMW PDMS may adversely affect outside-in signaling requires little imagination. Long chain hydrocarbon-like molecules such as these may impede the interactions of signaling complexes or improperly tether them. On the other hand, they may do nothing. Also possible as a source of cellular stress is the reduction of estrogen-mediated signaling.
Sequestration of hydrophobic molecules was demonstrated to have consequences for cells in culture. MCF-7 cells responsive to both PRL and estrogen exhibited selective induction of AP-1-driven luciferase expression in the presence of PDMS. PRL is a water-soluble hormone, and estrogen is a hydrophobic one; cells seeded directly on PDMS displayed significant luciferase expression in response to PRL but not estrogen. Furthermore, direct cell contact with PDMS was not necessary to witness its effects. MCF-7 MVLN cells responsive to estrogen showed reduced induction of luciferase substrate when small PDMS pieces were floated on top of the well medium. Extracted PDMS decreased induction further still, though only slightly. This trend was visible in cells cultured in serum-free medium, but not in cells cultured in 10% stripped serum. As ER is activated by other serum components besides estrogen, it is possible that water-soluble growth factors were capable of masking the additional hydrophobic capacity of the extracted PDMS pieces. An implication of this for microfluidic cell cultures utilizing serum-containing media is that as time passes after media replenishment, an increasing percentage of estrogen receptor activation may be attributable to non-estrogen-mediated pathways.
Preventing the hydrophobic absorption of small molecules into the PDMS bulk is not trivial. When microchannels were filled with estrogen-supplemented DMEM and sampled over the course of 24 hours, the estrogen content, as measured by ELISA, decreased consistently in channels to a level indicative of a high local partition ratio between DMEM and PDMS (Fig. 6). As expected, this pattern was matched in extracted microchannels. Neither type saw improvement after a 24-hour pretreatment with the same estrogen-supplemented DMEM. The latter observation implied that local saturation of PDMS with estrogen did not occur in the first 24 hours, and again did not occur within the next 24 hours after replacement with new media. An additional hypothesis, that the PDMS surface could be effectively blocked by serum proteins adsorbing to it, was tested on extracted PDMS channels. However, pretreatment for 24 hours with stripped serum performed no better, and arguably worse, than pre- treatments with estrogen-supplemented DMEM. In all cases, the concentration of estrogen fell by approximately an order of magnitude from 1 to 0.1 nM, a level not below physiological relevance; however, common cell culture media utilizing 10% serum would experience a reduction from the order of 0.1 to the order of 0.01 nM estrogen. While not culminating in total depletion, the reduction of medium estrogen was significant and consistent. Media collected from a 96-well plate over the same duration showed no time-dependent loss of estrogen, reflecting the inability of polystyrene to absorb hydrophobic molecules. Polystyrene microchannels likewise exhibited no reduction in estrogen concentration over 24 hours, despite a high SAV. Adsorption of estrogen to the inner surfaces of these microchannels was undoubtedly reduced by the hydrophilizing tissue culture treatment.
In summary, the leaching of uncrosslinked PDMS oligomers into culture media can potentially be combated in many ways. Among them, surface treatments may provide a reliable barrier against oligomer escape; the crosslinking reaction can be forced toward completion during curing, or uncrosslinked oligomers can be removed from the polymerized network post-cure. A combination of these may prove the most effective. This study took the last approach via Soxhlet extraction in ethanol, with the condition that noxious organic solvents be avoided. While devices undergoing this extraction procedure lost 4% of their mass, extraction was not complete, as oligomers were detected post-extraction in both microchannel media and cell membranes. EDS spatial mapping and spectral analysis of cell membranes indicated a decreased presence of silicon post-extraction, though the technique was not quantitative. The tradeoff of removing uncrosslinked oligomers from the polymer bulk is the subsequent increase in absorption of hydrophobic hormones. This was demonstrated as a decrease in luciferase induction by cells in response to supplemented estrogen in serum-free media; however, the effect was easily masked upon culturing in stripped serum. Two hypotheses for reducing estrogen absorption into PDMS were tested, both employing a 24-hour pretreatment of microchannels. Neither pre-saturation of the immediately surrounding PDMS nor blocking of the PDMS-liquid interface with serum proteins was able to prevent the rapid reduction of estrogen concentration. The microchannels with the highest estrogen retention were in fact not made of PDMS at all.
Experimental
PDMS device preparation and Soxhlet extraction
Polydimethylsiloxane (Sylgard 184, Dow) base and crosslinker were mixed manually at a 10:1 ratio and degassed for 45–60 minutes under vacuum at room temperature. The degassed PDMS was then poured over SU-8 master molds, cured at 80 °C for 4 hours, and allowed to cool to room temperature prior to removal from the master mold. PDMS devices to be extracted were then placed in the body of a 500 ml Soxhlet extractor. Extraction was performed overnight in ethanol (typically 14–20 hours) and resulted in the accumulation of uncrosslinked oligomers in the collection flask of the extractor apparatus. The PDMS devices were then removed from the extractor, and residual ethanol was evaporated. Finally, all devices, extracted or not, were sterilized via autoclave prior to adhesion to polystyrene omnitrays (Nunc) and subsequent cell seeding. Microchannels utilized in these experiments were 4.5 mm long from port center to port center, 670 μm wide, 250 μm tall in the channel, 750 μm tall at the port (Fig. 1).
Unlike dynamic, pump-based microfluidic systems, static microfluidic culture is continuously affected by evaporation. Evaporation was circumvented by the use of bioassay dishes containing an arbitrary but substantial volume of autoclaved water, into which the PDMS device-containing omnitrays were placed. As an additional measure, 5-μl sacrificial drops of sterile liquid (PBS or water) were placed liberally throughout each omnitray to minimize the contribution of the microchannels to saturating the airspace within the omnitray. These precautions were taken without exception whenever microchannels were incubated at 37 °C.
PDMS oligomer detection in microchannels
Two full plates of microchannels were prepared as above, one being subsequently extracted and one not. All 192 microchannels of each plate were filled with 5 μl of deionized ultra-filtered (DIUF) water and incubated for 24 hours at 37 °C. The contents of all microchannels were aspirated and pooled in a microcentrifuge tube, lyophilized, and analyzed by a MALDI-TOF mass spectrometer (REFLEX II, Bruker). X-axes report mass-to-charge ratio (m/z) and are interpreted as mass in daltons (Da), assuming single charges on ionized oligomers. Thus, peaks repeating at a regular interval indicate a distribution of oligomer lengths with the interval indicating the molecular weight of the monomer.
PDMS oligomer detection in cell membranes
Extracted and non-extracted PDMS microchannels were adhered to tissue culture treated polystyrene microscope slides and seeded with NMuMG epithelial cells. After 24 hours in culture at 37 °C, the cells were fixed with 4% paraformaldehyde for 20 minutes and washed 3× with PBS. The PDMS was removed, and cells were washed 3× in deionized water and allowed to dry at room temperature. The slides were then stored in slide holders until analysis.
Energy-dispersive X-ray spectroscopy (EDS) was employed to analyze the atomic composition of cell membranes. SEM images were acquired with an S-3400N scanning electron microscope (Thermo Electron Corp.). Energy-dispersive X-ray maps were created by scanning the field of view with an electron excitation source at 5 keV, collected at 11000 counts/sec, 40% dead time. The depth of penetration of the EDS mapping with the given applied energy was ∼1 μm. The Si signal maps were smoothed with a 7 × 7 filter, and the spectral background was automatically subtracted, resulting in the images of net signal above background given in the figure. Spectra represented in the figure are summations over the entire field of view, normalized to the phosphorus peak.
MCF-7 transfection and luciferase detection
PRL-deficient MCF-7 cells were maintained in phenol red-free RPMI 1640 (GIBCO) containing 10% horse serum and 50 μM ganciclovir as previously described.28 Prior to transfection, cells were grown in phenol red-free RPMI 1640 containing 5% charcoal-stripped fetal bovine serum for 3–4 days. Transient transfections were performed as previously described31 using the 4xAP-1-luciferase construct, which contains four GCN4 consensus AP-1 response elements upstream of a luciferase gene driven by a minimal promoter.32 Briefly, cells were transfected using Lipofectamine2000 (Invitrogen, Carlsbad, CA) for 4–6 hours prior to plating in wells ± circular PDMS inserts 21 mm in diameter overnight. Cells were serum-starved for 24 hours prior to treatment with 4 nM prolactin or 1 nM estrogen for 24 hours. Luciferase activity was detected using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) and normalized to beta-galactosidase activity. Induction of AP-1 was reported as fold increase in signal intensity over ethanol-treated control samples.
MCF-7 MVLN culture and luciferase detection
MCF-7 MVLN cells were created previously31 and consist of a stably transfected construct containing the consensus estrogen response element (ERE) upstream of the minimal HSV-thymidine kinase promoter and luciferase gene. MVLN cells were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Inc.) with phenol red and L-glutamine, supplemented with 10% fetal bovine serum (FBS; Biowest), 100 units/ml penicillin, and 100 μg/ml streptomycin (GIBCO/Invitrogen). For hormone treatments, cells were incubated for at least three days in phenol red-free DMEM (mediatech) supplemented with 4 mM L-glutamine (GIBCO), 10% charcoal dextran-stripped FBS, and penicillin/streptomycin. For experiments, ∼4,500 cells/well were seeded in white 96-well plates in either 2% or 10% stripped serum, phenol red-free DMEM. At 24 hours post-seeding, cells were treated for 30 minutes with 0.1% ethanol vehicle or the specified concentration of estrogen, diluted in 10% stripped serum, or serum free, in phenol red-free DMEM. For estrogen absorption experiments, a PDMS chunk (Soxhlet extracted or not, pre-weighed and autoclaved) of mass 10–14 mg was placed floating in each well (except for controls). For estrogen release experiments, the added PDMS pieces (10, 20, or 30 mg) were previously extracted and incubated individually for 24 hours inside a well with 100 μl of serum-free medium containing 10−7 M estrogen, after which time they were washed 3× with sterile water and transferred to cell-containing wells. 24 hours following estrogen treatment, PDMS pieces and media were removed and cells were analyzed for luciferase. Cell were lysed in wells by adding 50 μl of 1× GLO lysis buffer (Promega) for 20 minutes and then mixed with 50 μl/well of luciferase substrate (Promega). Luminescence read-outs were obtained by measuring total intensity (no filter) on a plate reader (Wallaq).
Time-resolved estrogen absorption and ELISA
PDMS devices were prepared as described above. Low-glucose phenol red-free DMEM containing 1 nM estrogen was added to microchannels and incubated at 37 °C for 0, 1, 3, 6, 12, or 24 hours. Approximately 30 microchannels were pooled for each time point. After the specified time interval, the estrogen solution was aspirated from the microchannels, transferred to a microcentrifuge tube, and stored at −80 °C. All microcentrifuge tubes were pre-incubated for 30 minutes with a 3% bovine serum albumin/0.1% Tween-20 solution to minimize any potential adsorption of serum estrogen to the sides of the tube. When all samples were collected, they were thawed to room temperature and assayed for the presence of estrogen with an ACE estradiol ELISA kit (#582251, Cayman Chemical). Standard curves were generated and linearized as instructed with the logit transform, and equations for deriving estrogen concentration were generated from the linearized curves.
Conclusion
PDMS remains a useful tool in the progression of microscale fluid handling. The leaching of uncured oligomers into culture media and the rapid partitioning of small hydrophobic molecules into the polymer bulk are two inherent characteristics of PDMS that should be carefully considered when interpreting data generated within PDMS-based microfluidic devices. As microfluidic principles are applied to the study of specific biological problems, it is important and useful to remain cognizant of the interaction between biological systems and the containers in which they are studied.
Supplementary Material
Acknowledgments
This research was supported by the following grants:NIH T32GM08349, NIH R21CA122672, and NIH K25CA104162
The authors wish to thank the CIC Mass Spectrometry Facility, Department of Chemistry, UW-Madison, and Dr. John Fournelle, SEM Laboratory, Department of Geology, UW-Madison, for their valuable input, as well as Bellbrook Labs for donating the polystyrene microchannels.
Footnotes
David J. Beebe has an ownership interest in Bellbrook Labs LLC, which has licensed technology reported in this publication.
References
- 1.Kane R, Takayama S, Ostuni E, Ingber D, Whitesides G. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 2.Jo B, Van Lerberghe L, Motsegood K, Beebe D. J Microelectomech Syst. 2000;9:76–81. [Google Scholar]
- 3.Leclerc E, Sakai Y, Fujii T. Biomedical Microdevices. 2003;5:109–114. [Google Scholar]
- 4.Nikkhah M, Strobl J, Agah M. Proc IEEE EMBS. 2007;29:6077–6080. [Google Scholar]
- 5.Gross P, Kartalov E, Scherer A, Weiner L. J Neurol Sci. 2007;252:135–143. doi: 10.1016/j.jns.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Yeon J, Park J. BioChip J. 2007;1(1):17–27. [Google Scholar]
- 7.Meyvantsson I, Beebe D. Annu Rev Anal Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Takayama S, Qian X, Ostuni E, Wu H, Bowden N, LeDuc P, Ingber D, Whitesides G. Langmuir. 2002;18:3273–3280. [Google Scholar]
- 9.Walker B, Zeringue H, Beebe D. Lab Chip. 2004;4:91–97. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 10.Rhee S, Taylor A, Tu C, Cribbs D, Cotman C, Jeon N. Lab Chip. 2005;5:102–107. doi: 10.1039/b403091e. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Alexander C, Beebe D. Lab Chip. 2007;7:726–730. doi: 10.1039/b618793e. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopdhyay R. Anal Chem. 2007;79(9):3248–3253. doi: 10.1021/ac071903e. [DOI] [PubMed] [Google Scholar]
- 13.Paguirigan A, Beebe D. Integr Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paguirigan A, Beebe D. BioEssays. 2008;30:811–821. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Park C, Whitesides G. Anal Chem. 2003;75:6544–6554. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Chaudhury M, Owen M. J Colloid Interphase Sci. 2000;226:231–236. [Google Scholar]
- 17.Eddington D, Puccinelli J, Beebe D. Sensors and Actuators B. 2006;114:170–172. [Google Scholar]
- 18.Bodas D, Khan-Malek C. Sensors and Actuators B. 2007;123:368–373. [Google Scholar]
- 19.Tóth A, Bertóti I, Blazsó M, Bánhegyi G, Bognar A, Szaplonczay P. J Appl Polymer Sci. 1994;52:1293–1307. [Google Scholar]
- 20.Makamba H, Hsieh Y, Sung W, Chen S. Anal Chem. 2005;77:3971–3978. doi: 10.1021/ac0502706. [DOI] [PubMed] [Google Scholar]
- 21.Millet J, Stewart M, Sweedler J, Nuzzo R, Gillette M. Lab Chip. 2007;7:987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez W, Hynard N, Evans J, George G. Silicon Chemistry. 2003;2:1–10. [Google Scholar]
- 23.Abate A, Lee D, Do T, Holtze C, Weitz D. Lab Chip. 2008;8:516–518. doi: 10.1039/b800001h. [DOI] [PubMed] [Google Scholar]
- 24.Roman G, Culbertson C. Langmuir. 2006;22:4445–4451. doi: 10.1021/la053085w. [DOI] [PubMed] [Google Scholar]
- 25.Rolland J, Van Dam M, Shcorzman D, Quake S, DeSimone J. J Am Chem Soc. 2003;126:2322–2323. doi: 10.1021/ja031657y. [DOI] [PubMed] [Google Scholar]
- 26.Toepke M, Beebe D. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 27.Smith N. Anal Biochem. 1979;94:100–104. doi: 10.1016/0003-2697(79)90796-6. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder M, Symowicz J, Schuler L. Mol Endocrinol. 2002;16:45–57. doi: 10.1210/mend.16.1.0762. [DOI] [PubMed] [Google Scholar]
- 29.Gutzman J, Nikolai S, Rugowski D, Watters J, Schuler L. Mol Endocrinol. 2005;19:1765–1778. doi: 10.1210/me.2004-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pons M, Gagne D, Nicolas J, Mehtali M. Biotechniques. 1990;9:450–459. [PubMed] [Google Scholar]
- 31.Carver K, Schuler L. Mol Cancer Res. 2008;6:634–643. doi: 10.1158/1541-7786.MCR-07-2069. [DOI] [PubMed] [Google Scholar]
- 32.Dong Z, Xu R, Kim J, Zhan S, Ma W, Colburn N, Kung H. J Biol Chem. 1996;271:9942–9946. doi: 10.1074/jbc.271.17.9942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


