Abstract
Single-layered embryonic skin either stratifies to form epidermis or responds to Wnt signaling (stabilized β-catenin) to form hair follicles. Postnatally, stem cells continue to differentially use Wnt signaling in long-term tissue homeostasis. We have discovered that embryonic progenitor cells and postnatal hair follicle stem cells coexpress Tcf3 and Tcf4, which can act as transcriptional activators or repressors. Using loss-of-function studies and transcriptional analyses, we uncovered consequences to the absence of Tcf3 and Tcf4 in skin that only partially overlap with those caused by β-catenin deficiency. We established roles for Tcf3 and Tcf4 in long-term maintenance and wound repair of both epidermis and hair follicles, suggesting that Tcf proteins have both Wnt-dependent and Wnt-independent roles in lineage determination.
Members of the Lef1 and Tcf family of DNA binding proteins bind β-catenin and transactivate Wnt target genes1. We previously discovered that Tcf3 is essential for gastrulation, where it differs from other Lef1 and Tcf binding proteins in acting not only in the presence but also in the apparent absence of Wnt signaling2–4. In embryonic skin, Tcf3 and Lef1 are expressed in basal progenitors4,5. As morphogenesis proceeds, Tcf3 becomes restricted to the early hair follicle ‘bulge’ region, a site where slow-cycling multipotent stem cells will reside, whereas Lef1 becomes confined to the base of the growing hair follicle, where committed, transit-amplifying matrix cells and precortical cells differentiate to produce the hair4,6. In postnatal skin, Tcf3 remains a faithful marker of hair follicle stem cells: its expression is maintained both in the bulge and in the outer root sheath (ORS) cells that trail down from the bulge. The ORS cells are thought to represent activated stem cells in transit to the hair follicle bulb, where they fuel production of the matrix cells, hair shaft and surrounding channel6–8. We have shown that bulge stem cell activation is dependent on Wnt signaling and stabilized β-catenin, whereas during the resting stage of the hair cycle, Tcf3 persists in the apparent absence of nuclear β-catenin2,5,9.
Lef1 and Tcf DNA binding proteins have not been identified in resident stem cells of epidermis or sebaceous glands. However, expression of a dominant-negative Lef1 promotes sebaceous gland cell proliferation and differentiation2,10, and ectopic induction of Tcf3 in the basal epidermal layer suppresses differentiation and changes the transcriptional profile to one resembling embryonic uncommitted epidermis4. Analogously, either through transgene expression9,11–13 or wounding14, elevated stabilized β-catenin induces the epidermis to undergo de novo hair follicle morphogenesis. Conversely, loss of β-catenin has been linked to defects in hair follicles and to skin tumor suppression15,16. It remains unresolved whether Wnt signaling affects epidermal and sebaceous gland stem cells, or only hair follicle stem cells. It is also unclear whether Tcf3 is essential for tissues that develop after gastrulation, and if so, whether Tcf3 has Wnt-independent and/or Wnt-dependent functions. Here we address these issues and uncover a hitherto unrecognized role for another Lef-Tcf member, Tcf4.
RESULTS
Tcf4 expression parallels that of Tcf3 in skin
To evaluate Tcf3 function in skin stem cells, we used the strategy outlined in Supplementary Figure 1 to engineer mice conditionally targeted for Tcf3. LoxP sites flanked exon 2 such that the keratin 14 (Krt14) promoter would activate Cre recombinase to excise the exon by embryonic day 15 (E15)17, introducing a frameshift mutation and yielding a nonfunctional mRNA product. We then used correctly targeted embryonic stem cell clones to generate two lines of mice, which we bred to generate Tcf3fl/fl; Tg(KRT14-cre)1Efu conditional knockout mice (transgene referred to here as ‘Krt14-cre’; mice referred to here as ‘Tcf3 cKO’). PCR genotyping confirmed the expected recombination events. Real-time PCR showed that Tcf3 mRNA was less abundant in Tcf3 heterozygotes than in wild-type skin epithelia and was absent from Tcf3 cKO skin epithelia. Immunoblotting and immunofluorescence with an antibody to the C-terminal region confirmed the absence of Tcf3 protein from Tcf3 cKO skin (Supplementary Fig. 1).
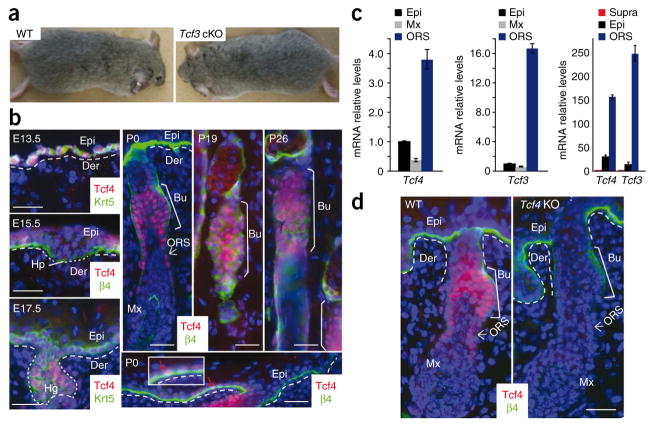
Notably, loss of Tcf3 had no obvious phenotypic or histological consequences to the skin (Fig. 1a and Supplementary Fig. 1). In searching for an underlying explanation, we found that Tcf4 shares a similar pattern of expression with Tcf3 in skin (Fig. 1b). At E13.5, Tcf4 was coexpressed with Tcf3 and Lef1 in the single layer of multipotent skin progenitors. By birth (postnatal day 0, P0), epidermal expression waned, showing nuclear Tcf4 and Tcf3 immunolabeling in only a few cells within the basal epidermal layer. Real-time PCR analyses confirmed the expression of low levels of Tcf3 and Tcf4 in neonatal skin (Fig. 1c). By contrast, all three Tcf genes became prominently expressed in the developing hair follicles, and postnatally, their hair follicle patterns were maintained. However, whereas Lef1 was concentrated at the leading front of downgrowing hair follicles5, Tcf4 immunolabeling paralleled that of Tcf3 (ref. 4) in becoming largely restricted to the bulge and lower ORS cells, thought to be direct bulge descendents6,7. As the lower hair follicle entered catagen and the region below the bulge degenerated at P19, Tcf4 was restricted to the bulge. During the next growth phase of the hair cycle (anagen, at P26), Tcf4 was largely expressed in the bulge but also in a trail of lower ORS cells (Fig. 1b). Real-time PCR analyses of mRNAs isolated from various P4 skin cell populations purified by flow cytometry18 were consistent with these data (Fig. 1c), and immunoblotting for Tcf4 revealed a Tcf4 protein in skin that corresponded in size to the short ‘B’ isoform found in intestinal epithelium (Supplementary Fig. 2).
Figure 1.
Tcf4 shares an expression pattern similar to that of Tcf3 in skin, where it becomes largely restricted to the slow-cycling hair follicle (bulge) stem cells and their early ORS progeny, (a) Tcf3 cKO mouse and wild-type (WT) littermate have indistinguishable phenotypes. (b) Immunolocalization of Tcf4 reveals temporal expression analogous to that of Tcf3 (ref. 4). Inset shows higher magnification of occasional Tcf4+ cell (red arrow) in postnatal epidermis. The most prominent postnatal labeling of Tcf4 is in hair follicle bulge stem cells and their early descendents in the ORS6,7 (white arrow). Hp, hair placode; Hg, hair germ; Bu, bulge; Mx, matrix; Epi, epidermis; Der, dermis. (c) Real-time PCR analyses of mRNAs from flow cytometry–purified populations of cells from P4 skin18. When normalized against the epidermis, both Tcf4 and Tcf3 are enriched in ORS cells that include the bulge and early bulge progeny. When normalized against suprabasal epidermal cells, Tcf3 and Tcf4 mRNAs are clearly detected in basal cells. Error bars indicate s.d. (d) Immunofluorescence of PO skin confirms that Tcf4 expression is ablated in Tcf4 knockout (KO) skin and that the antibody to Tcf4 is specific. Scale bars represent 20 μm.
Tcf3 and Tcf4 function in skin is Wnt dependent and independent
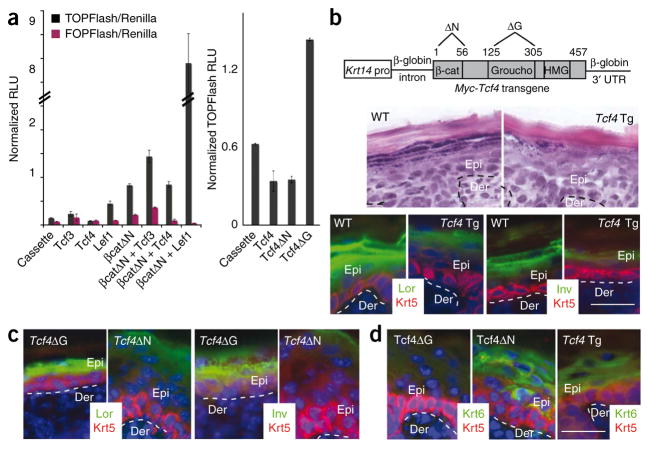
When Tcf4 is absent from mouse intestinal epithelial stem cells, which lack Tcf3, tissue development is severely impaired, and the mice die shortly after birth19. By contrast, skin grafts from these mice revealed no obvious visual, histological abnormalities (Fig. 1d and Supplementary Fig. 3). The lack of an overt hair phenotype in skin singly targeted for Tcf3 or Tcf4 suggested that the two proteins have redundant functions in bulge skin stem cells. To test this possibility, we first conducted Wnt assays in cultured keratinocytes using the TOPFlash Tcf reporter plasmid. As previously shown for Tcf3 (ref. 2), the Tcf4B isoform requires its N-terminal domain, which associates with stabilized β-catenin to transactivate TOPFlash (Fig. 2a and Supplementary Fig. 2). Additionally, removal of Groucho binding domain of Tcf4 elevated basal levels of TOPFlash, suggesting that in keratinocytes, Tcf4 has repressor activity when Wnt signaling is low or absent (Fig. 2a). Moreover, when we engineered transgenic mice to express Myc epitope–tagged Tcf4B isoform under the control of the basal epidermal Krt14 promoter, the mice showed an open-eye phenotype and altered epidermal differentiation (Fig. 2b), a phenotype markedly similar to that of our previously generated Krt14-Myc-Tcf3 transgenic mice2. A similar phenotype was obtained with Krt14-Myc-Tcf4ΔN mice, whereas Krt14-Myc-Tcf4ΔGroucho mice appeared normal (Fig. 2c,d). Given these observations, we attribute these effects on the epidermis to Tcf4 repressor activity that seems to be independent of β-catenin. Such functions have been previously ascribed to Tcf3, but not Tcf4.
Figure 2.
Sustained epidermal expression suggests repressor and activator functions for Tcf4. (a) Left, the B isoform of Tcf4 (referred to here as Tcf4) represses basal TOPFlash Wnt reporter expression, as previously reported for Tcf3 (ref. 2). Right, Tcf4ΔG, which lacks the Groucho binding domain, does not repress TOPFlash, whereas Tcf4ΔN, which lacks the β-catenin-interacting domain, does. Error bars indicate s.d. RLU, relative light units. (b)Top, Tcf4 transgenes used to generate mice. Bottom, Krt14-Tcf4 transgenic (Tcf4Tg) mice die at birth and show repression of the epidermal differentiation phenotype, as previously described for Krt14-Tcf3 transgenic mice2. Lor, loricrin; Inv, involucrin. (c,d) This phenotype is independent of the β-catenin-interacting domain but dependent on the Groucho repressor binding domain, (c) Expression of Lor and Inv is reduced in Tcf4ΔN, but not Tcf4ΔG, mice, (d) Krt6 expression is induced in Tcf4ΔN and Tcf4ΔTg, but not Tcf4ΔG, mice. Scale bars represent 20 μm.
Defects from loss of Tcf3 and Tcf4 in skin epithelium
To further test the possibility of functional redundancy between Tcf3 and Tcf4, we generated Tcf3fl/fl; Tcf4−/−; Krt14-cre mice, herein referred to as Tcf3/4 null. Tcf3/4-null newborn pups showed visibly thinner skin and often lacked whiskers (Fig. 3a). Histology revealed that hair follicles were initiated, but defects in subsequent follicle down-growth appeared (Fig. 3b,c and Supplementary Fig. 3). Additionally, although epidermis was replete with spinous, granular and stratum corneum layers, it was typically thinner than normal, and basal cells were flattened rather than columnar. The degree of these early phenotypic and histological abnormalities in the skin varied among the Tcf3/4-null newborn pups. However, because later-stage defects were consistently and uniformly severe, this seemed more likely to reflect variation in the timing of Cre-mediated excision rather than genetic strain differences. The Krt14 promoter is typically strongly active by E15.5 (ref. 17).
Figure 3.
Characterization of skin of newborn mice lacking Tcf3 and Tcf4. (a) Tcf3/4-null PO pups differ from wild-type and singly null mice in having thinner skin. (b) H&E staining shows that hair follicle downgrowth in skin from a severely affected PO Tcf3/4-null pup is defective, and the epidermis is abnormally thin compared to control wild-type skin (shown) or singly targeted Tcf3 cKO or Tcf4-null skins (see also Supplementary Fig. 3). (c) Histological analysis of toluidine blue–stained semithin sections reveals that Tcf3/4-null epidermis shows differentiating spinous, granular and stratum corneum layers, but cells in the basal layer are flattened, and differentiation appears morphologically less well developed, (d–h) Immunofluorescence analysis for the indicated markers, (d) Tcf3 and Tcf4 are absent from Tcf3/4-null skin relative to control skin. (e,f) PO Tcf3/4-null hair follicles still express Sox9, an early bulge stem cell marker, and Lhx2, which at this stage marks the leading edge of the downgrowing follicle. (g) Tcf3/4-null skin is also still proliferating at P, as shown by Ki67 immunostaining. (h) However, follicle regions show some TUNEL-positive cells, suggestive of a failure to survive (arrows, middle panel), (i) Quantification of data in g and h. Overall, the percentage of apoptotic cells was still very low (~5%). Error bars indicate s.d. HF, hair follicle. Scale bars represent 10 μm in c and 20 μm in b,d–h.
Immunostaining for Tcf3 and Tcf4 verified that both were ablated in the Tcf3/4-null epidermis (Fig. 3d). Immunolabeling patterns of two other follicle stem cell transcription factors, Sox9 and Lhx2, did not seem to be affected by loss of Tcf3 and Tcf4, at least at early stages of development (Fig. 3e,f). Ki67 immunolabeling was also comparable in newborn (P0) skin, suggesting that epithelial proliferation is intact at this time (Fig. 3g). By contrast, P0 Tcf3/4-null skin showed greater TUNEL-positive staining, suggestive of enhanced cell death (Fig. 3h). Quantifications of these data (Fig. 3i) confirmed an imbalance in epidermal homeostasis.
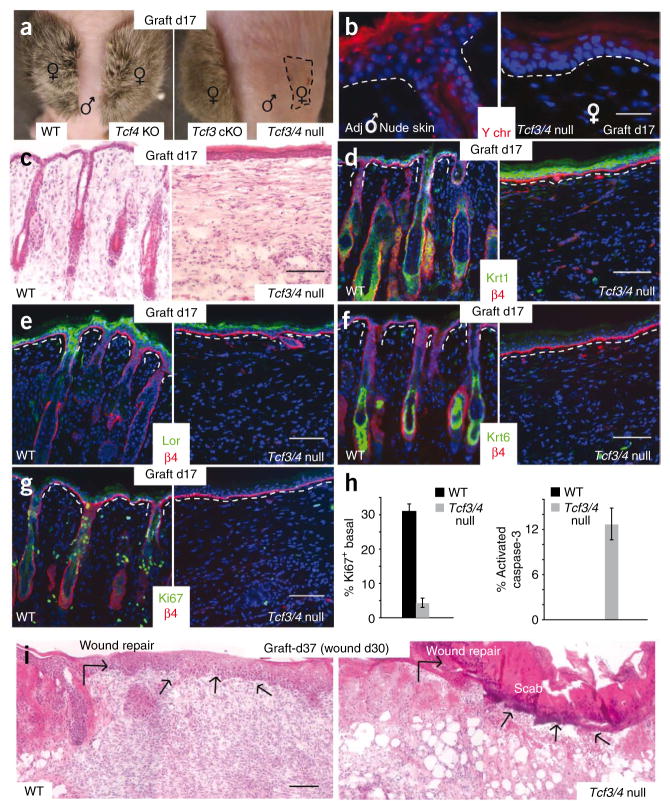
To evaluate the long-term consequences of Tcf3 and Tcf4 deficiency in skin, we engrafted female skins onto the backs of male immunocompromised nude mice, or male skin onto female nude mice. Seventeen days after grafting, a full coat of hair was readily detected in wild-type, Tcf3 cKO and Tcf4−/− skins (Fig. 4a). In contrast, Tcf3/4-null skin grafts consistently showed no visible hairs and had shrunk in overall area relative to the other grafts (Fig. 4a). Like the control grafts, however, day 17 Tcf3/4-null female skin grafts showed a Y-chromosome pattern opposite from that of the adjacent nude mouse skin, indicating that the grafts had taken and that the skin epidermis overlying the graft at this time had not arisen from wound repair by the surrounding skin (Fig. 4b). Histological analysis confirmed that hair follicles were completely absent from the Tcf3/4-null graft (Fig. 4c). Below we address possible reasons for the markedly enhanced severity in hair follicle phenotype after engraftment of Tcf3/4-null skin.
Figure 4.
Skin grafting permits evaluation of the long-term consequences of Tcf3 and Tcf4 ablation in skin. (a) After 17 d of engraftment of female skins onto the backs of male nude mice, Tcf3 cKO and Tcf4 KO skins are indistinguishable from the wild type, whereas Tcf3/4-null grafts (area indicated by dashed line) shrink and show no hair. (b) Y-chromosome FISH shows that the engrafted Tcf3/4-null epidermis is still female and is not derived from the male nude epidermis surrounding the graft. (c) H&E staining reveals absence of hair follicles from Tcf3/4-null skin grafts. (d–g) Differentiation of the epidermis still occurs in Tcf3/4-null skin, as shown by immunolabeling for Krt1 (spinous) and loricrin (granular) markers. (f) Although epidermis of Tcf3/4-null grafts appears thicker, no immunolabeling was detected for Krt6, which marks the hair follicle ORS (companion layer) of normal skin and the suprabasal epidermis of hyperproliferative skin. (g) Ki67 immunolabeling reveals a paucity of proliferative keratinocytes within day 17 Tcf3/4-null skin grafts. (h) Quantification of Ki67 and activated caspase 3 staining. Error bars indicate s.d. (i) Tcf3/4-null engrafted skin is defective in reepithelialization after wounding at day 30 after engraftment. Shown are H&E-stained skins 7 d after wounding. Arrows indicate sites where reepithelialization has occurred to repair the wound, in control but not in Tcf3/4-null skin. Scale bars indicate 20 μm in b and 100 μm in c–g and i.
Although hair follicles were absent, epidermal differentiation seemed to be intact, as judged by immunostaining for markers including Krt1 (spinous layer) and loricrin (granular layer) (Fig. 4d,e; see also Supplementary Fig. 3). Additionally, despite a slightly thickened epidermis, immunolabeling was negative for Krt6, a keratin naturally expressed in hair follicles but aberrantly induced in epidermis under hyperproliferative conditions (Fig. 4f). Notably, Ki67 labeling was barely detected in P17 Tcf3/4-null grafts (Fig. 4g,h). This differed from the P0 Tcf3/4-null state or the P17 grafts of wild-type or singly null skin. Finally, consistent with signs of cell death observed in P0 Tcf3/4-null interfollicular epidermis, immunolabeling for the apoptotic marker activated caspase 3 was exclusive to the Tcf3/4-null state (Fig. 4h).
The defects we observed in Tcf3/4-null grafts were not accompanied by inflammation (Supplementary Fig. 4), but rather seemed to be rooted in an inability to maintain long-term self- renewing populations of skin epithelia. Further evidence in support for this view was provided by introducing full-thickness wounds into the grafts. In contrast to control skin grafts, which rapidly regenerated epidermis to replace the scab, Tcf3/4-null grafts showed no signs of epithelial regeneration even after a week of monitoring (Fig. 4i).
After grafting, unwounded Tcf3/4-null grafts continued to shrink in size, whereas wild-type and singly null grafts remained constant. By 2 months, the shrunken areas of Tcf3/4-null skin grafts appeared crusted and decayed. By conducting Y-chromosome in situ hybridizations of male grafts onto female nude mice, we learned that whereas the underlying dermis of grafts of all genotypes seemed to maintain its normal size, most of the overlying Tcf3/4-null (male) conditional-knockout epidermis had been lost and replaced by nude (female) epidermis moving in from the border (Supplementary Fig. 5). Thus, all skin epithelial cells with a capacity for long-term tissue maintenance seemed to be progressively lost.
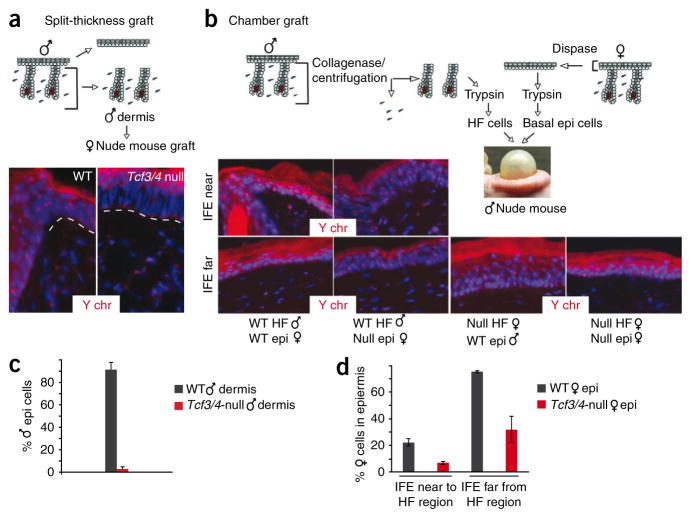
A priori, the unexpected loss of epithelium from long-term Tcf3/4-null grafts could be explained in part by loss of Tcf3 and Tcf4 in the embryonic bulge, which was recently shown to be required for completion of hair follicle morphogenesis, sebaceous gland development and efficient epidermal repair6. To explore this possibility, we used split-thickness grafts of male P0 dermis, in which cells from the dermal hair follicles are challenged to repair the missing overlying epidermis (Fig. 5). Whereas epidermal repair in control grafts was largely derived from hair follicle cell progenitors, repair in Tcf3/4-null grafts was derived from the host nude mouse (Fig. 5a,c).
Figure 5.
Measuring the stem cell potential within hair follicles and epidermis in the absence of Tcf3 and Tcf4. (a) Hair follicles in Tcf3/4-null dermis seem to lack the potential to regenerate epidermis. After dispase treatment to remove and discard epidermis, entire wild-type or Tcf3/4-null male skin dermis (containing hair follicles) was engrafted onto female nude mice. After 30 d, control (wild-type) grafts showed Y chromosome–positive epidermal cells, whereas epidermis from Tcf3/4-null dermis was derived from nude epidermal reepithelialization. (b) Chamber graft mixing experiments with purified, single-cell suspensions of sex-marked epidermal or hair follicle cells show that, in contrast to wild type, neither Tcf3/4-null epidermal cells nor hair follicle cells contribute properly to reestablishment of skin epidermis. IFE, interfollicular epidermis, either near to or far from hair follicles. (c) Quantification of male IFE cells from split-thickness grafts. (d) Quantification of female epidermal cells in chamber grafts detected in near or far IFE. Error bars (c,d), s.d.
We next tested the efficacy of the epidermal fraction using chamber grafts, in which we combined female cells from wild-type or Tcf3/4-null epidermis with wild-type hair follicle–enriched dermal cells (Fig. 5b). Even though overlying epidermal repair resulted largely from wild-type dermal cells, we were still able to readily identify contributions from wild-type epidermal cells. By contrast, we detected far fewer signs of Tcf3/4-null epidermal contribution in these grafts (Fig. 5d).
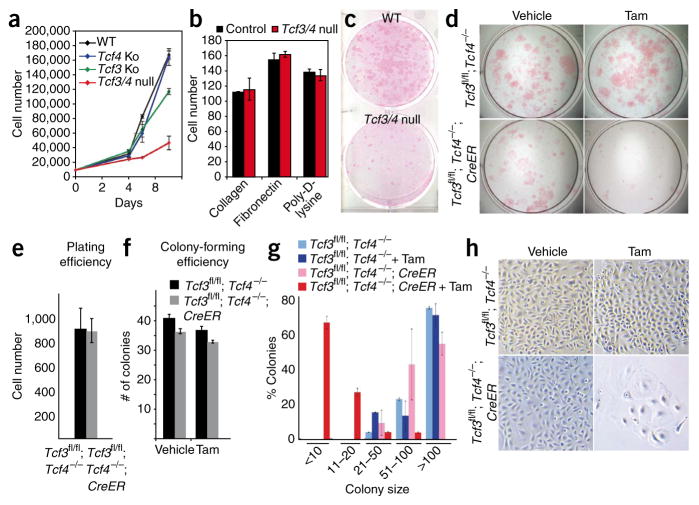
To better understand why proliferation persists only short-term in Tcf3/4-null embryos and P0 grafts, we cultured primary mouse keratinocytes from P0 skins and monitored their growth potential over time. During the first 4 d in culture, cells from the different genotypes showed similar growth rates (Fig. 6a–c). Shortly thereafter, Tcf3/4-null primary mouse keratinocytes substantially slowed in growth, whereas singly targeted cells continued to expand exponentially. When plated onto various extracellular matrix substrata, Tcf3/4-null and wild-type cells adhered similarly (Fig. 6b), making it unlikely that cell-substratum adhesion defects accounted for the observed growth differences.
Figure 6.
Loss of Tcf3 and Tcf4 results in inability of cultured epidermal keratinocytes to undergo long-term self-renewal. (a) Tcf3/4-null primary mouse keratinocyte cultures show a growth defect. (b) No differences in cell-substratum adhesion are seen when equal numbers of primary mouse keratinocytes are plated onto various matrices and quantified after 1 h. (c) Rhodamine B staining of primary mouse keratinocytes cultured for 10 d reveals a growth defect in the absence of Tcf3 and Tcf4. (d–h) Inducible deletion of Tcf3 from Tcf4−/− cells affects their growth potential. Cultured primary mouse keratinocytes from epidermis of Tcf3fl/fl; Tcf4−/− P0 mice were passaged twice and then infected with retrovirus expressing either green fluorescent protein (GFP) alone or GFP with tamoxifen-inducible Cre (transgene cre-Esr1; referred to here as ‘CreER’). Infected cells were isolated by flow cytometry based on GFP level, and 1 d after plating, cells were treated with tamoxifen or vehicle control to induce Cre recombinase activity. Twelve days after plating, cells were fixed and stained with rhodamine B to visualize and quantify the numbers and sizes of colonies (those with at least four cells, 72 h after plating). Plating (e,f) and colony-forming (f) efficiencies were comparable between wild-type and Tcf3/4-null cells, but the average size of Tcf3/4-null colonies was markedly smaller than that of wild-type colonies (g) and the morphology of the mutant cells revealed signs of premature differentiation (h). All error bars indicate s.d.
Under conditions where singly targeted and wild-type cultures grew and could be passaged well, our Tcf3/4-null cultures continued to grow poorly and did not survive passaging. To better monitor the process, we cultured control keratinocytes and keratinocytes expressing tamoxifen-inducible Cre from Tcf3fl/fl; Tcf4−/− mice. Irrespective of tamoxifen treatment or Cre status, keratinocytes adhered and formed small colonies (at least four cells) with similar efficiencies (Fig. 6d–h). However after 12 d, only the Tcf3/4-null colonies were arrested, showing morphological signs of differentiation (Fig. 6d,g,h). The inability of Tcf3/4-null cultures to generate large colonies was consistent with our in vivo data and underscored a paucity of long-term self-renewal capacity in cultures lacking these transcription factors.
Effects of Tcf3/4 versus Ctnnb1 loss overlap only partially
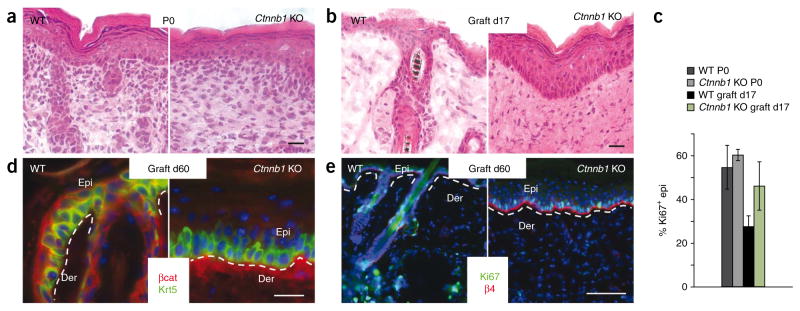
The ability of Tcf3 and Tcf4 deficiency to compromise long-term tissue homeostasis in the skin suggests that either Wnt signaling affects lineages besides the hair follicle, or Tcf3 and Tcf4 exert additional Wnt-independent effects in establishing and/or regulating epidermal progenitors. To distinguish between these possibilities, we mated our Krt14-cre mice to mice conditionally targeted for β-catenin (Ctnnb1)15. Similar to previous findings15 and supporting the established role of β-catenin in hair follicle stem cell activation and lineage determination2,11, β-catenin-deficient skin did not develop hair follicles (Fig. 7a and Supplementary Fig. 6). The developmental phenotype caused by β-catenin loss was more severe than that seen in Tcf3/4-null skin. One likely explanation is that Lef1, an additional β-catenin-interacting partner still expressed in embryonic skin, is presumably able to transmit the early Wnt signals that initiate and regulate follicle downgrowth2,11.
Figure 7.
Differences between Tcf3/4-null and Ctnnb1-null skin. (a) Conditional ablation of Ctnnb1 by Krt14-Cre results in death shortly after birth, when hair follicle morphogenesis has already been blocked15. (b–e) Notably, epidermis is hyper-thickened (b), and proliferation, as judged by Ki67 (c,e), is not compromised, even when skins are grafted and examined 60 d after engraftment. Overlying epidermis is clearly derived from the engrafted skin, as revealed by lack of β-catenin immunolabeling (d). Scale bars represent 20 μm in a,b,d and 100 μm in e. Error bars in c indicate s.d.
If Tcf3 and Tcf4 always require β-catenin to exert their function, then Tcf3/4-null skin engraftments should either resemble or be less severe than Ctnnb1; Krt14-cre conditionally null (referred to here as Ctnnb1-null) engraftments. However, under conditions where Tcf3/4-null grafts showed a marked decline in epidermal survival, Ctnnb1-null grafts showed a hyper-thickened, healthy epidermis (Fig. 7b). Further analyses confirmed the maintenance of engrafted Ctnnb1-null epidermis and its hyperproliferative state (Fig. 7c). Even after 2 months, the epidermis was still hyper-thickened, proliferative and negative for β-catenin (Fig. 7d,e). Thus, although loss of either Tcf3 and 4 or β-catenin compromised long-term maintenance of the hair follicle lineage, in which Wnt signaling is known to be crucial for bulge stem cell activation, the inability to maintain epidermis seemed to be specific to the loss of Tcf3 and Tcf4, for which an essential function for Wnt proteins has not been reported.
A full-scale dissection of the molecular differences that cause Tcf3/4-null epidermis to be unable to maintain long-term homeostasis is beyond the scope of the present study. To gain some insights, however, we conducted microarray analysis on mRNAs purified from freshly isolated Tcf3/4-null and wild-type back-skin epidermis. We conducted our analyses on flow cytometry–purified, α6 integrin–expressing cells from the epidermal fractions of dispase-treated E17.5 skin (a time typically preceding gross phenotypic changes)6.
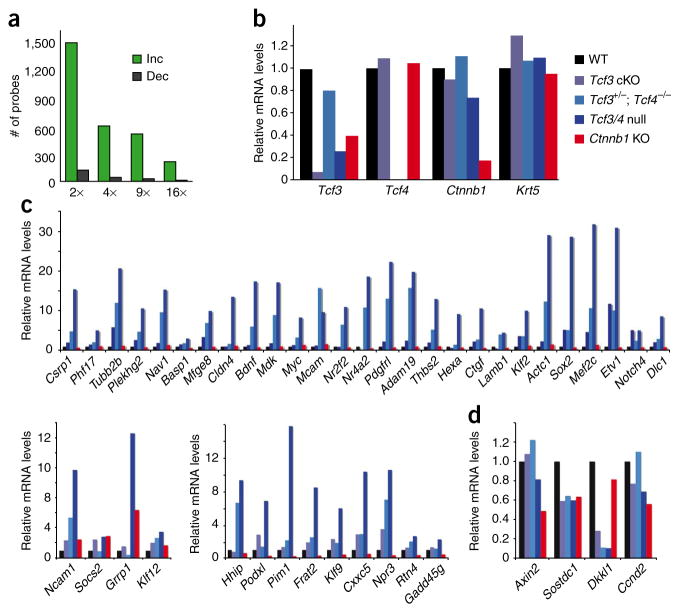
The underlying pathways are likely to be complex, but we restricted our focus to two points. First, the majority of probes scored as upregulated, rather than downregulated, in Tcf3/4-null compared to control cells (Fig. 8a). Second, >150 genes that scored as upregulated in Tcf3/4-null embryonic epidermal cells were recently shown to bind Tcf3 directly in embryonic stem cells20. To gain insights into the possible significance of this finding, we compared the expression of this group of genes in the context of Tcf3, Tcf4 and β-catenin status in the epidermis.
Figure 8.
Ablation of Tcf3 and Tcf4 in skin leads to an upregulation of gene expression. (a) Basal epidermal keratinocytes were purified by flow cytometry from E17.5 skins of Tcf3/4-null and wild-type mice, and their mRNAs were subjected to microarray analyses as outlined in Online Methods. Comparative analysis scored genes as being increased (‘Inc’) or decreased (‘Dec’) by the fold indicated. Most genes scored as upregulated upon loss of Tcf3 and Tcf4. (b–d) Real-time PCR analyses of mRNAs from flow cytometry–purified basal epidermal keratinocytes of wild-type, Tcf3 cKO, Tcf3+/−; Tcf4−/−, Tcf3/4-null and Ctnnb1 cKO E17.5 embryos. Genes chosen for analyses in b are controls to verify the status of Tcf3, Tcf4 and Ctnnb1 relative to comparable levels of the basal marker Krt5. Genes chosen for analyses in c scored as upregulated in Tcf3/4-null compared to wild-type epidermis and are known to directly bind to Tcf3 in embryonic stem cells20. Genes upregulated in E17.5 Tcf3/4-null basal epidermis were mostly unchanged (top) in Ctnnb1-null basal cells. A minority were either slightly increased or decreased (bottom panels). (d) A few genes scoring as decreased in the Tcf3/4-null samples also show reductions in expression when β-catenin is lost. Some of these are Wnt target genes, including Dkkl1 and Sostdc1.
For this purpose, we isolated α6 integrin–expressing cells from dispase-removed E17.5 epidermises of Tcf3 cKO, Tcf3+/−; Tcf4−/−, Tcf3/4-null and Ctnnb1-null embryos. After isolating and confirming the quality of their mRNAs (Fig. 8b), we compared the expression status of the abovementioned group of genes, which possess established Tcf3 binding sites and were upregulated in the absence of Tcf3 and Tcf4. Most of the mRNAs encoded by these genes showed little change in the absence of β-catenin relative to the wild-type control, whereas a minority were downregulated (Fig. 8c,d). These data are consistent with a model in which Tcf3 and Tcf4 can repress genes when β-catenin is absent. However, the results may also reflect the residual Wnt signaling that might occur in embryonic epidermis lacking Tcf3 and Tcf4 but not β-catenin. Resolution of this issue and other aspects of these distinct phenotypes await more detailed studies, including characterization of the genome occupancy of Lef1, Tcf3, Tcf4 and β-catenin proteins in embryonic epidermal chromatin.
Discussion
Canonical Wnt signaling has been implicated in a diverse array of cellular functions, including stem cell proliferation and self-renewal21–23; stem cell activation, fate determination and differentiation9,11–13; and aging and senescence24–28. Stabilized β-catenin acts as a transcriptional cofactor for Tcf3, Tcf4, Tcf1 and Lef1, as well as other DNA binding proteins1,21. In this regard, the early signs of hair follicle morphogenesis seen in Tcf3/4-null skin, but not in the β-catenin conditional-null counterpart, were notable and consistent with the known presence of Lef1 in the embryonic skin. The established role of Lef1 in the Wnt signaling cross-talk that contributes to the initiation steps in hair follicle morphogenesis29 provides an explanation for the more severe consequences of β-catenin loss compared to Tcf3 and Tcf4 loss during these early stages of skin development2,11,30
Toward the end of embryogenesis, however, the expression of Lef1 diverges from that of Tcf3 and Tcf4, exposing essential functions for Tcf3 and Tcf4. These roles were relatively slow to emerge, and in some cases only became obvious after grafting, when hair follicles disappeared altogether. That said, the mildest and more frequently observed Tcf3- and Tcf4-deficient hair follicle phenotype was remarkably similar to the most severe Sox9-deficient phenotype, in which hair follicle morphogenesis progresses until about P4 and then arrests, resulting in a complete absence of hair follicles in the ensuing post-natal hair cycle6. Notably, all three transcription factors mark the long-term, self-renewing stem cells of the hair follicle. These stem cells are essential to postnatal completion of hair follicle morphogenesis as well as the subsequent cyclic bouts of hair regeneration throughout the life of the animal. The coexpression of these genes and the similarities in Tcf3/4-null and Sox9-deficient hair follicle phenotypes points to a crucial role for Tcf3 and Tcf4 in hair follicle stem cell maintenance. Additionally, these new data suggest that Tcf3 and Tcf4 are the key transcription factors that collaborate with β-catenin in its established role in hair follicle stem cell activation9,12,13.
A surprising feature of Tcf3/4-null skin was its inability to maintain long-term epidermal homeostasis. Although we did detect low levels of Tcf3 and Tcf4 transcripts and Tcf4 protein in some resident postnatal epidermal cells, Tcf3 and Tcf4 were much more prominent in embryonic than postnatal epidermis. This raises the question of whether long-term epidermal progenitors are specified before birth, as we recently showed for hair follicle stem cells6. Another intriguing aspect of these epidermal defects is that they are not seen in mice15 or in skin grafts (described above) conditionally targeted for β-catenin. These observations, coupled with our transgenic and transcriptional comparisons, suggest that these differences might be partly rooted in the ability of these Tcf proteins to act not only in Wnt signaling, but also as repressors when β-catenin is low or absent. Overall, our data support the notion that Tcf3 and Tcf4 may be essential for establishing and maintaining all skin epithelial stem cells through Wnt-dependent and Wnt-independent roles.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Generation of constructs
The Tcf3 targeting vector used the pGKneobpAloxX2 PGKDTA vector kindly provided by P. Soriano and was previously described3.
Tcf4 B-isoform cDNA was cloned out from E17.5 total skin cDNA using forward primer Tcf4F1-BamH1 and reverse primer Tcf4bR-BamH1R to amplify the full-length Tcf4 B isoform. Sequences of cloning primers are listed in Supplementary Table 1. The PCR product was digested with BamH1 and ligated to a Krt14 promoter cassette that was digested with BamH1 and phosphatase treated with shrimp alkaline phosphatase (SAP; Roche). After its sequences were verified, the Krt14-Tcf4 template was used to amplify the Tcf4 gene with a BglII restriction site introduced in its 5′ end so as to be in-frame when ligated into the pCMVmyc vector (Clontech). The BglII- and BamH1-digested insert was then ligated to a pCMVmyc vector that was digested with BlgII and treated with SAP. We then used primers to amplify the Myc epitope–tagged, full-length Tcf4 (Myc-Tcf4) gene, digested the DNA with BamH1, and ligated it a to Krt14 promoter cassette that was cut with BamH1 and treated with SAP.
To generate Krt14-Tcf4ΔN, Krt14-Tcf4 was used as a template for PCR with forward primer Tcf4F166-BamH1 and reverse primer Tcf4bR-BamH1. The PCR product was digested with BamH1 and ligated to a Krt14 promoter cassette that was cut with BamH1 and treated with SAP.
To generate Krt14-Tcf4ΔC, the template Krt14-Tcf4 was amplified using forward primer Tcf4F1-BamH1 and reverse primer Tcf4R948-stopXba1. The PCR product was cut with BamH1 and Xba1 and ligated to a Krt14 promoter cassette that was cut with BamH1 and Xba1. To generate Krt14- Tcf4ΔG, the Krt14-Tcf4 template was amplified using forward primer Tcf4F1-BamH1 and reverse primer Tcf4R385 to generate fragment A, and primers Tcf4R-BamH1 and Tcf4F359-aattII to generate fragment B. Fragment B was cut with AattII to delete nucleotides 920–1,400 and then ligated to fragment A, which was digested with AattII and amplified using primers Tcf4F1BamH1 and Tcf4b-RBamH1. The PCR product was cut with BamH1 and ligated to a Krt14 cassette that was digested with BamH1 and treated with SAP.
To generate pCMVmyc vectors expressing Tcf4 constructs (full-length, ΔN, ΔG and ΔC), Krt14-Tcf4 constructs (full-length, ΔN, ΔG and ΔC) were amplified with forward primer BglII-Tcf4F1 and reverse primer BglII-stop-Tcf4 R12889. The PCR products were cut with BglII and ligated to a pCMVmyc vector that was digested with BglII and treated with SAP. To generate Krt14-Myc-Tcf4 transgenes (full-length, ΔN, ΔG and ΔC), the respective pCMVmyc vectors were used as templates with forward primer mycBamH1F824 and reverse primer mycBamH1R939 to amplify the Myc-tagged versions. All Myc-tagged inserts were digested with BamH1 and ligated to a Krt14 promoter vector cassette that was digested with BamH1 and treated with SAP. The SacI and HindIII fragments of Krt14-Myc-Tcf4, Krt14-Myc-Tcf4ΔN and Krt14-Myc-Tcf4ΔG were then used for injection to engineer transgenic mice as previously described17.
Generation and analysis of Tcf3/4-null and Krt14-Tcf4 transgenic mice
To conditionally inactivate the Tcf3 gene, a targeting strategy was designed to allow Cre-mediated deletion of exon 2. NotI-linearized Tcf3 targeting construct DNA (30 μg; Supplementary Fig. 1) was electroporated into GS1 embryonic stem cells. After 6 d of G418 selection, ~300 individual colonies were screened for homologous recombination by PCR and Southern blotting (Supplementary Fig. 1). A second electroporation was done to introduce a CMV-Cre expression plasmid into a G418-resistant clone that had undergone homologous recombination with the targeting construct DNA. After 5 d of culture in the absence of G418 selection, ~300 individual colonies were screened for G418 sensitivity and screened by PCR for loss of the floxed neo cassette between exons 2 and 3. An embryonic stem cell clone containing a floxed Tcf3 allele, as confirmed by Southern blotting and PCR (Supplementary Fig. 1), was injected in blastocysts of 57Bl/6 mice to generate chimeric mice carrying the Tcf3 targeted allele.
We bred the chimeric mice containing the Tcf3fl/+ allele to germline transmission and then mated them to Krt14-Cre mice. Both Tcf3fl/fl; Krt14-Cre mouse lines, which appeared phenotypically wild type, were then bred to Tcf4+/−mice until mice doubly null in their skin epithelium for Tcf3 (conditional) and Tcf4 (straight) were generated. Because Tcf4−/− mice die at birth, we used skin grafting as previously described31 for long-term skin monitoring.
Split-thickness and chamber graft assays were done as described previously6,32. For cell mixing chamber graft experiments, 5 × 105 cells from dermal fractions enriched for hair follicles18 were combined with 5 × 105 keratinocytes from dispase- and trypsin-treated pure epidermis. Cell mixtures were then added to chambers that had been inserted onto the back skin of nude mice. Chambers were removed after healing (7–10 d), and grafted skins were collected and analyzed after 30 d. Full-thickness wounds were produced with a 3-mm Miltex biopsy punch administered to grafted skins at day 30, and skins were analyzed 7 d later.
Cell isolation and in vitro culture
Primary mouse keratinocytes were isolated, cultured and assayed as described32. Cell proliferation was assessed using a Beckman Z2 Coulter counter. Cell adhesion assays were done with equal numbers of freshly isolated primary mouse keratinocytes placed on glass cover slips coated with 10 μg ml−1 fibronectin (Upstate Biotechnology), 10 μg ml−1 laminin (BD Biosciences) or 100 μg ml−1 poly-D-lysine (BD Biosciences). After 1 h, nonadherent cells were removed with PBS, and adherent cells were fixed, stained and counted using ImageJ software.
Colony formation efficiency was analyzed as follows: 2 × 104 live, freshly isolated primary mouse keratinocytes were plated onto mitomycin-treated fibroblast feeders, and fresh medium was changed every other day. Feeders were removed 10 or 12 d later, and colonies were stained with rhodamine B. TOPFlash Wnt reporter assay was done as previously described2, with FOPFlash (mutant Tcf-Lef binding sites) as a negative control.
Immunoblot, immunofluorescence and histology
To analyze Tcf3 expression, 40 μg protein from flow cytometry–sorted E17.5 epidermal cell lysates were resolved by SDS-PAGE, immunoblotted, probed with primary antibodies to Tcf3 (guinea pig, 1:3,000; Fuchs laboratory) and β-actin (mouse, 1:1,000; Sigma), and detected by chemiluminescence (ECL reagents, Amersham Biosciences).
Immunofluorescence was done on 10-μm OCT-frozen back skin sections as described4 using a block-diluent solution of 5% normal donkey serum, 2% gelatin and 0.2% Triton X-100 in PBS. We used primary antibodies to Tcf3 (guinea pig, 1:300; Fuchs laboratory), Tcf4 (goat, 1:100; Santa Cruz Biotechnology), Krt5 (rabbit, 1:300; Fuchs laboratory), Krt1 (rabbit, 1:200; Fuchs laboratory), loricrin (rabbit, 1:200; Fuchs laboratory), β4 integrin (rat, 1:200; BD Biosciences), Krt6 (rabbit, 1:500; Fuchs laboratory), Ki67 (rabbit, 1:500; Novocastra Laboratories), Cd3 (1:100; Chemicon), F4/80 (rat, 1:100; Serotec), Gr1 (mouse, 1:100; R&D Systems), β-catenin (mouse, 1:500; Sigma), active caspase 3 (rabbit, 1:1,000; R&D). DNA fragmentation analyses were done using a TUNEL kit (Chemicon). Y-FISH chromosome analyses were done using a Cy3 Star★FISH detection kit (Cambio).
Histological analyses were done on tissues that were fixed for ≥1 h in 0.05 M sodium cacodylate buffer containing 2% glutaraldehyde, 4% formaldehyde and 2 mM CaCl2, and then processed for Epon embedding. One-micron sections were cut, stained with toluidine blue and examined with a Zeiss Axioplan microscope.
Real-time PCR gene expression analysis
Total RNAs were purified with an Absolutely RNA kit (Stratagene) and reverse-transcribed using oligo(dT) primers (Superscript III First-Strand Synthesis System, Invitrogen). The Roche LightCycler system and software and DNA Master SYBR Green II reagents were used for real-time PCR. Differences between samples and controls were normalized to the level of Gapd and calculated based on the 2−Δ ΔCP method. Primer sequences are listed in Supplementary Table 1.
Sample preparation for microarrays
After enzymatic treatment with dispase, back-skin epidermis was removed from E17.5 embryos and incubated in 0.05% trypsin for 10 min at 37 °C. After straining to remove debris, dissociated cells were exposed to phycoerythrin-conjugated antibody to α6 integrin (BD Biosciences), and basal cells were flow cytometry–sorted by surface α6 integrin levels. RNAs were isolated with an RNeasy Micro kit (Qiagen) and fluorometrically quantified (RiboGreen, Molecular Probes). Quality was assessed by RNA 6000 Pico assay (Agilent), and 800 ng were primed with oligo(dT)-T7 primer and reverse-transcribed (Superscript III cDNA synthesis kit; Invitrogen). One round of amplification and labeling was done to obtain biotinylated cRNA (MessageAmp aRNA kit, Ambion), and 10 μg of labeled cRNA was hybridized at 45 °C for 16 h to Affymetrix GeneChip Mouse Genome 430 2.0 arrays. Processed chips were read by an argon-ion laser confocal scanner (Genomics Core Facility, Memorial Sloan-Kettering Cancer Center).
Supplementary Material
Acknowledgments
We thank H. Clevers for providing Tcf4 knockout mice, Fuchs lab members for their help and critical discussions of the work and N. Stokes for assistance in the Comparative Biosciences Center animal facility at Rockefeller University. E.F. is an Investigator of the Howard Hughes Medical Institute. H.N. was the recipient of a Ruth Kirschstein postdoctoral fellowship from the National Institutes of Health. This work was supported by the Howard Hughes Medical Institute and a grant from the National Institutes of Health (R01-AR31737).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
AUTHOR CONTRIBUTIONS
H.N. designed and conducted experiments, analyzed data and wrote the paper. B.J.M. generated the Tcf3 cKO mice. M.R. conducted the microarray analysis. L.P. conducted the skin grafting. M.N. and T.M.S. provided technical assistance. H.A.P. conducted the histological analysis. E.F. designed experiments, analyzed data and wrote the paper. All authors read and contributed to the manuscript.
References
- 1.Korinek V, et al. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrill BJ, et al. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 5.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 6.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 8.Alonso L, et al. Sgk3 links growth factor signaling to maintenance of progenitor cells in the hair follicle. J Cell Biol. 2005;170:559–570. doi: 10.1083/jcb.200504131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowry WE, et al. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- 11.Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 12.Van Mater D, Kolligs FT, Dlugosz AA, Fearon ER. Transient activation of beta-catenin signaling in cutaneous keratinocytes is sufficient to trigger the active growth phase of the hair cycle in mice. Genes Dev. 2003;17:1219–1224. doi: 10.1101/gad.1076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 14.Ito M, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 15.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 16.Malanchi I, et al. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 17.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 20.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 22.Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Fleming HE, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2008;2:274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X, et al. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell. 2007;27:183–196. doi: 10.1016/j.molcel.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida M, Han L, Bellido T, Manolagas SC, Kousteni S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin-dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol Chem. 2005;280:41342–41351. doi: 10.1074/jbc.M502168200. [DOI] [PubMed] [Google Scholar]
- 26.Essers MAG, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase RaI and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 29.van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 30.Liu TX, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman CK, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.