Abstract
Transposons populate the landscape of all eukaryotic genomes. Often considered purely genomic parasites, transposons can also benefit their hosts, playing roles in gene regulation and in genome organization and evolution. Peaceful coexistence with mobile elements depends upon adaptive control mechanisms, since unchecked transposon activity can impact long-term fitness and acutely reduce the fertility of progeny. Here, we review the conserved roles played by small RNAs in the adaptation of eukaryotes to coexist with their genomic colonists. An understanding of transposon-defense pathways has uncovered recurring themes in the mechanisms by which genomes distinguish “self” from “non-self” and selectively silence the latter.
Introduction
Transposons thrive as parasites of host genomes. When mobilized, they can disrupt protein-coding genes, alter transcriptional regulatory networks, and cause chromosomal breakage and large-scale genomic rearrangement (McClintock, 1951). Cells must therefore engage in an ongoing struggle to protect genomic integrity by guarding cellular DNA from the activity of mobile elements. Discriminating these parasites from a cell's own protein-coding genes is no small task. Individual transposons fall into many classes and bear little overall resemblance to each other. They employ myriad movement strategies, thus confounding any attempt to target a specific and distinguishable replication intermediate. Instead, our still emerging understanding points to a transposon defense that requires a working memory of each individual element. That memory appears to arise after initial colonization and a period of largely unregulated activity during which the mobility of the element, per se, is the Achilles’ heel that insures its downfall. By jumping into specific loci, transposons become trapped in a silencing program that instructs a small RNA-based immune system to selectively silence homologous elements in germ cells, thus guarding the genetic integrity of the species.
On the whole, transposon families can be categorized into a few broad classes of elements that differ in both their structure and movement strategies. The principal division separates retrotransposons (class I) from DNA transposons (class II). Retrotransposons replicate via an RNA intermediate that is reverse transcribed prior to its integration into the host genome. This class is further segregated into elements that are bounded by long terminal repeats (LTR), similar to those of retroviruses, and those that are not (non-LTR).
Non-LTR elements are subdivided into long interspersed nucleotide elements (LINEs) and short interspersed nucleotide elements (SINEs), depending upon their size and origin. Their expression is invariably driven by the combination of internal promoter and 3′ end formation signals that travel with each new full-length insertion. The autonomous members of this group, those not requiring a helper element for mobility, characteristically contain two internal open reading frames (ORFs): one directing synthesis of a DNA binding protein and the other encoding endonuclease and reverse transcriptase enzymes, which are separated posttranslationally (reviewed in Kazazian, 2004).
LTR elements resemble the retroviruses from which they are apparently derived. They encode gag and pol proteins, which can mediate their replicative transfer to new sites in the genome. Consistent with their viral origins, some LTR elements can move not only within genomes but also from cell to cell. Examples are found within the gypsy family in Drosophila. These elements, termed infectious retroviruses or errantiviruses, possess an envelope (env) gene that enables infection of neighboring cells and even horizontal transfer among species (Kim et al., 1994; Song et al., 1994).
Unlike retrotransposons, for which each transposition event generates an additional copy of itself elsewhere in the genome, class II DNA transposons mobilize via a “cut-and-paste” mechanism. Thus, each transposition event is a zero-sum game wherein one site loses transposon information while another gains it. However, because sequences are duplicated upon element integration and because the excision site must be repaired as the element leaves, most transposition events leave scars in the form of short repeats. Autonomous DNA transposons harbor a transposase gene that recognizes the element's flanking terminal inverted repeats (TIRs) and that catalyzes both excision and reintegration. There are also nonautonomous DNA transposons that require the donation of a transposase protein from another functional element.
The diversity of transposable elements and the degree to which they burden eukaryotic genomes is remarkably variable. In mammals, transposons constitute up to 50% of the genome (reviewed in Kazazian, 2004). In comparison, only ~5% of the Drosophila genome is composed of mobile elements (Bergman et al., 2006). While the Arabidopsis genome maintains numerous members of all classes of transposable elements, the budding yeast S. cerevisiae contains only members related to a single LTR retrotransposon family. Drosophila harbors roughly 150 different element families. These comprise a wide variety of LTR and non-LTR retrotransposons, each of which is present in limited number within the genome. The transposon content of the mouse genome is also dominated by retroelements, but in this case by very large numbers of only a few related elements from the IAP (LTR), LINE1, and SINE B1 (non-LTR) retrotransposon families.
The Nature of Host-Transposon Interactions
For decades, researchers have sought to understand our relationship to and coexistence with the mobile elements that colonize our genomes. Genetic studies have sought to probe mechanisms of transposon control by understanding circumstances in which it is lost. These studies tended to underscore the deleterious effects of unregulated activity. However, it has been apparent from the moment of their discovery, inherent in their being dubbed “control elements” by McClintock (1951), that the relationship between host genomes and transposons might be more mutualistic. Proposed positive roles for transposons have taken many forms, and a few selected case studies serve as examples.
Transposons as Drivers of Speciation
Observations of their underlying role in mating incompatibilities (i.e., hybrid dysgenesis, see below) have led to the proposal that transposable elements might help to promote speciation events (Bingham et al., 1982). This could occur if a particular transposon were to colonize a geographically isolated population of a species. After a period of adaptation, the element would be brought under control. However, if the species were to attempt to re-establish interbreeding, its parental, naive population would be unable to control the transposon and would, therefore, fail to produce fertile offspring (reviewed in Rose and Doolittle, 1983). Many other mechanisms, including single gene-interaction-incompatibilities, are sufficient to induce inviability in F1 progeny (Brideau et al., 2006), so it is not presently clear how major a role transposons play in driving speciation. However, transposon incompatibilities present a validated mechanism for producing reproductive isolation that might be reinforced by any number of additional or subsequent genomic alterations.
Heterochromatin and Genome Organization
Transposable elements disproportionately populate heterochromatic genomic domains, including centromeres and, in some organisms, telomeres (reviewed in Pardue and DeBaryshe, 1999). In the Drosophila genus, telomeres lack the simple repeat structure found in many eukaryotes. This is correlated with a lack of identifiable telomerase components or detectable telomerase activity. Instead, chromosomal ends are maintained by the preferential insertion of the non-LTR retrotransposons, HeT-A, Tart, and TAHRE, in head-to-tail arrays at telomere ends (Levis et al., 1993). This is likely a general property of Dipterans, since no studied member of this order contains a functional telomerase. This implies that in a broad and complex group of animals, transposons have been domesticated and harnessed to solve the end-replication problem (reviewed Villasante et al., 2008). This must require subtle control over both transposon activity and insertion preferences to maintain telomeres that neither shrink nor grow to unacceptable extents. This is an ironic example of how transposons, usually thought to create genome instability, can serve precisely the opposite function.
The Evolution of Genes and Genomes
Sequence composition, expression levels, and tissue and cellular expression patterns are the critical and functional features of eukaryotic genes. Interestingly, transposons are able to drive genic evolution and diversity by impacting nearly every one of these properties. Not only is the transposition machinery capable of duplicating processed genes and generating novel pseudogenes (Esnault et al., 2000), but human endogenous retroviruses (HERVs) also appear to have caused genomic deletions and rearrangements during human evolution (Hughes and Coffin, 2001). Transposons also modify genic sequence composition. In fact, approximately 4% of human genes possess some transposon-derived coding sequences. Such events potentially generate genetic divergences that could drive evolution (Nekrutenko and Li, 2001). Additionally, since many transposons contain their own transcriptional regulatory elements, their mobilization can influence the expression patterns (White et al., 1994) or translational efficiency of neighboring genes (Landry et al., 2001). Finally, immunoglobulin and T cell receptor maturation require RAG1/RAG2-initiated V(D)J recombination. Remarkably, this process is functionally related to transposon excision pathways, providing an example of how cells may have co-opted transposon components and transposition strategies to generate an extremely complex system of genic diversity (Agrawal et al., 1998; Hiom et al., 1998).
On the whole, these properties point to complex relationships between transposable elements and their hosts.
Small RNAs and Transposon Control Pathways
Despite some clear benefits of colonization, any symbiotic relationship between a transposon and its host depends heavily on the ability of the host to tame an element's more aggressive tendencies. The heterogeneous nature of transposon families requires flexible recognition and control mechanisms. That niche has been filled in many eukaryotic organisms by pathways that use small RNAs to guide silencing, which we discuss below. Though we focus here on the dominant role of small RNA pathways in transposon control, it is important to note that other mechanisms may also contribute to element regulation. For example, regulated splicing patterns can impact P element movement, and sequence-specific binding proteins can impact the methylation state of some elements (Laski et al., 1986; Schlappi et al., 1994).
RNA Interference
The discovery of RNA interference (RNAi) has transformed our understanding of gene regulation, mechanisms of heterochromatin formation, and transposon control (Fire et al., 1998). The term RNAi has come to encompass an increasingly broad family of related pathways, in which small RNAs from ~20–30 nucleotides in length serve as guides to target recognition and regulation. In the canonical RNAi pathway, small RNAs are generated from double-stranded precursors by a ribonuclease enzyme termed, Dicer (Bernstein et al., 2001). Small RNAs act in complex with a second defining component of RNAi-related pathways, the Argonaute (AGO) proteins, together forming the RNA-induced silencing complex (RISC) (Hammond et al., 2000; Tuschl et al., 1999). AGO proteins are characterized by the presence of a PAZ and PIWI domain, which fold to form a channel in which a single-stranded small RNA guide is held at each end by one of its constituent domains (Lingel et al., 2003; Song et al., 2003, 2004; Yan et al., 2003). The PIWI domain also harbors nuclease activity. This is formed from a ribonuclease H-like motif and is capable of cleaving RNA transcripts as directed by the small RNA. In addition to target cleavage, RISC can also inhibit protein synthesis and direct chromatin modifications that ultimately lead to transcriptional repression (reviewed in Slotkin and Martienssen, 2007).
Studies of the biological roles of the canonical RNAi pathway have focused largely on the regulation of gene expression. MicroRNAs (see Reviews by R.W. Carthew and E.J. Sontheimer on page 642 and O. Voinnet on page 669 of this issue, and Essay by A. Ventura and T. Jacks in this issue of Cell) serve as endogenous guides of the RNAi pathway and are found broadly throughout plant and animal kingdoms, in which this general regulatory paradigm appears to have separately evolved. MicroRNAs act as key components of gene regulatory circuits, essentially as the posttranscriptional equivalent of transcription factors, impacting nearly all types of biological pathways. However, even before the connection between microRNAs (then called small temporal RNAs) and the RNAi pathway was appreciated, early studies pointed to links between RNAi and the control of selfish genetic elements (Ketting et al., 1999; Reinhart and Bartel, 2002; Tabara et al., 1999; Wu-Scharf et al., 2000). Early mutational hunts for RNAi pathway components pointed to clear overlaps with so-called “mutator” genes, whose alteration mobilized certain class II C. elegans transposons. As catalogs of small RNA species began to emerge from several organisms, a surprisingly large family of microRNAs emerged along with small RNAs that mapped to repetitive, heterochromatic regions or to specific transposable elements (Aravin et al., 2003; Llave et al., 2002; Reinhart and Bartel, 2002). Repeat-associated small-interfering RNAs (rasiRNAs) were particularly abundant in Drosophila germline tissues but seemed to be absent from most larval stages. rasiRNAs are approximately 23–26 nt in length, several nucleotides longer than the 20–24 nt small-interfering RNAs (siRNAs) and micro-RNAs. This pointed to potential differences in the biogenesis mechanisms that generate these two small RNA classes. Subsequently, rasiRNAs were also detected in zebrafish (Chen et al., 2005), presaging the discovery of dominant and conserved roles for small RNA pathways in transposon control across large evolutionary distances.
Piwi Proteins, piRNAs, and Germline Transposon Regulation
It seems almost fitting in retrospect that the discovery of a signature component of small RNA-directed silencing pathways came initially from studies of Drosophila gametogenesis (Lin and Spradling, 1997). The broader class of Argonaute proteins can be divided, in most animals, into two clades. Those most similar to Arabidopsis Argonaute-1 (the AGO clade) generally bind double-stranded RNA (dsRNA)-derived small RNAs, such as microRNAs and siRNAs. These proteins and their binding partners (as a class) show largely ubiquitous expression patterns. The second clade of Argonaute proteins, the Piwi clade, was named after a founding Drosophila family member, which was initially studied because of its effects on gonadal development.
Mutations in Piwi lead to defects in oogenesis and a depletion of germline stem cells (Cox et al., 1998, 2000). The Drosophila genome encodes two additional Piwi family proteins, Aubergine (Aub) and Argonaute 3 (AGO3), which are also expressed primarily within gonadal tissues. While lesions in AGO3 have not yet been analyzed, mutations in Aub disrupt gametogenesis, leading to embryonic axis specification defects and an accumulation of dsDNA breaks in germ cell chromosomes (Harris and Macdonald, 2001; Klattenhoff et al., 2007; Theurkauf et al., 2006). Numerous genetic studies pointed toward these phenotypes being linked to roles of Piwi family members in controlling transposons. For example, piwi mutant animals mobilize the gypsy retrotransposon (Sarot et al., 2004), and aubergine mutations derepress TART (Savitsky et al., 2006) and the P element (Reiss et al., 2004).
Considered together, these studies raised expectations that Piwi proteins might bind to small RNAs that would direct them to silence mobile genetic elements. However, the characterization of Piwi-interacting RNAs (now known as piRNAs) from mammals provided a confusing surprise. In mouse, rat, and human testes, Piwi orthologs indeed bind to small RNA species that were larger than microRNAs and siRNAs, reminiscent of Drosophila rasiRNAs. However, unlike rasiRNAs, mammalian piRNAs are selectively depleted of repeat and transposons sequences, with more than 90% of piRNAs mapping uniquely within mammalian genomes (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006). piRNAs show an inexplicable and overwhelming bias for a 5′ uridine (U) residue but share no other distinguishing sequence features. In adult testis, mammalian piRNAs arise from large genomic clusters whose position but not sequence content is evolutionarily conserved. Similarly, in C. elegans, 21U RNAs are derived from large, continuous genomic tracts and bind to worm Piwi orthologs (Batista et al., 2008; Das et al., 2008; Ruby et al., 2006; Wang and Reinke, 2008). The functions of these tremendously abundant RNA species remains obscure, but some crystallization of genetic and molecular data occurred with the analysis of Piwi-associated RNA populations from Drosophila gonads.
Hybridization to microarrays and small-scale sequencing detected Drosophila piRNAs with complementarity to a variety of mobile genetic elements. These represented several transposons and transposon classes, including roo, the I element, gypsy, and the testis-specific Su(Ste) locus (Saito et al., 2006; Vagin et al., 2006). Overall, Drosophila piRNAs are enriched for species that are antisense to transposons, consistent with the link to transposon control implied by genetic studies. Importantly, the production of piRNAs is independent of Dicer, strongly suggesting that a distinct biogenesis mechanism accompanied their difference in size from canonical small RNAs (Vagin et al., 2006).
While these studies were key indicators of the direct roles of Piwi proteins and piRNAs in transposon control, the underlying construction of the transposon silencing pathway, and even the source of the transposon-targeting piRNAs, remained a mystery. Illumination came from the application of next-generation sequencing technologies and a detailed cataloging of small RNAs bound to each of the three Drosophila Piwi proteins (Brennecke et al., 2007; Gunawardane et al., 2007).
Despite their differences in sequence content, fly and mammalian piRNAs share many features (reviewed in Klattenhoff and Theurkauf, 2008). Strikingly, Drosophila piRNAs also arise from chromosomal clusters, though both the content and organization of these differed from their mammalian counterparts. Virtually all Drosophila piRNA clusters lie in heterochromatin, with the most prominent sitting at heterochromatin/euchromatin boundaries near the centromeres of each chromosome. piRNA clusters reside in the most repeat-rich regions of the Drosophila genome and are composed of ancient fragmented transposon copies that are significantly diverged from active transposon consensus sequences. Thus, they give rise to piRNA populations that can be matched to Drosophila transposons, representing all major classes and element families.
The hypothesis that piRNAs directly control transposons was virtually confirmed by the observation that two major piRNA clusters had already been identified as transposon regulatory loci without any underlying molecular explanation of how control was exerted. One such locus was X-TAS at cytological position 1A that conferred the ability to silence the P element (Biémont et al., 1990; Ronsseray et al., 1991). A second locus was flamenco, situated near the centromere of the X chromosome, which had been identified as a master regulator of several LTR retrotransposons of the gypsy family, including gypsy itself, ZAM, and Idefix (Mével-Ninio et al., 2007; Prud'homme et al., 1995).
piRNA Ping-Pong
Overall, Drosophila piRNA populations are strongly enriched for sequences antisense to transposons, consistent with their recognition and silencing of transposon mRNAs (Brennecke et al., 2007; Gunawardane et al., 2007). This occurs despite most clusters containing randomly oriented transposon fragments and giving rise to piRNAs from both strands. Piwi and Aub complexes mirrored the overall antisense bias; however, AGO3 behaved differently, harboring mainly sense-oriented small RNAs. In the few cases wherein AGO3 complexes were enriched for antisense species, the orientation of the Piwi and Aub-bound species also flipped, suggesting a mechanistic relationship between these complexes (Brennecke et al., 2007), which is supported by the physical interaction seen between Piwi proteins in zebrafish (Houwing et al., 2008).
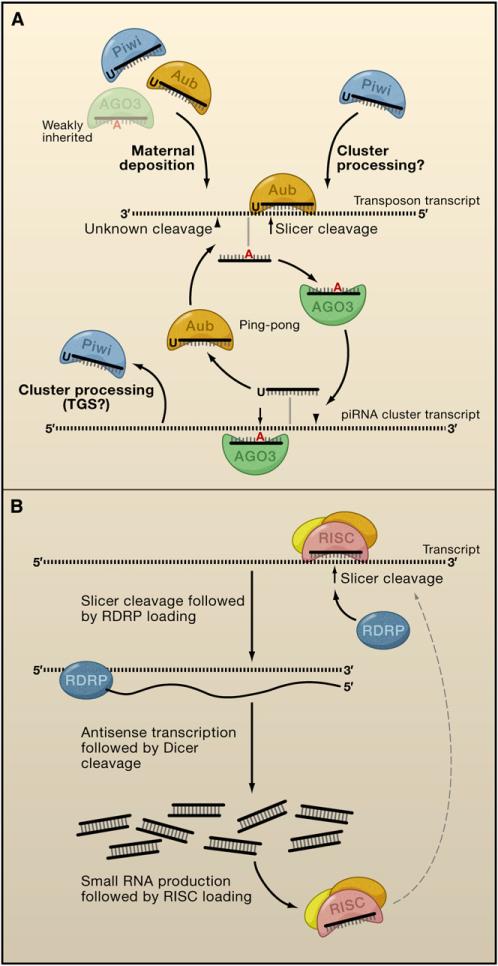
Indeed, sense and antisense piRNAs targeting individual transposons tended to have overlapping 5′ ends separated by precisely 10 nt (Brennecke et al., 2007; Gunawardane et al., 2007). This relationship was consistent with prior demonstrations that piRNAs were not produced by a Dicer-dependent mechanism but did suggest an alternative. Many studies of Argonaute activity demonstrated that it cleaves its target 10 nt from the 5′ end of the guide (Hammond et al., 2000; Zamore et al., 2000). Piwi proteins share this property (Saito et al., 2006), suggesting that Piwi-mediated cleavage could have a role in producing the 5′ ends of sense and antisense piRNAs. These studies led two groups to propose a model for piRNA biogenesis and amplification now known as the ping-pong cycle (Brennecke et al., 2007; Gunawardane et al., 2007) (Figure 1A).
Figure 1. Strategies for Shaping Small RNA Populations.
Ping-pong and RNA-dependent RNA polymerase (RDRP) cycles amplify small RNA populations against target molecules.
(A) Maternal, and possibly primary, cluster-derived piRNAs initiate the ping-pong cycle by targeting the cleavage of transposon transcripts. A secondary piRNA is then produced after a 3′ cleavage of unknown source. AGO3 loads secondary piRNAs, which then target the cleavage of antisense transposon transcripts from piRNA clusters. A novel piRNA is subsequently produced and loaded into Aub, which can then target an additional transposon transcript, restarting the amplification cycle. (TGS, transcriptional gene silencing.)
(B) Transcripts targeted by small RNAs are cleaved by the RNA-induced silencing complex (RISC). RDRP subsequently loads and reverse-transcribes the transcript. The new dsRNA molecule is recognized and processed into small RNAs by a Dicer protein, followed by loading of single-stranded small RNAs into RISC. Finally, RISC is able to target additional cellular transcripts and restart the cycle.
In this model, the cycle is initiated by generating what we refer to here as primary piRNAs, which are sampled from piRNA clusters. The set of cluster-derived small RNAs that are antisense to expressed transposons identify and cleave their targets. This results in the genesis of a new, sense piRNA in an AGO3 complex, termed a secondary piRNA. The AGO3-bound sense piRNA then seeks a target, likely a transposon-cluster transcript that contains antisense transposon sequences. AGO3-directed cleavage generates additional antisense piRNAs capable both of actively silencing their target element and reinforcing the cycle through the creation of additional sense piRNAs (Figure 1A). Since Argonaute proteins are catalytic, the activities and abundances of individual family members can be balanced to bias the system toward antisense species.
The combination of transposon-rich piRNA clusters and the ping-pong amplification cycle creates an elegant small RNA based immune system with both genetically encoded and adaptive phases. The piRNA clusters themselves form a genetic record of transposon exposure and control. Clusters also supply primary piRNAs and antisense transcripts as a substrate to the adaptive phase. The ping-pong cycle can make use of primary piRNAs, combining these with mRNA transcripts from active transposons to optimize the activity of the pathway against the mobile elements that challenge any individual organism. In the long term, transposon control is gained by transposition of an element into a piRNA cluster, as has been observed for insertion of the P element into X-TAS (Ronsseray et al., 1991). Thus, the system provides a means to discriminate diverse transposon classes from endogenous genes based upon the one unique property that defines these genomic parasites, their mobility. Signatures of the ping-pong cycle have been detected and confirmed in a number of organisms, including zebrafish (Houwing et al., 2007, 2008) and mouse (Aravin et al., 2007, 2008), suggesting the conservation of this mechanism to combat transposons in germline tissues.
The ping-pong cycle is functionally analogous to the production of secondary siRNAs via RNA-dependent RNA polymerase (RDRP) activity in plants, worms, and S. pombe, in the sense that it leads to an amplification of small RNAs (reviewed in Hartig et al., 2007) (Cogoni and Macino, 1999; Dalmay et al., 2000; Mourrain et al., 2000; Smardon et al., 2000) (Figure 1B). However, unlike secondary siRNA production, ping-pong appears to have no ability to spread small RNA production along target sequences outside of the boundaries of the original trigger (Brennecke et al., 2008).
Small RNAs as Vectors for Epigenetic Inheritance
While piRNA clusters and their participation with transposon mRNAs in the ping-pong model accounted for many aspects of transposon silencing in Drosophila, several observations went unexplained. When strains of wild-caught Drosophila melanogaster were crossed to laboratory strains, a surprising incompatibility was observed. Progeny from laboratory males and wild females developed normally and were fertile. However, progeny of wild males and laboratory females displayed both gonadal hypertrophy and sterility (termed dysgenic), despite being genetically identical to those produced in the reciprocal cross (Kidwell et al., 1977; Picard, 1976). This phenotype, hybrid dysgenesis, was accompanied by chromosome breakage and an unusual accumulation of germline mutations (Pélisson, 1981; Rubin et al., 1982).
The underlying cause of hybrid dysgenesis was traced to transposon mobilization in the progeny of intercrossed strains (Bucheton et al., 1984; Kidwell, 1983; Pélisson, 1981; Rubin et al., 1982). In the two best-studied models, either the P or I element had colonized wild populations, but these animals had adapted to effectively silence the element. Laboratory strains had been sequestered before either P or I entered D. melanogaster populations, and thus laboratory strains had no innate immunity to either element. The differential behavior of reciprocal crosses strongly implied the existence of a maternal factor that could influence the ability of progeny to silence inherited elements (Bregliano et al., 1980).
Early clues to the nature of the maternal factor came from observations that Piwi proteins are essential for transposon silencing in the context of several models of hybrid dysgenesis (Reiss et al., 2004; Sarot et al., 2004). Moreover, both Piwi and Aub are maternally deposited and accumulate in the pole plasm, the specialized cytoplasm at the posterior end of the developing embryo that will give rise to the future germline. Small RNAs present in maternal germ cells are also faithfully transmitted to progeny (Blumenstiel and Hartl, 2005); however, since the sperm discards most of its cytoplasm postmeiotically, similar species are likely not paternally inherited. This gave rise to clear differences in the embryonic content of piRNAs, depending upon whether an element was maternally or paternally inherited, and these differences correlated perfectly with the ability of progeny to silence the dysgenesis-inducing transposon (Brennecke et al., 2008). These studies demonstrated that differences in the inheritance of maternal small RNA populations underlie hybrid dysgenesis. They also highlighted the broader conclusion that maternally inherited small RNAs are required to prime resistance pathways at each generation in order to effectively silence at least some elements, and the presence of sequences within a piRNA cluster corresponding to a particular element may not alone be sufficient to achieve effective silencing in the absence of maternal small RNAs (Brennecke et al., 2008).
In most dysgenesis systems, fertile progeny can emerge from dysgenic crosses at a very low frequency. This allows populations to eventually adapt to exposure to a new element. This has been modeled with the I element, which required up to 15 generations for a sensitive population to gain full control (Pélisson and Bregliano, 1987). Interestingly, this outcome required continuity of the maternal lineage, consistent with a successive accumulation of maternally transmitted immunity. The penetrance of the dysgenic phenotype can also be influenced by external factors, including the temperature at which the mother is reared and her age (Bucheton, 1978). While it remains to be proven, one hypothesis is that environment can influence the content of maternal small RNA populations and thus alter the phenotype of progeny in a heritable manner using small RNAs as the vector to transmit epigenetic information.
Thus far, the transmission of traits via small RNAs has only been observed in Drosophila. However, small RNAs, or their binding partners, accumulate in the oocytes of other species (Houwing et al., 2007; Watanabe et al., 2006), suggesting the possibility of widespread roles for small RNA pathways in exerting maternal effects on the phenotypes of their progeny.
RNAs and Transposon Defense in Mammals
Observations emerging from Drosophila fueled a reevaluation of the roles of piRNAs in mammalian transposon control. When transposon expression was looked at directly, it became clear that in mutations of the two mammalian Piwi proteins, mili and miwi2, both LINE-1 (non-LTR) and IAP (LTR) retrotransposons showed increased expression (Aravin et al., 2007; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008). This strongly predicted the existence of piRNA populations that could target transposons in mammals. Here, transposon control occurs by transcriptional gene silencing, where DNA methylation patterns maintain the state set during embryogenesis in developing male germ cells, called prospermatogonia (Kato et al., 2007). The expression of Mili and Miwi2 could be detected in this cell type, and these bound to populations of piRNAs that were indeed enriched for transposons (Aravin et al., 2007, 2008; Carmell et al., 2007). Like AGO3, Mili shows a preference for piRNAs corresponding to transposon sense strands, while Miwi2 contains mainly antisense piRNAs (Aravin et al., 2006, 2008; Girard et al., 2006). Also paralleling the fly system, there is a strong signature of the ping-pong amplification cycle, with sense and antisense species showing the distinctive 10 nt 5′ overlap.
piRNAs in prospermatogonia are derived from transposon-rich piRNA clusters, much as is observed in Drosophila (Aravin et al., 2007, 2008; Brennecke et al., 2007). There are both one-stranded clusters, similar to those first seen in mammals, and two-stranded clusters that mirror the majority seen in Drosophila. An appreciable difference between the mammalian and fly systems can be seen in that sense-oriented piRNAs are enriched for primary species (1U, no 10A), whereas antisense species are mainly secondary (no 1U, 10A). Thus, isolated transposons seem to initiate the piRNA pathway in mammals and use the ping-pong pathway to engage cluster-derived transcripts as a source of antisense information (Aravin et al., 2008). The importance of the ping-pong cycle and the obligate link between Mili and Miwi2 is emphasized by the observation that the Miwi2 protein both fails to bind small RNAs and is lost from the nucleus in Mili mutants (Aravin et al., 2008).
Transcriptional Gene Silencing
The ping-pong cycle, with its piRNA-directed consumption of transposon transcripts, has the capacity to silence transposons solely at the posttranscriptional level. However, studies in the male germline of mammals, considered together with a vast literature in plants and fungi, with earlier hints from Drosophila, indicated that small RNAs could also silence repeat elements at the transcriptional level.
Although plants lack Piwi proteins, they have evolved specialized RNAi systems that generate distinct small RNA classes. This is accomplished through the use of specialized Dicer and Argonaute proteins (reviewed in Slotkin and Martienssen, 2007). In particular, a relatively larger class of 24–26 nt siRNA has been linked to both transposon silencing and DNA methylation (Kasschau et al., 2007). While the structure of this silencing pathway is presently less clear than are the transposon silencing mechanisms in flies, it involves specific recognition of repeat elements by specialized RNA polymerases, RNA pol IV and pol V (Herr et al., 2005; Onodera et al., 2005; Wierzbicki et al., 2008). The activity of these enzymes seems to mark transcripts for recognition by the RNA-dependent RNA polymerase 2 (RDR2) complex for conversion to double-stranded RNA (Dalmay et al., 2000). The resulting dsRNA is processed by dicer-like 3 (DCL3) into ~24 nt siRNAs (Xie et al., 2004), which join one of the plant's 12 Argonaute proteins (AGO4), whose bound small RNA populations are heavily enriched for repeats (Qi et al., 2006). Therefore, disruption of any component of this pathway leads to at least partial loss of DNA methylation on many transposons (reviewed in Matzke and Birchler, 2005). Additionally, centromeric repeats and retrotransposons act to mutually reinforce silencing (May et al., 2005). While it is clear that small RNAs act at the transcriptional level in plants to silence mobile elements, it is not at all apparent how plants distinguish these elements (non-self) from their protein coding genes (self) or precisely what interactions lead from recognition of targets by AGO4 complexes to the deposition of DNA methylation marks.
Some understanding of how RNA-dependent RNA polymerase (RDRP) -dependent systems, like those found in plants, create stable and selective silencing may be gained by comparison to Schizosaccharomyces pombe, which has proven tremendously important to our understanding of the biochemical features of small-RNA directed chromatin modification. S. pombe centromeres are generally related in structure to those of vertebrates (Clarke et al., 1986). While such constitutive heterochromatin is thought to be transcriptionally inert, S. pombe centromeres are in fact transcribed, with this transcription important both for their packaging into heterochromatin and for their function in chromosome segregation (reviewed in Kloc and Martienssen, 2008). S. pombe possesses only a single Argonaute and a single Dicer gene, and disruption of either leads to defects in the formation of centromeric heterochromatin (Volpe et al., 2002). It has been proposed that combined sense and antisense transcription of centromeric repeats gives rise to an initial siRNA population, which directs AGO to cleave transcripts associated with this locus (Irvine et al., 2006). Through a coupling whose biochemical basis is not understood, but which is also observed in plants, cleavage activates the RNA-dependent RNA polymerase complex (RDRC) to generate antisense RNA from targeted transcripts. This produces additional dsRNA, which is subsequently processed into 21–24 nt siRNAs by Dicer (Colmenares et al., 2007). Additionally, RDRCs in C. elegans appear to be capable of directly generating secondary siRNAs, as a result of unprimed RNA synthesis (Sijen et al., 2007).
Again, this reinforcing amplification loop provides the analog of the ping-pong cycle from Drosophila (Figure 1). RDRC generated centromeric siRNAs act through the RITS complex (Verdel et al., 2004), in collaboration with the SHREC complex (Sugiyama et al., 2007), to direct the deposition of histone modifications and to establish a silent chromatin state. The initial dichotomy, how a locus could be both active and silent, was solved by examining the functional output of the small RNA pathway through the cell cycle. During interphase, the locus is indeed silent and small RNA pathways lack substrates from centromeric repeats. However during cell division, when histones re-assort to newly replicated chromosomes, the centromeric repeats are freed from their heterochromatic context and are transcribed. This initiates the silencing cycle and allows the formation of expression-dependent heterochromatin at these sites with each division (Chen et al., 2008; Kloc et al., 2008).
In mammals, the precise biochemical mechanisms that lead to deposition of small RNA-directed methylation marks are unclear. However, epistasis relationships with canonical DNA methylation pathways have been established. Mice with mutations affecting Dnmt3L, the primary initiator of de novo DNA methylation in the mouse germline, display a global loss of DNA methylation (Bourc'his and Bestor, 2004; Kato et al., 2007). Dnmt3L acts downstream of the piRNA pathway since disruption of dnmt3L has minor effects on piRNA populations, consistent with increased expression of the elements, which these mutants fail to silence. In Neurospora, DNA methylation appears only after the deposition of histone modifications, likely pointing to chromatin modifying enzymes as intermediaries between AGO complexes and DNA methyltransferases (Tamaru et al., 2003). In Drosophila, Piwi, Aub and another piRNA pathway component, Spindle-E (spn-E), are essential for transcriptional gene silencing (Haynes et al., 2006; Pal-Bhadra et al., 2004), based on their impacts on the expression of variably silenced markers in Drosophila somatic tissues. Here, the pathway must act through its effects on histone modifications since flies lack DNA cytosine methylation. While effects on marker genes in the soma have been abundantly validated, the impact of transcriptional silencing on transposons is less clear. Indeed, nuclear run-on experiments show that mutations in piRNA pathway components have no impact on the transcription of these elements in ovaries (Sigova et al., 2006).
Endogenous RNAi Pathways in Germline and Somatic Tissues
Despite genetic evidence connecting the Piwi pathway to adult, somatic transposon suppression (Pal-Bhadra et al., 2004), piRNAs have not been detected in somatic tissues. As a result, the mechanisms underlying somatic transposon silencing have remained elusive. One hypothesis is that piRNA-directed patterns of heterochromatin set during embryogenesis could be maintained throughout the life of the organism. However, it has become clear that canonical RNAi pathways also produce endogenous small RNAs, some of which correspond to repeat elements.
Endogenous siRNA pathways have been uncovered in both germline and somatic tissues of Drosophila (Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008). In both contexts, siRNAs are derived from overlapping convergent transcription units and from structured genomic loci, which seem to be dedicated to small RNA generation. Thus, inter- or intramolecular interactions can form dsRNAs that serve as substrates for Dicer-2. Repeat elements also give rise to abundant endo-siRNAs. While the source of dsRNA triggers is less clear in this case, analyses of unambiguously mapping species demonstrates that piRNA clusters and probably dispersed transposon copies also participate in siRNA generation. Whether siRNAs are formed by hybridization of precursor transcripts from both cluster strands or whether they arise from the interaction of cluster transcripts with transposon mRNAs remains to be determined.
Flies with mutations in proteins essential for the endo-siRNA pathway are viable and fertile (Förstemann et al., 2005; Lee et al., 2004; Liu et al., 2003; Okamura et al., 2004), although experiments in cell culture have demonstrated the requirement of the pathway to silence a variety of transposons (Rehwinkel et al., 2006). Thus, in the germline, the endo-siRNA pathway must cooperate with the piRNA pathway in a manner in which the latter can compensate for loss of the former. Interestingly, in flies the reverse is not true, but the piRNA pathway does appear to be dispensable in the female germ cells of mammals, which contain a rich endo-siRNA population corresponding to both genes and repeats (Tam et al., 2008; Watanabe et al., 2008). This raises the possibility that piRNA and endo-siRNA pathways may play more equal, possibly redundant, roles in transposon control in oocytes. Interestingly, one transposon family, MT, is heavily targeted by the endo-siRNA pathway but generates virtually no homologous piRNAs. Loss of Dicer but not of Piwi family proteins in growing oocytes dramatically elevates MT levels (Murchison et al., 2007), demonstrating the active and dominant role of siRNAs in restraining this element.
Endo-siRNAs in both mice and flies also target protein-coding genes. In Drosophila, genic endo-siRNAs are derived either from convergently transcribed, overlapping 3′ UTRs or from dedicated structured loci. A similar situation is observed in plants, where, under certain conditions, siRNAs are preferentially generated from transcription units with overlapping 3′ untranslated regions (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). In flies, loss of AGO2 or Dicer-2 results in measurable but modest effects on the expression of targeted genes. In mouse, the mechanisms which give rise to dsRNA proved more unusual (Tam et al., 2008; Watanabe et al., 2008). An examination of unambiguously mapping small RNA species indicated that the sense-oriented siRNAs came from protein-coding transcripts. However, the antisense species arose from pseudogene copies of the corresponding loci. Deletion of Dicer showed strong impacts on the genes that could be targeted by these small RNAs, suggesting that a subset of mammalian pseudogenes had evolved into antisense regulators, at least in this specialized cell type.
Studies of plants and animals have revealed common themes in repeat silencing, with each relying to different degrees on compartmentalized piRNA or endo-siRNA pathways to repress transposons at the transcriptional or posttranscriptional level. However, in few places has repeat silencing been carried to as ultimate an endpoint as is seen in ciliates.
DNA Rearrangements in Ciliates
Ciliates employ remarkable repeat silencing and heterochromatin formation systems, where repetitive DNA is actually eliminated from their genome during sexual development (reviewed in Yao and Chao, 2005). Despite being a single-celled organism, ciliated protozoans possess both a germline micronucleus and a somatically active macronucleus. During sexual development, the developing macronuclear genome undergoes extensive chromosomal breakage and DNA elimination. Eliminated elements are typically transposon derived, representing between 6,000 and 100,000 individual elements, and comprise between 10% and 95% of the germline genome (reviewed in Coyne et al., 1996). In the germline of spirotrichous ciliates, some exons are even scrambled and must be reordered during formation of the somatic genome. This unscrambling utilizes functional RNA transcripts to template proper reassembly of coding sequences in the developing macronucleus (Nowacki et al., 2008). This represents one of many studies that have indicated that RNA plays a key role in directing DNA rearrangements.
RNAi-related mechanisms are critical for excision of the germline-limited DNA in Tetrahymena and Paramecium. Prior to elimination, these sequences are bidirectionally transcribed, giving rise to dsRNA (Chalker and Yao, 2001; Lepère et al., 2008a), which is processed by Dicer (Lepère et al., 2008b; Malone et al., 2005; Mochizuki and Gorovsky, 2005) to generate 25–32 nt small RNAs, called scan RNAs (scnRNAs). Although these are produced by Dicer processing, they join a Piwi family protein, Twi1p (in Tetrahymena), which is also required for elimination (Mochizuki et al., 2002). Thus, scnRNAs are likely the ciliate equivalent of piRNAs.
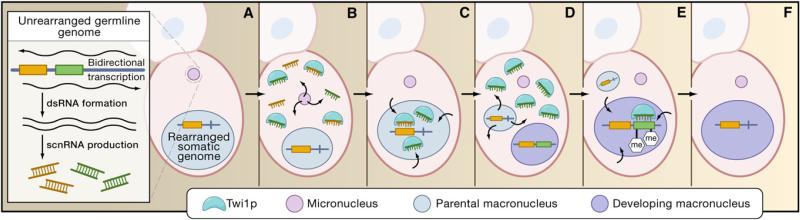
Twi1p appears to load scnRNA populations and then “scans” the parental, rearranged, macronuclear genome (Figure 2). The scnRNA population develops a memory of the rearranged sequences, in the current model, by depleting scnRNAs corresponding to all elements that persist within the parental somatic nucleus (Figure 2C). The population is then transferred into the new macronucleus, where the remaining scnRNAs target homologous DNA for elimination (Figure 2E). Throughout this process, Twi1p directly interacts with both parental and zygotic transcripts via the activity of an RNA helicase Ema1p (Aronica et al., 2008), and in paramecium these transcripts are essential for faithful sequence elimination (Lepère et al., 2008a).
Figure 2. Model of RNAi-Based Scanning and Sequence Elimination in Tetrahymena thermophila.
Simplified view of Twi1p scanning and targeting of sequence elimination. Nuclear development is not depicted (see Matzke and Birchler, 2005, for a detailed overview of nuclear progression during development). Each depicted cell is one of a mating pair (shaded cells).
(A) Generation of scan RNAs (scnRNAs) from the eliminated (green) and noneliminated (orange) germline sequences.
(B) Micronuclear export and loading of scnRNAs into cytoplasmic Twi1p.
(C) Twi1p import into the parental macronucleus, where Twi1p with bound scnRNAs “scans” the parental genome and are depleted from the population if matching to the parental, rearranged genome.
(D) Export of Twi1p, bound with eliminated-element-enriched scnRNAs, to the cytoplasm.
(E) Twi1p import into the developing macronucleus followed by targeting of elements to be eliminated with histone modifications (me, methylation).
(F) The newly rearranged macronuclear genome after recognition and elimination of elements targeted by the rearrangement machinery.
Chromatin remodeling enzymes and histone modifications appear to act as the intermediate guides to DNA elimination (Liu et al., 2004, 2007; Taverna et al., 2002). Thus, the process seems analogous to small RNA-guided heterochromatin formation in plants and animals, though with a dramatically different outcome. The shaping of functional small RNA populations via comparison to the parental macronuclear genome also seems to parallel the amplification of piRNA populations via ping-pong in multicellular animals, though the mechanisms used to accomplish the goal are clearly different.
It will be critical to deepen our understanding of DNA elimination, as comparative studies of plant, metazoan, and ciliate systems will likely reveal the core underlying properties of transposon recognition and control systems. However, ciliates must also solve several more specialized problems. Once a sequence is eliminated from the macronucleus, it is also eliminated in subsequent generations (Garnier et al., 2004; Meyer, 1992). Thus, errors are propagated in general but can be reversed by mating to a parent that retains a particular sequence, which dominantly instructs retention in both daughter macronuclei (Chalker and Yao, 1996; Duharcourt et al., 1995). It is tempting to speculate that this type of reversible (on a multigenerational time scale) remodeling of the genome might serve as a form of epigenetic memory and inheritance or as a catalyst of genome evolution; however, evidence supporting such a thesis has not yet emerged. Despite the necessary machinery, it seems particularly odd that Tetrahymena chooses to delete selfish DNA only from the somatic nucleus. This could indicate that, as in the examples posed above, ciliates may derive some benefit from the conservation of repetitive DNA in their germline DNA. Answers to such questions may come from the sequence of ciliate micronuclear genomes, which are currently being determined.
Perspective
The control of mobile genetic elements boils down to two major problems. The first is one of self versus non-self recognition. This requires that transposons be somehow distinguished from endogenous genes and selectively targeted for silencing. The second problem centers on an ability to recognize and repress such a diversity of element types. In the evolutionary one-upmanship that must drive the evolution of selfish genetic elements, an organism is not only unable to anticipate the sequences of elements which might newly invade their genome but also cannot anticipate that their structure or replication strategy would be one encountered by that species previously. Small RNA pathways represent an ideal approach to compensate for such uncertainty. They provide a flexible system for recognizing nucleic acids and for inhibiting their activity at virtually every level of gene expression. Thus, a class II transposon might be targeted by altering its chromatin structure, by destroying the mRNAs, which it encodes, or by preventing the translation of factors required for its movement. The evolution of recognition requires only that the sequence of the element be converted into small RNAs through one of a variety of mechanisms. The key is to establish specificity toward “foreign” elements, and studies of transposon control, particularly in Drosophila, have begun to provide insights into how this occurs. In at least a few cases, it has been demonstrated that the acquisition of transposon resistance correlates with the insertion of an element into a piRNA cluster (Ronsseray et al., 1991). This immediately incorporates the sequence of that element into the basic silencing program of the organism, and it does so by exploiting the only conserved and universal property of transposons—that they move. Clearly, in at least some cases, being part of the basic silencing repertoire is insufficient for fully effective silencing (Brennecke et al., 2008). Instead, there must be an additional step to magnify responses against active elements. In metazoans, this appears to be accomplished by the ping-pong amplification cycle, which pairs piRNA cluster transcripts and transposon mRNAs in a mutually reinforcing small RNA biogenesis loop. In S. pombe, Argonaute-mediated cleavage of transcripts from centromeric repeats provokes the creation of additional double-stranded RNA, and thus small RNAs, by recruiting an RNA-dependent RNA polymerase. In ciliates, the precise mechanism that optimizes small RNA populations is unclear; however, there appears to be an element of genome comparison that hones small RNA populations for the efficient elimination of targeted repeats. Plants may also optimize their small RNA populations, perhaps by linking recruitment of polIV and polV to target loci with some prior small RNA-directed modification.
Given the variety of pathogens and parasites that plague every living organism, it is likely that genomic parasites evolved hot on the heels of the emergence of replicating genomes. The deeply conserved use of small RNAs as mechanisms to defend genomes against mobile elements points to this being a very early, or perhaps even the ancestral, role for RNAi-related pathways. In this regard, Argonaute proteins themselves are related to transposase enzymes (Song et al., 2004), suggesting that perhaps transposons and their control mechanisms evolved in concert, allowing the parasite to efficiently colonize without destroying its host. Irrespective of the evolutionary roots of transposon control, these species have come to form substantial components of our genomes, including up to 99% of genomic DNA in some species of lily. By taming these elements, organisms have achieved not only some measure of détente with transposons but also the ability to use socialized elements to organize genomes, promote the evolution of genome structure and gene regulation, and in some cases play essential roles in maintaining genome integrity.
ACKNOWLEDGMENTS
We thank Douglas Chalker for critical reading of the manuscript and members of the Hannon Laboratory for helpful discussion. C.D.M. is a Beckman fellow of the Watson School of Biological Sciences and is supported by a National Science Foundation Graduate Research Fellowship. This work was supported in part by grants from the National Institutes of Health to G.J.H. and a kind gift from Kathryn W. Davis (G.J.H.).
REFERENCES
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, Desouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabéhère D, Ashburner M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Biémont C, Ronsseray S, Anxolabéhère D, Izaabel H, Gautier C. Localization of P elements, copy number regulation, and cytotype determination in Drosophila melanogaster. Genet. Res. 1990;56:3–14. doi: 10.1017/s0016672300028822. [DOI] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc. Natl. Acad. Sci. USA. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bregliano JC, Picard G, Bucheton A, Pelisson A, Lavige JM, L'Heritier P. Hybrid dysgenesis in Drosophila melanogaster. Science. 1980;207:606–611. doi: 10.1126/science.6766221. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Bucheton A. Non-Mendelian female sterility in Drosophila melanogaster: influence of ageing and thermic treatments. I. Evidence for a partly inheritable effect of these two factors. Heredity. 1978;41:357–369. doi: 10.1038/hdy.1978.106. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Amstutz H, Fishel B, Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao MC. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hartig JV, Tomari Y, Förstemann K. piRNAs–the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–1713. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 2001;29:487–489. doi: 10.1038/ng775. [DOI] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr., Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: Factors Affecting Chromosomal Contamination in the P-M System. Genetics. 1983;104:317–341. doi: 10.1093/genetics/104.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Terzian C, Santamaria P, Pélisson A, Purd'homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kloc A, Martienssen RA. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–517. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Medstrand P, Mager DL. Repetitive elements in the 5′ untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–116. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lepère G, Bétermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008a;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2008b doi: 10.1093/nar/gkn1018. in press. Published online December 22, 2008. 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 1992;6:211–222. doi: 10.1101/gad.6.2.211. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Drosophila telomeres: two transposable elements with important roles in chromosomes. Genetica. 1999;107:189–196. [PubMed] [Google Scholar]
- Pélisson A. The I–R system of hybrid dysgenesis in Drosophila melanogaster: are I factor insertions responsible for the mutator effect of the I–R interaction? Mol. Gen. Genet. 1981;183:123–129. doi: 10.1007/BF00270149. [DOI] [PubMed] [Google Scholar]
- Pélisson A, Bregliano JC. Evidence for rapid limitation of the I element copy number in a genome submitted to several generations of I-R hybrid dysgenesis in Drosophila melanogaster. Mol. Gen. Genet. 1987;207:306–313. [Google Scholar]
- Picard G. Non-mendelian female sterility in Drosophila melanogaster: hereditary transmission of I factor. Genetics. 1976;83:107–123. doi: 10.1093/genetics/83.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanagaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reiss D, Josse T, Anxolabéhère D, Ronsseray S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol. Genet. Genomics. 2004;272:336–343. doi: 10.1007/s00438-004-1061-1. [DOI] [PubMed] [Google Scholar]
- Ronsseray S, Lehmann M, Anxolabéhère D. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics. 1991;129:501–512. doi: 10.1093/genetics/129.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR, Doolittle WF. Molecular biological mechanisms of speciation. Science. 1983;220:157–162. doi: 10.1126/science.220.4593.157. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retro-virus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlappi M, Raina R, Fedoroff N. Epigenetic regulation of the maize Spm transposable element: novel activation of a methylated promoter by TnpA. Cell. 1994;77:427–437. doi: 10.1016/0092-8674(94)90157-0. [DOI] [PubMed] [Google Scholar]
- Sigova A, Vagin V, Zamore PD. Measuring the rates of transcriptional elongation in the female Drosophila melanogaster germ line by nuclear run-on. Cold Spring Harb. Symp. Quant. Biol. 2006;71:335–341. doi: 10.1101/sqb.2006.71.031. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA. rasiRNAs, DNA damage, and embryonic axis specification. Cold Spring Harb. Symp. Quant. Biol. 2006;71:171–180. doi: 10.1101/sqb.2006.71.066. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A, de Pablos B, Méndez-Lago M, Abad JP. Telomere maintenance in Drosophila: rapid transposon evolution at chromosome ends. Cell Cycle. 2008;7:2134–2138. doi: 10.4161/cc.7.14.6275. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- White SE, Habera LF, Wessler SR. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc. Natl. Acad. Sci. USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]