Abstract
The effects of 1,25-dihydroxyvitamin D3 [1,25-(OH)2D3] are mediated by the vitamin D receptor (VDR), a member of the nuclear receptor superfamily of transcriptional regulators. We have identified upstream exons of the human (h) VDR gene that are incorporated into variant transcripts, two of which encode N-terminal variant receptor proteins. Expression of the hVDR gene, which spans more than 60 kb and consists of at least 14 exons, is directed by two distinct promoters. A tissue-specific distal promoter generates unique transcripts in tissues involved in calcium regulation by 1,25-(OH)2D3 and can direct the expression of a luciferase reporter gene in a cell line-specific manner. These major N-terminal differences in hVDR transcripts, potentially resulting in structural differences in the expressed receptor, may contribute to cellular responsiveness to 1,25-(OH)2D3 through tissue differences in the regulation of VDR expression.
Keywords: 1,25-dihydroxyvitamin D/gene structure
The active hormonal form of vitamin D, 1,25-dihydroxyvitamin D3 [1,25-(OH)2D3], has a central role in calcium and phosphate homeostasis and the maintenance of bone. Apart from its calcitropic effects, 1,25-(OH)2D3 regulates cell growth and differentiation in many target tissues (1, 2). The effects of 1,25-(OH)2D3 are mediated by the vitamin D receptor (VDR), a member of the nuclear receptor superfamily of transcriptional regulators that also includes steroid, thyroid, retinoid, and many orphan receptors. Upon binding hormone the VDR regulates gene expression by direct interaction with specific sequence elements in the promoter region of hormone-responsive target genes. This transactivation or repression involves multiple interactions with protein cofactors, heterodimerization partners, and the transcription machinery (3, 4).
Complementary DNAs for human and rat VDRs were cloned in 1988 (5, 6) and more recently from quail (7), chicken (8), mouse (9), and Xenopus (10). The gene structure and pattern of transcription have not been analyzed thoroughly, although a recent study (11) reported alternatively spliced human (h) VDR transcripts in the kidney that vary only in their 5′ untranslated regions (5′ UTRs). While the regulation of VDR abundance is important for modulating 1,25-(OH)2D3 responsiveness in target cells (12–14), the mechanisms by which VDR levels are regulated remain unclear. It is evident that ligand-induced stabilization of the VDR protein is one mechanism by which VDR levels are regulated (15–17); however, the relative importance of transcriptional effects is unclear. Our aim was to analyze the structure and pattern of transcription of the hVDR gene and to assess transcriptional control by identifying and characterizing the promoter region(s) of the gene.
In this study, upstream exons of the hVDR gene that are incorporated into 5′ variant transcripts have been identified, suggesting that expression of the hVDR gene is directed by more than one promoter. A subset of these transcripts is expressed in a tissue-specific pattern, and further variant transcripts have the potential to encode N-terminal variant receptor proteins. Multiple, functionally distinct isoforms mediate tissue- and/or developmental-specific effects of many other members of the nuclear receptor superfamily, and we suggest that the hVDR also has multiple mRNA and protein isoforms.
MATERIALS AND METHODS
Isolation and Characterization of Genomic Clones.
A human lymphocyte cosmid library (Stratagene) was screened by using three separate hVDR cDNA fragments: a 2.1-kb fragment encompassing the entire coding region but lacking most of the 3′ UTR, a 241-bp PCR product spanning exons 1–3 and a 303-bp PCR product spanning exons 3 and 4. Each probe was labeled by nick translation (Life Technologies, Gaithersburg, MD) with [α-32P]dCTP. Positively hybridizing colonies were selected, and secondary and tertiary screens were carried out until complete purification. Cosmid DNA from positive clones was purified (Qiagen), digested with different restriction enzymes, and characterized by Southern blot analysis by using specific [γ-32P]ATP-labeled oligonucleotides as probes. Cosmid clones were sequenced directly by using dye-termination chemistry and automated fluorescent sequencing on an ABI Prism 377 DNA Sequencer (Perkin–Elmer).
Rapid Amplification of cDNA 5′ Ends (5′-RACE).
Alternative 5′ variants of the human VDR gene were identified by 5′-RACE using commercially prepared, anchor-ligated cDNA from human kidney (CLONTECH). Two rounds of PCR using nested reverse primers in exons 3 and 2 (P1: 5′-CCGCTTCATGCTTCGCCTGAAGAAGCC-3′; P2: 5′-TGCAGAATTCACAGGTCATAGCATTGAAG-3′) were carried out on a Corbett FTS-4000 Capillary Thermal Sequencer (Corbett Research, Sydney, NSW, Australia). After 26 cycles of PCR, 2% of the primary reaction was reamplified for 31 cycles. PCR products were cloned into PUC18 and sequenced.
Cell Culture.
The embryonal kidney cell line, HEK-293, an embryonal intestine cell line, Intestine-407, and WS1, a fetal skin fibroblast cell line were all cultured in MEM with Earle’s salts and supplemented with either 10% heat-inactivated FBS, 15% FBS, or 10% FBS with nonessential amino acids, respectively. The osteosarcoma cell lines MG-63 and Saos-2 were cultured in MEM with nonessential amino acids and 10% heat-inactivated FBS and McCoy’s 5a medium with 15% FBS, respectively. The breast carcinoma cell line T47D and the colon carcinoma cell lines LIM 1863 [a gift from R. H. Whitehead (18)] and COLO 206F were cultured in RPMI 1640 medium supplemented with 0.2 units bovine insulin/ml and 10% FBS, 5% FBS, or 10% FBS, respectively. HK-2 kidney proximal tubule cells were grown in keratinocyte serum-free medium supplemented with 5 ng/ml recombinant EGF/40 μg/ml bovine pituitary extract. BC1 primary fetal osteoblast-like cells [a gift from R. Mason, (19)] were grown in MEM with 5% FBS and 5 mg/liter vitamin C. For transfection studies, the mouse embryonic fibroblast-like cell line, NIH 3T3, and the monkey kidney-derived fibroblast-like cell line, COS 7, were maintained in DMEM with 10% FBS. Unless otherwise stated all cell lines were obtained from the American Type Culture Collection (Manassas, VA).
Reverse Transcriptase–PCR (RT-PCR).
Total RNA extracted from approximately 1.5 × 107 cells, from leukocytes prepared from 40 ml blood, or from human tissue using acid-phenol extraction was purified by using a guanidium isothiocyanate-cesium chloride step gradient. First-strand cDNA was synthesized from 5 μg of total RNA primed with random hexamers (Promega) using Superscript II reverse transcriptase (Life Technologies). One-tenth of the cDNA (2 μl) was used for subsequent PCR, with 36 cycles of amplification, using exon-specific forward primers (exon 1a: corresponding to nucleotides 1–21 of hVDR cDNA; exon 1d: 5′-GGCTGTCGATGGTGCTCAGAAC-3′; exon 1f: 5′-AAGTTCCTCCGAGGAGCCTGCC-3′) and a common reverse primer in exon 3 [corresponding to nucleotides 301–280 of hVDR cDNA (5)]. All RT-PCRs were repeated multiple times by using RNA/cDNA prepared at different times from multiple sources. Each PCR included an appropriate cDNA-negative control, and additional controls included RT-negative controls prepared alongside cDNA and RNA/cDNA prepared from VDR-negative cell lines. PCR products were separated on 2% agarose and visualized with ethidium bromide staining.
Functional Analysis of hVDR Gene Promoters.
Sequences flanking exons 1a, 1d, and 1f (see Fig. 1A) were PCR-amplified by using Pfu polymerase (Stratagene) and cloned into the pGL3basic vector (Promega) upstream of the luciferase gene reporter. Promoter–reporter constructs were transfected into NIH 3T3 and COS 7 cells by using the standard calcium phosphate-precipitation method. Cells were seeded at 2.3–2.5 × 106 per 150-cm2 flask the day before transfection. Several hours before the precipitates were added the medium was changed to DMEM with 2% charcoal-stripped FBS. Cells were exposed to precipitate for 16 h before subculturing and were harvested 24 h later. The parent vector pGL3basic was used as a promoterless control in these experiments and a simian virus 40 promoter–chloramphenicol acetyltransferase (CAT) gene reporter construct was cotransfected as an internal control for transfection efficiency in each case. The activity of each construct was corrected for transfection efficiency and for the activity of the pGL3 basic empty vector control and expressed as a percentage of the activity of the construct 1a(−488,+75). Luciferase and CAT assays were carried out in triplicate, and each construct was tested in transfection at least three times.
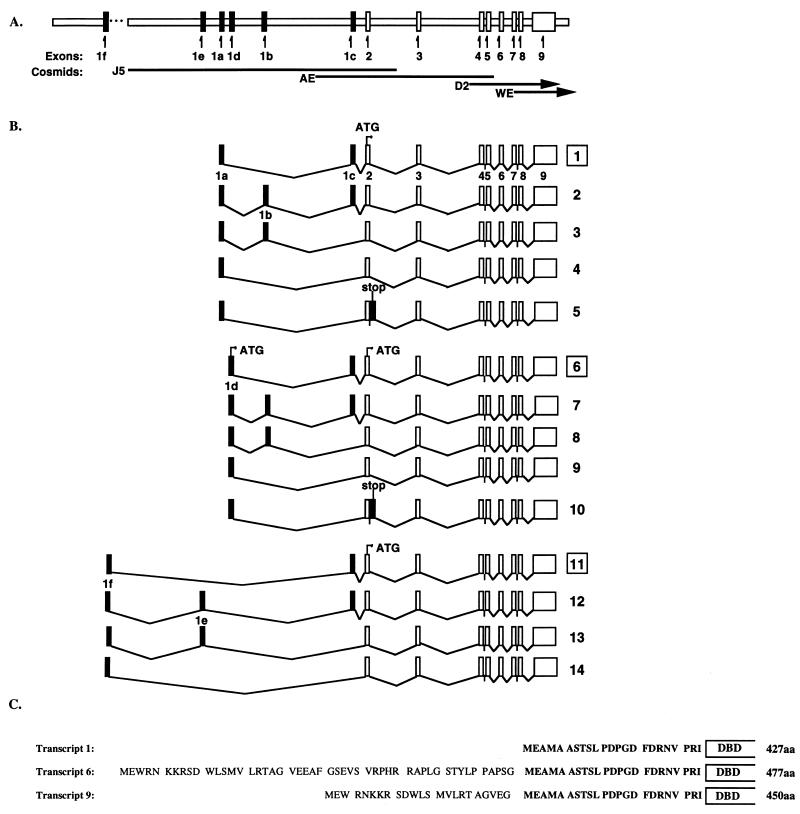
Figure 1.
(A) Human VDR gene locus. Four overlapping cosmid clones were isolated from a human lymphocyte genomic library (Stratagene) and directly sequenced. Clone J5 extends from the 5′ flanking region to intron 2; AE, from intron 1b to intron 5; D2, from intron 3 to the 3′ UTR; WE, from intron 6 through the 3′ flanking region. Sequence upstream of exon 1f was obtained by anchored PCR from genomic DNA. (B) Structure of hVDR transcripts. Transcripts 1–5 originate from exon 1a. Transcript 1 corresponds to the published cDNA (5). Transcripts 6–10 originate from exon 1d and transcripts 11–14 originate from exon 1f. Boxed numbers indicate the major transcript (based on the relative intensities of the multiple PCR products) within each exon-specific group of transcripts generated with a single primer set. While all transcripts have a translation initiation codon in exon 2, exon 1d transcripts have the potential to initiate translation upstream in exon 1d, with transcripts 6 and 9 encoding VDR proteins with extended N termini. (C) N-terminal variant proteins encoded by novel hVDR transcripts. Transcript 1 corresponds to the published cDNA sequence (5) and encodes the 427-aa hVDR protein. Transcripts 6 and 9 code for a protein with an extra 50 aa or 23 aa, respectively, at the N-terminal. The 23 aa of the hVDR A/B domain are shown in bold.
RESULTS
Identification of Alternative 5′ Variants of the hVDR Gene.
Upstream exons were identified in human kidney VDR transcripts by 5′ RACE (exons 1f, 1e, 1d, and 1b) and localized by sequencing of cosmid clones (Fig. 1A). To verify these results and to characterize the structure of the 5′ end of the VDR gene, exon-specific forward primers were used with a common reverse primer in exon 3 to amplify specific VDR transcripts from human tissue and cell line RNA (Fig. 1B). The identity of these PCR products was verified by Southern blot and by cloning and sequencing. Five different VDR transcripts originating from exon 1a were identified. The major transcript (transcript 1 in Fig. 1B) corresponds to the published cDNA sequence (5). Three less-abundant forms (2, 3, and 4 in Fig. 1B) arise from alternative splicing of exon 1c and a novel 122-bp exon 1b into or out of the final transcript. These three variant transcripts were described recently by Pike and colleagues (11). A fifth minor variant was identified (5 in Fig. 1B) that lacks exons 1b and 1c, but includes an extra 152 bp of intronic sequence immediately 3′ to exon 2, potentially encoding a truncated protein as a result of an in-frame termination codon in intron 2.
Four more transcripts were characterized that originate from exon 1f, a novel 207-bp exon more than 9 kb upstream from exon 1a. The major 1f-containing transcript (11 in Fig. 1B) consists of exon 1f spliced immediately adjacent to exon 1c. Three less-abundant variants (12, 13, and 14 in Fig. 1B) arise from alternative splicing of exon 1c and a novel 159-bp exon 1e into or out of the final transcript. All these hVDR variants differ only in their 5′ UTRs and encode identical proteins from translation initiation in exon 2.
Of considerable interest, another five hVDR transcripts were identified that originate from exon 1d, a novel 96-bp exon located 296 bp downstream from exon 1a. The major exon 1d-containing transcript (6 in Fig. 1B) utilizes exon 1d in place of exon 1a of the hVDR cDNA. Three minor variants (7, 8, and 9 in Fig. 1B) arise from alternative splicing of exons 1b and 1c into or out of the transcript, analogous to the exon 1a-containing variants 2, 3, and 4. A fifth minor variant transcript (10 in Fig. 1B) lacks exons 1b and 1c, but includes 152 bp of intron 2 analogous to the exon 1a-containing transcript 5, and also potentially encodes a truncated protein. Two of these exon 1d-containing hVDR transcripts encode an N-terminal variant form of the hVDR protein. Utilization of an ATG codon in exon 1d, which is in a favorable context and in-frame with the major translation start site in exon 2, would generate a protein with an additional 50 aa N-terminal to the ATG codon in exon 2 in the case of variant 6 or 23 aa in the case of variant 9 (Fig. 1C).
The relative level of expression of the different transcripts is difficult to address with PCR since relatively minor transcripts may be amplified. However, Southern blots of PCR products from the linear range of PCR amplification indicated that equivalent amounts of PCR product were accumulated after 26 cycles for exon 1a transcripts compared with 30 cycles for exon 1d transcripts, suggesting that 1d abundance is about 5% of that of 1a transcripts. This is consistent with the frequency of clones selected and sequenced from RACE analysis of two separate samples of kidney RNA: 1a (21/27; 78%), 1d (2/27; 7%), and 1f (4/27; 15%). RT-PCR with exon 1a-, 1d-, or 1f-specific forward primers and reverse primers in exons 7 or 9, followed by cloning and sequencing, suggests that these 5′ variant transcripts are not associated with differences at the 3′ end of the transcript.
Exon–Intron Organization of the hVDR Gene.
Overlapping cosmid clones were isolated from a human lymphocyte genomic library and characterized by hybridization to exon-specific oligonucleotide probes (Fig. 1A). The exon–intron boundaries of the hVDR gene were determined by comparison of the genomic sequence from cosmid clones with the cDNA sequence. Upstream exons were localized in the VDR gene by sequencing cosmid clones, which extend approximately 7 kb into the intron between exons 1e and 1f, enabling verification of both their sequence and the presence of consensus splice donor/acceptor sites. Sequence upstream of exon 1f was obtained by anchored PCR from genomic DNA by using commercially available anchor-ligated DNA (CLONTECH). In total, the hVDR gene spans more than 60 kb and consists of at least 14 exons (Fig. 1A).
Tissue-Specific Expression of hVDR Transcripts.
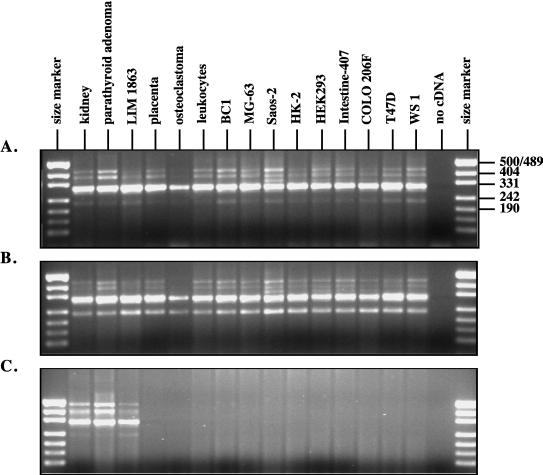
The pattern of expression of variant hVDR transcripts was examined by RT-PCR in a variety of cell lines and tissues with exon 1a-, 1d-, or 1f-specific forward primers and a common reverse primer in exon 3. Exon 1a and 1d transcripts (Fig. 1B, variants 1–10) were coordinately expressed in all RNA samples analyzed (Fig. 2 A and B). Exon 1f transcripts (Fig. 1B, variants 11–14), however, were detected only in RNA from human kidney tissue (two separate samples), human parathyroid adenoma tissue, and an intestinal carcinoma cell line, LIM 1863 (Fig. 2C). Interestingly, these represent major target tissues for the calcitropic effects of vitamin D.
Figure 2.
RT-PCR analysis of expression of variant hVDR transcripts. (A) Exon 1a transcripts (220 bp, 301 bp, 342 bp, 372 bp, and 423 bp). (B) Exon 1d transcripts (224 bp, 305 bp, 346 bp, 376 bp, and 427 bp). (C) Exon 1f transcripts (228 bp, 309 bp, 387 bp, and 468 bp). RT-PCR was carried out with exon 1a-, 1d-, or 1f-specific forward primers and a common reverse primer in exon 3. The sizes of the PCR products and the pattern of bands are similar in A and B by virtue of the identical splicing pattern of exon 1a and 1d transcripts and the fact that primers were designed to generate PCR products of comparable sizes. All tissues and cell lines are human in origin.
Functional Analysis of hVDR Gene Promoters.
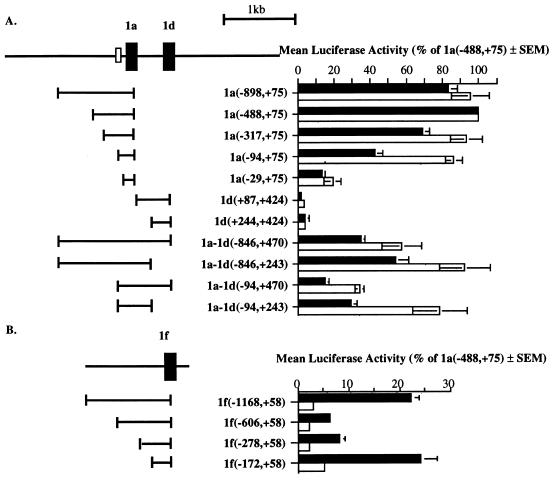
Promoter activities of the 5′ flanking regions of exons 1a, 1d, and 1f were examined in NIH 3T3 and COS 7 cells (Fig. 3). Sequences flanking exon 1a exhibited high promoter activity in both cell lines (Fig. 3A). Maximum luciferase expression of 36- and 54-fold over the empty vector was attained for construct 1a(−488,+75) in NIH 3T3 and COS 7 cells, respectively. This activity could be attributed largely to a GC-rich region containing multiple consensus Sp1-binding motifs lying within 100 bp immediately adjacent to the transcription start site. This region alone, upstream of a luciferase reporter [construct 1a(−94,+75)], accounted for 43% of the maximum activity observed in NIH 3T3 cells and 86% of the maximum observed in COS 7 cells. The removal of this GC-rich region [construct 1a(−29,+75)] reduced luciferase activity to only 13% of the maximum in NIH 3T3 and 19% in COS 7 cells. These results are comparable with those reported recently (11).
Figure 3.
Functional analysis of sequence-flanking exons 1a and 1d (A) and exon 1f (B) in NIH 3T3 (solid bars) and COS 7 cells (open bars). The parent vector pGL3basic was used as a promoterless control, and a promoter-chloramphenicol acetyltransferase (CAT) gene reporter construct was cotransfected as an internal control for transfection efficiency in each case. The activity of each construct was corrected for transfection efficiency and for the activity of the pGL3basic empty vector control and expressed as a percentage of the activity of the construct 1a(−488,+75) ± SEM of at least three separate transfections. Exon 1a and 1d flanking constructs are defined in relation to the transcription start site of exon 1a, designated +1, which lies 54 nt upstream of the published cDNA (5). Exon 1f flanking constructs are defined relative to the exon 1f transcription start site, designated +1. Transcription start sites were determined by the 5′ termini of the longest RACE clones. The open box corresponds to the GC-rich region.
Despite the fact that VDR transcripts that originated from exon 1d were identified, distinct promoter activity was not associated with sequences within 300 bp of exon 1d [constructs 1d(+87,+424) and 1d(+244,+424)]; rather, the sequence immediately adjacent to exon 1d may contain a suppressor element (Fig. 3A). Construct 1a-1d(−846,+470), spanning the 5′ flanking regions of both exons 1a and 1d, resulted in only 42% and 60% of the activity of 1a(−898,+75) in NIH 3T3 and COS 7 cells, whereas the 3′ deletion of 227 bp restored luciferase activity to 65% and 97% of the activity of 1a(−898,+75), respectively. Similarly, the 5′ truncated construct 1a-1d(−94,+470), spanning the 5′ flanking regions of both 1a and 1d, resulted in only 35% and 40% of the activity of 1a(−94,+75), while a further 3′ deletion of 227 bp restored luciferase activity to 69% and 91% of the activity of 1a(−94,+75) in NIH 3T3 and COS 7 cells. It is possible that transcription from exons 1a and 1d is driven by overlapping promoter regions rather than from two distinct promoters, as has been described for the mouse androgen receptor gene (20).
Sequence upstream of exon 1f showed significant promoter activity in NIH 3T3 cells of 22% of that of the most active construct, 1a(−488,+75), or 9-fold over pGL3basic [construct 1f(−1168,+58)] (Fig. 3B). A shorter construct [1f(−172,+58)] had similar activity, with evidence of a suppressor element (between nucleotides −278 and −172) able to repress luciferase activity by 70%. Interestingly, the same constructs were not active in COS 7 cells. This cell line-specific activity of exon 1f flanking sequences may reflect a requirement for tissue- or cell-specific protein factors.
DISCUSSION
We have identified 5′ variant transcripts of the hVDR that suggest the existence of alternative promoters. These transcripts may not have been discriminated in previous Northern analyses because of their similarity in size. Transcription initiation from exons 1a or 1f and alternative splicing generate VDR transcripts that vary in their 5′ UTRs but encode the same 427-aa protein. Transcription initiation from exon 1d and alternative splicing generate hVDR transcripts with the potential to encode variant proteins with an additional 50 or 23 aa at the N terminus. There was no evidence that these 5′ variants are associated with differences at the 3′ end of the transcript. Although isoforms are common in other members of the nuclear receptor superfamily, the only evidence for isoforms of the hVDR is a common polymorphism in the triplet encoding the initiating methionine of the 427-aa form of the VDR that results in initiation of translation at an alternative start codon beginning at the 10th nucleotide downstream, encoding a protein truncated by 3 aa at the N terminus (21). Similarly, two forms of the avian VDR, differing in size by 14 aa, are generated from a single transcript by alternative translation initiation (8), and in the rat a dominant-negative VDR is generated by intron retention (22).
Heterogeneity in the 5′ region is a common feature of other nuclear receptor genes. Tissue-specific alternative-promoter usage generates multiple transcripts of the human estrogen receptor α (ERα), the human and rat mineralocorticoid receptors, and the mouse glucocorticoid receptor (GR), which differ in their 5′ UTRs but code for identical proteins (23–26). However, other members of the nuclear receptor superfamily have multiple, functionally distinct isoforms arising from differential promoter usage and/or alternative splicing. The generation of N-terminal variant protein isoforms has been described for the progesterone receptor (PR), peroxisome proliferator-activated receptor (PPARγ), and the retinoid and thyroid receptors (27–32). Some receptor isoforms exhibit differential promoter-specific transactivation activity. The N-terminal A/B regions of many nuclear receptor proteins possess a ligand-independent transactivation function (AF1). An AF1 domain has been demonstrated for the thyroid receptor β1 (TRβ1), ER, GR, PR, PPARγ, and the retinoid receptors (33–38). The activity of the AF1 domain has been shown to vary in both a tissue- and promoter-specific manner (39–42). The N-terminal A/B region of nuclear receptors is the least-conserved domain across the family and between receptor subtypes, varying considerably both in length and sequence. The VDR, however, is unusual as its N-terminal A/B region is much shorter than that of other nuclear receptors, with only 23 aa N-terminal to the DNA-binding domain, and deletion of these residues seems to have no effect on VDR function (43). This region in other receptors is associated with optimal ligand-dependent transactivation and can interact directly with components of the basal transcription complex (33, 44–46). Two stretches of basic amino acid residues, RNKKR and RPHRR, in the predicted amino acid sequences of the variant hVDR N termini (Fig. 1C) resemble nuclear localization signals (47). An N-terminal variant VDR protein therefore might exhibit different transactivation potential, possibly mediated by different protein interactions, or may specify a different subcellular localization.
The tissue-specific expression of exon 1f-containing transcripts is mediated by a distal promoter more than 9 kb upstream of exons 1a and 1d. Exon 1f transcripts were detected only in kidney tissue, parathyroid adenoma tissue, and an intestinal cell line, LIM 1863. It is interesting that these tissues represent major target tissues for the calcitropic effects of vitamin D. The absence of 1f-containing transcripts in two other kidney cell lines, HK-2 (proximal tubule) and HEK-293 (embryonal kidney), as well as one other embryonal intestinal cell line, Intestine-407, suggests that the expression of 1f transcripts is cell type-specific. The cell line-specific activity of exon 1f flanking sequences in promoter reporter assays may reflect a requirement for tissue- or cell-specific protein factors to mediate expression from this promoter.
This study has demonstrated that expression of the human VDR gene, which spans more than 60 kb and consists of 14 exons, is under complex transcriptional control by multiple promoters. The expression of multiple exon 1f transcripts is mediated by utilization of a distal tissue-specific promoter. Transcription from a proximal promoter, or promoters, generates multiple variant hVDR transcripts, two of which code for N-terminal variant proteins. Multiple, functionally distinct isoforms mediate the tissue- and/or developmental-specific effects of many members of the nuclear receptor superfamily. Although the actual relative abundance of the various transcripts and their levels of translation in vivo have not yet been characterized, this paper suggests that major variant isoforms of the hVDR exist. Differential regulation of these hVDR gene promoters and of alternative splicing of variant VDR transcripts may have implications for understanding the various actions of 1,25-(OH)2D3 in different cell types, and variant VDR transcripts may play a role in tissue-specific VDR actions in bone and calcium homeostasis.
Acknowledgments
Cosmid clone D2 was a gift from Dr. C. Hahn of Adelaide University, Australia. We thank E. Buttrose and T. Cock for their technical assistance and Dr. C. White for obtaining human tissues. This work was supported by the National Health and Medical Research Council of Australia.
ABBREVIATIONS
- VDR
vitamin D receptor
- hVDR
human VDR
- 1
25-(OH)2D3, 1,25-dihydroxyvitamin D3
- 5′ UTR
5′ untranslated region
- 5′ RACE
rapid amplification of cDNA 5′ ends
Footnotes
References
- 1.Ross T K, Darwish H M, DeLuca H F. Vitam Horm. 1994;49:281–326. doi: 10.1016/s0083-6729(08)61149-8. [DOI] [PubMed] [Google Scholar]
- 2.Christakos S, Raval-Pandya M, Wernyj R P, Yang W. Biochem J. 1996;316:361–371. doi: 10.1042/bj3160361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haussler M R, Jurutka P W, Hsieh J C, Thompson P D, Selznick S H, Haussler C A, Whitfield G K. Bone. 1995;17:33S–38S. doi: 10.1016/8756-3282(95)00205-r. [DOI] [PubMed] [Google Scholar]
- 4.Strugnell S A, Deluca H F. Proc Soc Exp Biol Med. 1997;215:223–228. doi: 10.3181/00379727-215-44131. [DOI] [PubMed] [Google Scholar]
- 5.Baker A R, McDonnell D P, Hughes M, Crisp T M, Mangelsdorf D J, Haussler M R, Pike J W, Shine J, O’Malley B W. Proc Natl Acad Sci USA. 1988;85:3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmester J K, Wiese R J, Maeda N, DeLuca H F. Proc Natl Acad Sci USA. 1988;85:9499–9502. doi: 10.1073/pnas.85.24.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elaroussi M A, Prahl J M, DeLuca H F. Proc Natl Acad Sci USA. 1994;91:11596–11600. doi: 10.1073/pnas.91.24.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Hanson K, DeLuca H F. Arch Biochem Biophys. 1997;339:99–106. doi: 10.1006/abbi.1996.9864. [DOI] [PubMed] [Google Scholar]
- 9.Kamei Y, Kawada T, Fukuwatari T, Ono T, Kato S, Sugimoto E. Gene. 1995;152:281–282. doi: 10.1016/0378-1119(94)00735-b. [DOI] [PubMed] [Google Scholar]
- 10.Li Y C, Bergwitz C, Juppner H, Demay M B. Endocrinology. 1997;138:2347–2353. doi: 10.1210/endo.138.6.5210. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto K, Kesterson R A, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S, Inoue Y, Morita K, Takeda E, Pike J W. Mol Endocrinol. 1997;11:1165–1179. doi: 10.1210/mend.11.8.9951. [DOI] [PubMed] [Google Scholar]
- 12.Dokoh S, Donaldson C A, Haussler M R. Cancer Res. 1984;44:2103–2109. [PubMed] [Google Scholar]
- 13.Chen T L, Li J M, Ye T V, Cone C M, Feldman D. J Cell Physiol. 1986;126:21–28. doi: 10.1002/jcp.1041260104. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan A V, Feldman D. Mol Endocrinol. 1992;6:198–206. doi: 10.1210/mend.6.2.1314957. [DOI] [PubMed] [Google Scholar]
- 15.Costa E M, Feldman D. Endocrinology. 1987;120:1173–1178. doi: 10.1210/endo-120-3-1173. [DOI] [PubMed] [Google Scholar]
- 16.Wiese R J, Uhland-Smith A, Ross T K, Prahl J M, DeLuca H F. J Biol Chem. 1992;267:20082–20086. [PubMed] [Google Scholar]
- 17.Arbour N C, Prahl J M, DeLuca H F. Mol Endocrinol. 1993;7:1307–1312. doi: 10.1210/mend.7.10.8264662. [DOI] [PubMed] [Google Scholar]
- 18.Whitehead R H, Jones J K, Gabriel A, Lukies R E. Cancer Res. 1987;47:2683–2689. [PubMed] [Google Scholar]
- 19.Slater M, Patava J, Kingham K, Mason R S. Am J Phys. 1994;267:E990–E1001. doi: 10.1152/ajpendo.1994.267.6.E990. [DOI] [PubMed] [Google Scholar]
- 20.Grossmann M E, Lindzey J, Blok L, Perry J E, Kumar M V, Tindall D J. Biochemistry. 1994;33:14594–14600. doi: 10.1021/bi00252a027. [DOI] [PubMed] [Google Scholar]
- 21.Saijo T, Ito M, Takeda E, Huq A H, Naito E, Yokota I, Sone T, Pike J W, Kuroda Y. Am J Hum Genet. 1991;49:668–673. [PMC free article] [PubMed] [Google Scholar]
- 22.Ebihara K, Masuhiro Y, Kitamoto T, Suzawa M, Uematsu Y, Yoshizawa T, Ono T, Harada H, Matsuda K, Hasegawa T, et al. Mol Cell Biol. 1996;16:3393–3400. doi: 10.1128/mcb.16.7.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keaveney M, Klug J, Dawson M T, Nestor P V, Neilan J G, Forde R C, Gannon F. J Mol Endocrinol. 1991;6:111–115. doi: 10.1677/jme.0.0060111. [DOI] [PubMed] [Google Scholar]
- 24.Zennaro M C, Keightley M C, Kotelevtsev Y, Conway G S, Soubrier F, Fuller P J. J Biol Chem. 1995;270:21016–21020. doi: 10.1074/jbc.270.36.21016. [DOI] [PubMed] [Google Scholar]
- 25.Castren M, Damm K. J Neuroendocrinol. 1993;5:461–466. doi: 10.1111/j.1365-2826.1993.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 26.Strahle U, Schmidt A, Kelsey G, Stewart A F, Cole T J, Schmid W, Schutz G. Proc Natl Acad Sci USA. 1992;89:6731–6735. doi: 10.1073/pnas.89.15.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeltsch J M, Turcotte B, Garnier J M, Lerouge T, Krozowski Z, Gronemeyer H, Chambon P. J Biol Chem. 1990;265:3967–3974. [PubMed] [Google Scholar]
- 29.Elbrecht A, Chen Y, Cullinan C A, Hayes N, Leibowitz M, Moller D E, Berger J. Biochem Biophys Res Commun. 1996;224:431–437. doi: 10.1006/bbrc.1996.1044. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Qi C, Korenberg J R, Chen X N, Noya D, Rao M S, Reddy J K. Proc Natl Acad Sci USA. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leid M, Kastner P, Chambon P. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 32.Lazar M A. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 33.Tomura H, Lazar J, Phyillaier M, Nikodem V M. Proc Natl Acad Sci USA. 1995;92:5600–5604. doi: 10.1073/pnas.92.12.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 35.Hollenberg S M, Evans R M. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 36.Meyer M E, Pornon A, Ji J W, Bocquel M T, Chambon P, Gronemeyer H. EMBO J. 1990;9:3923–3932. doi: 10.1002/j.1460-2075.1990.tb07613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werman A, Hollenberg A, Solanes G, Bjorbaek C, Vidal-Puig A J, Flier J S. J Biol Chem. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 38.Nagpal S, Friant S, Nakshatri H, Chambon P. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 40.Bocquel M T, Kumar V, Stricker C, Chambon P, Gronemeyer H. Nucleic Acids Res. 1989;17:2581–2595. doi: 10.1093/nar/17.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagpal S, Saunders M, Kastner P, Durand B, Nakshatri H, Chambon P. Cell. 1992;70:1007–1019. doi: 10.1016/0092-8674(92)90250-g. [DOI] [PubMed] [Google Scholar]
- 42.Dowhan D H, Muscat G E. Nucleic Acids Res. 1996;24:264–271. doi: 10.1093/nar/24.2.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sone T, Kerner S, Pike J W. J Biol Chem. 1991;266:23296–23305. [PubMed] [Google Scholar]
- 44.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ford J, McEwan I J, Wright A P, Gustafsson J A. Mol Endocrinol. 1997;11:1467–1475. doi: 10.1210/mend.11.10.9995. [DOI] [PubMed] [Google Scholar]
- 47.Boulikas T. Crit Rev Eukaryotic Gene Expression. 1993;3:193–227. [PubMed] [Google Scholar]