Understanding how base pairing and stacking mediate the dissipation of electronic energy in DNA is essential for understanding the initial steps in UV photodamage. Excited states of individual DNA bases decay in solution to the ground state either directly by ultrafast internal conversion or, in the case of pyrimidine bases, indirectly via longer-lived triplet and 1nπ*states.1 The base stacking present in single- and double-stranded DNA causes these systems to have dramatically longer excited-state lifetimes compared to monomeric bases.2-5 Femtosecond transient absorption experiments have detected long-lived excited states in numerous π-stacked systems from dinucleosides5 to G-quadruplexes.1 These long-lived states are formed in high yields only when π-stacking is present and are observed in stacks comprised of both AT and GC base pairs.3,4,6 A recent model assigns these states to charge-transfer (CT) excited states or exciplexes formed between π-stacked bases that arise from initially populated Frenkel exciton states.3-5 Although base stacking is clearly a requirement for the formation of these long-lived states, the consequences of base pairing on DNA excited-state dynamics is still highly uncertain and is the motivation for this study.
The possibility that UV mutagenicity is a consequence of proton transfer between paired bases was proposed many years ago.7 More recently, proton transfer was suggested to be responsible for the photostability of DNA.8-10 Using IR-UV hole-burning spectroscopy, Abo-Riziq et al. observed a broad UV spectrum for isolated Watson Crick (WC) GC base pairs in the gas phase, whereas sharp UV spectra were observed for non-WC GC base pairing combinations.11 The broad UV spectrum unique to GC base pairs in the WC conformation was suggested to result from lifetime shortening due to a proton transfer mechanism. Subsequent ab initio calculations implicated an ultrafast deactivation pathway between the excited 1ππ* state and ground state mediated by proton transfer.9,12 Recently, Schwalb and Temps reported shortened fluorescence lifetimes in isolated WC GC base pair analogs relative to the monomers in chloroform using fluorescence up-conversion spectroscopy.13 However, because these model systems lack π-stacking interactions, their relevance to duplex DNA is uncertain.
Here, we investigate the effect of base pairing on excited-state dynamics in GC-containing duplexes when base stacking is also present. This study is timely given recent reports of inter- and intrastrand charge transfer states in computational studies of excited states in double-stranded DNA.9,12,14-16 We report the discovery of a pronounced isotope effect on excited-state lifetimes in an alternating GC-oligonucleotide, demonstrating that interstrand hydrogen bonds can significantly affect excited-state dynamics in double-stranded DNA.
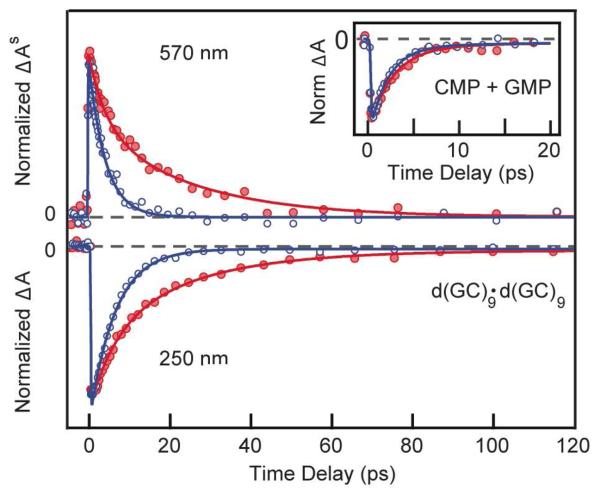
Transient absorption signals were recorded for d(GC)9·d(GC)9 at an excitation wavelength of 266 nm and probe wavelengths of 250 nm and 570 nm in H2O and D2O, as shown in Figure 1. Figure 1 shows that there is significantly faster ground-state recovery for d(GC)9·d(GC)9 in H2O compared to D2O. In contrast, only a minor isotope effect (Figure 1 inset) is observed on the more rapidly decaying signals for an equimolar mixture of the 5′-mononucleotides, CMP and GMP. Circular dichroism spectra for d(GC)9·d(GC)9 are identical in H2O and D2O (Figure S1), indicating that replacement of exchangeable hydrogens by deuterium atoms does not measurably perturb the duplex structure. The observed isotope effect is thus not the result of a change in secondary structure.
Figure 1.
Normalized transient absorption signals showing excited-state absorption (upper panel) and ground-state bleach recovery (lower panel) of d(GC)9·d(GC)9 in H2O (blue circles) and D2O (red circles). The inset shows the 250 nm transient for an equimolar mixture of the monomers CMP and GMP in H2O (blue circles) and D2O (red circles). Signals at 570 nm were corrected for solvated electrons using the procedure described previously.2 Solid curves are nonlinear least-squares fits to the data points.
In H2O, the transient signals decay monoexponentially with time constants of 4.1 ± 0.3 ps and 6.3 ± 0.4 ps for probe wavelengths of 570 and 250 nm, respectively (Table 1). Measurements at 250 nm monitor ground-state repopulation after excitation; whereas, probing at 570 nm reports on excited-state populations.17 The longer time constant observed at 250 nm compared to the 570 nm probe wavelength likely results from vibrational cooling following fast relaxation from an excited state to the ground state.18 These lifetimes, which are approximately an order of magnitude longer than the fluorescence lifetimes of CMP and GMP,1 are assigned to long-lived exciplex states.4 A small constant offset is seen at longer delay times at 250 nm and is assigned to a minor amount of photobleaching.
Table 1.
Time constants and percent amplitudes (in parentheses) from global fits to transient signals of d(GC)9·d(GC)9 in H2O and D2O.a,b Amplitudes do not sum to one hundred percent due to a small amount of residual photobleaching.
| Solvent | Probe λ (nm) |
τ1 (ps) | τ2 (ps) |
|---|---|---|---|
| H2O | 570 | 4.1 ± 0.3 (100) | -- |
| 250 | 6.3 ± 0.4 (98) | -- | |
| D2O | 570 | 4.1 ± 0.3 (41) | 22 ± 4 (58) |
| 250 | 6.3 ± 0.4 (41) | 22 ± 4 (56) |
Stated uncertainties are twice the standard error.
Parameters with identical values and uncertainties were globally linked during fitting.
In D2O, the signals decay initially with the same time constants seen in H2O (τ1 in Table 1), but an additional, approximately equal-amplitude component with a lifetime of 22 ± 4 ps is also detected (Table 1). In UV/IR experiments on the closely related alternating copolymer poly[d(GC)]·poly[d(GC)] in D2O, Doorley et al.19 observed biexponentially decaying signals with lifetimes of 7 ± 1 and 30 ± 4 ps and approximately equal amplitudes. These values agree well with our observations for d(GC)9·d(GC)9 in D2O probed at 250 nm. The similarity between UV/UV and UV/IR bleach recovery signals for DNA systems was noted earlier.20
Doorley et al.19 argued that the slow component is due to a 1nπ* state of 2′-deoxycytidine. They suggested further that the absence of a slow decay in our earlier UV/UV measurement4 for d(GC)9·d(GC)9 in H2O could have been due to difficulties detecting a 1nπ* state by transient absorption spectroscopy. However, the signals in Fig. 1 clearly show that the 22 ps component is easily observed in D2O, but is absent in H2O. Furthermore, the presence of the 22 ps component at both 250 and 570 nm probe wavelengths for d(GC)9·d(GC)9 in D2O rules out assignment of the long-lived state to a 1nπ* state because nucleobase 1nπ* states do not absorb at visible wavelengths.17
A deuterium kinetic isotope effect is observed in pump-probe experiments on single nucleotides due to different rates of vibrational cooling in H2O vs. D2O following ultrafast ground state repopulation.18 This is the reason for the modest kinetic isotope effect (τD / τH) of 1.3 seen in the CMP and GMP equimolar mixture (Table 2). However, vibrational cooling of a hot ground state is not detectable at a probe wavelength of 570 nm, and the isotope effect seen in d(GC)9·d(GC)9 must have a different origin.
Table 2.
Time constants and percent amplitudes (in parentheses) from global fits to transient absorption signals (266 nm pump / 250 nm probe) for various DNAs.a, b Amplitudes do not sum to one hundred percent due to a small amount of residual photobleaching.
| System | Solvent | τ1 (ps) | τ2 (ps) |
|---|---|---|---|
| CMP + GMP | H2O | 2.0 ± 0.3 (92) | 31 ± 26 (8) |
| D2O | 2.6 ± 0.4 (91) | 31 ± 26 (9) | |
| d(C4G4)·d(C4G4) | H2O | 3.1 ± 0.7 (61) | 22 ± 5 (32) |
| D2O | 3.1 ± 0.7 (60) | 22 ± 5 (33) | |
| d[(GX)9GC] | H2O | 5.1 ± 1.0 (72) | 25 ± 5 (25) |
| D2O | 6.6 ± 2.0 (55) | 25 ± 5 (43) |
Stated uncertainties are twice the standard error.
Parameters with identical values and uncertainties were globally linked during fitting.
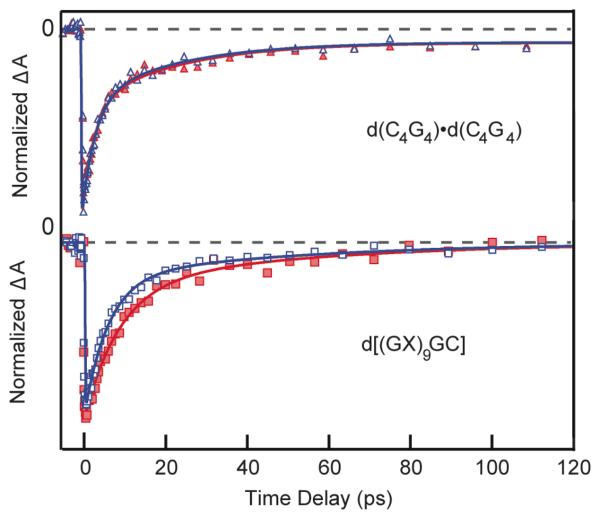
In order to test the hypothesis that the isotope effect is due to interstrand proton transfer, experiments were carried out on d[(GX)9GC], where X is 3-methylcytidine. Methylation of C at N3 prevents WC base pairing with G.21 Bleach-recovery signals of single-stranded d[(GX)9GC], in H2O and D2O are shown in the lower panel of Figure 2. Figure 2 shows that suppressing base pairing between strands with alternating GC bases eliminates the pronounced dynamical differences seen for d(GC)9·d(GC)9 in H2O and D2O.
Figure 2.
Normalized transient absorption signals at 266 nm pump / 250 nm probe of d(C4G4)·d(C4G4) (upper panel) and d[(GX)9GC] (lower panel) in H2O (open markers) and D2O (closed markers).
The d[(GX)9GC] signals at 570 nm (Figure S2) and 250 nm were fit to two exponentials plus a time-independent offset. As shown in Table 2, a 5 to 7 ps component is found along with an additional 25 ± 5 ps component that agrees well with the 22 ± 4 ps lifetime found for d(GC)9·d(GC)9 in D2O. This lifetime, which is similar to the value of 12 ± 8 ps reported for the RNA dinucleoside monophosphate CpG,5 is assigned to an intrastrand exciplex state. The kinetic isotope effect of 1.3 observed for the fast decay component of d[(GX)9GC] is consistent with vibrational cooling, which could be a consequence of rapid deactivation of monomer-like excited states formed where bases are less well stacked in this single-stranded form.
The lack of an isotope effect in d[(GX)9GC] where base pairing is absent, strongly suggests that an interstrand process contributes to the excited-state dynamics of d(GC)9·d(GC)9. Significantly, no isotope effect is observed for the non-alternating GC duplex d(C4G4)·d(C4G4) (Figure 2, upper panel, and Table 2). These findings parallel previous results on AT-containing DNAs where a solvent kinetic isotope effect was observed in aqueous solutions of d(AT)9·d(AT)9, but none was seen for the non-alternating duplex d(A)18×d(T)18.3
The fact that an isotope effect is seen in d(GC)9·d(GC)9, but not in d(C4G4)·d(C4G4), which has the identical base pairing motif, indicates that the quenching mechanism is not restricted to interactions within a single base pair, but instead must involve a pathway that is additionally mediated by base stacking. Crespo-Hernández et al. proposed that the isotope effect observed in alternating d(AT)9·d(AT)9 results from interstrand proton transfer initiated by the formation of an intrastrand exciplex state.3 The present results lend support to the concept that exciplex states with significant CT character enable proton transfer across base pairs. The exciplex state formed in alternating G and C bases in d(GC)9·d(GC)9 is expected to have stronger CT character than in d(C4G4)·d(C4G4). In d(C4G4)·d(C4G4), which contains just a single 5′-CpG-3′ step, the signal is dominated by excimer states formed in CC and/or GG stacks. The latter states may lack sufficient charge separation to drive interstrand proton transfer. Alternatively, as suggested by a reviewer, charge delocalization over like bases in each strand of d(C4G4)·d(C4G4) could prevent the degree of charge localization needed to induce proton transfer.
The formation of an intrastrand exciplex state with strong CT character between stacked cytosine and guanine is expected to give guanine cationic character and cytosine anionic character, while each remains base paired to its neutral complementary base. Importantly, the barriers for proton transfer have been predicted to be lowered in both the one-electron oxidized and one-electron reduced GC base pairs.22-24 Calculations by Li and Sevilla23 predict that proton transfer for the GC radical anion base pair is energetically favorable with a free energy change of −3 kcal/mol and a small activation barrier of 1 kcal/mol. However, proton transfer in the isolated GC radical cation base pair is predicted to be slightly energetically unfavorable.23 Earlier, Bertran et al. predicted that single proton-transfer reaction is endergonic for the GC radical cation base pair, with a calculated barrier of 4.3 kcal mol−1.22
Kumar and Sevilla found that including water molecules to mimic the hydration environment in duplex DNA lowers the predicted free energy change to −0.65 kcal mol−1 with an activation energy of 1.42 kcal mol−1 for the GC radical cation base pair, making proton transfer favorable.24 Recent experimental work by Sevilla and coworkers on double-stranded DNA provides evidence that one-electron oxidized GC base pairs exist in the deprotonated neutral radical form following interbase proton transfer at 77K.25 These studies supporting the feasibility of proton transfer in radical ion base pairs make it plausible that interstrand proton transfer could occur either in concert with or sequential to intrastrand electron transfer initiated by UV absorption.
In summary, a pronounced isotope effect is observed in d(GC)9·d(GC)9, but not in non-alternating or single-stranded GC-containing DNAs. The dynamics suggest that proton-coupled electron transfer is an important decay pathway in duplex DNAs of appropriate base sequence. Proton transfer is thus capable of mediating excited-state decay (i.e. electron-hole recombination) in DNA as well as the rate of hole transport through DNA.26,27 Further work is needed to obtain direct evidence for proton transfer, to determine any long-time photoproducts that may be produced, and to ascertain the timescales of electron and proton motion.
Supplementary Material
Acknowledgment
This research was supported by grants from the National Institutes of Health (R01 GM64563) and the National Science Foundation (CHE-0809754). Measurements were performed in Ohio State's Center for Chemical and Biophysical Dynamics, using equipment funded by the National Science Foundation and the Ohio Board of Regents.
Footnotes
Supporting Information Available: Experimental methods, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Middleton CT, de La Harpe K, Su C, Law YK, Crespo-Hernández CE, Kohler B. Annu. Rev. Phys. Chem. 2009;60:217–239. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- 2.Crespo-Hernández CE, Kohler B. J. Phys. Chem. B. 2004;108:11182–11188. [Google Scholar]
- 3.Crespo-Hernández CE, Cohen B, Kohler B. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 4.Crespo-Hernández CE, de La Harpe K, Kohler B. J. Am. Chem. Soc. 2008;130:10844–10845. doi: 10.1021/ja802183s. [DOI] [PubMed] [Google Scholar]
- 5.Takaya T, Su C, de La Harpe K, Crespo-Hernández CE, Kohler B. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10285–10290. doi: 10.1073/pnas.0802079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de La Harpe K, Crespo-Hernández CE, Kohler B. ChemPhysChem. 2009;10:1421–1425. doi: 10.1002/cphc.200900004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Löwdin PO. Rev. Mod. Phys. 1963;35:724–732. [Google Scholar]
- 8.Schultz T, Samoylova E, Radloff W, Hertel IV, Sobolewski AL, Domcke W. Science. 2004;306:1765–1768. doi: 10.1126/science.1104038. [DOI] [PubMed] [Google Scholar]
- 9.Sobolewski AL, Domcke W, Hättig C. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17903–17906. doi: 10.1073/pnas.0504087102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markwick PRL, Doltsinis NL. J. Chem. Phys. 2007;126:175102/1–175102/1-6. doi: 10.1063/1.2728897. [DOI] [PubMed] [Google Scholar]
- 11.Abo-Riziq A, Grace L, Nir E, Kabelac M, Hobza P, de Vries MS. Proc. Natl. Acad. Sci. U.S.A. 2005;102:20–23. doi: 10.1073/pnas.0408574102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobolewski AL, Domcke W. Phys. Chem. Chem. Phys. 2004;6:2763–2771. [Google Scholar]
- 13.Schwalb NK, Temps F. J. Am. Chem. Soc. 2007;129:9272–9273. doi: 10.1021/ja073448+. [DOI] [PubMed] [Google Scholar]
- 14.Perun S, Sobolewski AL, Domcke W. J. Phys. Chem. A. 2006;110:9031–9038. doi: 10.1021/jp061945r. [DOI] [PubMed] [Google Scholar]
- 15.Lange AW, Herbert JM. J. Am. Chem. Soc. 2009;131:3913–3922. doi: 10.1021/ja808998q. [DOI] [PubMed] [Google Scholar]
- 16.Olaso-González G, Merchán M, Serrano-Andres L. J. Am. Chem. Soc. 2009;131:4368–4377. doi: 10.1021/ja808280j. [DOI] [PubMed] [Google Scholar]
- 17.Hare PM, Crespo-Hernández CE, Kohler B. Proc. Natl. Acad. Sci. U.S.A. 2007;104:435–440. doi: 10.1073/pnas.0608055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middleton CT, Cohen B, Kohler B. J. Phys. Chem. A. 2007;111:10460–10467. doi: 10.1021/jp0740595. [DOI] [PubMed] [Google Scholar]
- 19.Doorley GW, McGovern DA, George MW, Towrie M, Parker AW, Kelly JM, Quinn SJ. Angew. Chem. Int. Ed. 2009;48:123–127. doi: 10.1002/anie.200803904. [DOI] [PubMed] [Google Scholar]
- 20.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernández CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nir E, Janzen C, Imhof P, Kleinermanns K, de Vries MS. Phys. Chem. Chem. Phys. 2002;4:732–739. [Google Scholar]
- 22.Bertran J, Oliva A, Rodríguez-Santiago L, Sodupe M. J. Am. Chem. Soc. 1998;120:8159–8167. [Google Scholar]
- 23.Li X, Cai Z, Sevilla MD. J. Phys. Chem. B. 2001;105:10115–10123. [Google Scholar]
- 24.Kumar A, Sevilla MD. J. Phys. Chem. B. 2009;113:11359–11361. doi: 10.1021/jp903403d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adhikary A, Khanduri D, Sevilla MD. J. Am. Chem. Soc. 2009;131:8614–8619. doi: 10.1021/ja9014869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giese B, Wessely S. Chem. Commun. 2001:2108–2109. doi: 10.1039/b106059g. [DOI] [PubMed] [Google Scholar]
- 27.Kawai K, Osakada Y, Majima T. ChemPhysChem. 2009;10:1766–1769. doi: 10.1002/cphc.200900148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.