Abstract
The ability of a single protein to interact with multiple protein partners is central to many biological processes. However, the physical chemical and structural basis of the multispecificity is not understood. In Escherichia coli the protein, BirA, can self-associate to a homo-dimer or form a hetero-dimer with the biotin carboxyl carrier protein of the biotin-dependent carboxylase, acetyl CoA carboxylase. The first interaction results in binding of BirA to the biotin operator sequence to repress transcription initiation at the biotin biosynthetic operon and the second is a prerequisite to post-translational biotin addition to the carrier protein for use in metabolism. A single surface on BirA is used for both interactions and previous studies indicate that, despite the structural differences between the alternative partners, the two dimerization reactions are isoenergetic. In this work the underlying thermodynamic driving forces and the sequence determinants of the two interactions were investigated in order to elucidate the energetic and structural underpinnings of the dual specificity. Combined measurements of the temperature and salt dependencies of hetero-dimerization indicate a modest unfavorable enthalpy and no dependence on salt concentration. By contrast, homo-dimerization is characterized by a very large unfavorable enthalpy and a modest dependence on salt concentration. Measurements of the function of BirA variants with single amino acid replacements in the alternative dimerization reactions indicate that although considerable overlap in structural determinants for both interactions exists, hotspots specific for one but not the other were detected.
Keywords: Protein-protein interactions, thermodynamics, bi-specificity, analytical ultracentrifugation, surface loops
Introduction
In complex biological circuitry a single protein may interact with alternative protein partners to produce distinct biological outcomes. For example, the high mobility group A (HMGA) protein, an architectural transcription factor, can bind to at least 18 distinct transcription factors to regulate transcription at a number of different genes(1,2). Many of these multi-specific proteins use a single protein surface to interact with the different partners. Among these is the nuclear transport protein Ran GTPase that interacts with at least five distinct partners in nuclear trafficking(3). Despite application of bioinformatics, computational, and experimental tools, the underlying physical-chemical basis of this multi-specificity in protein-protein interactions is not understood.
The multifunctional protein BirA is both a metabolic enzyme and a transcription factor. BirA catalyzes biotin addition to a biotin-dependent carboxylase and represses transcription initiation at the biotin biosynthetic operon(4,5). In its metabolic function, the protein binds to substrates biotin and ATP to catalyze synthesis of bio-5’-AMP with release of pyrophosphate(6). The resulting enzyme adenylate complex, holoBirA, interacts with apoBCCP, the unbiotinylated biotin carboxyl carrier protein (apoBCCP) of acetyl CoA carboxylase, and transfers biotin to a single lysine residue on the protein. Alternatively, holoBirA forms a homo-dimer that binds sequence-specifically to the biotin operator (bioO) sequence to repress transcription initiation at the biotin biosynthetic operon(7,8). The switch of BirA from metabolic enzyme to transcription repressor is controlled by the intracellular biotin demand, a demand that is signaled by the intracellular apoBCCP concentration(9,10).
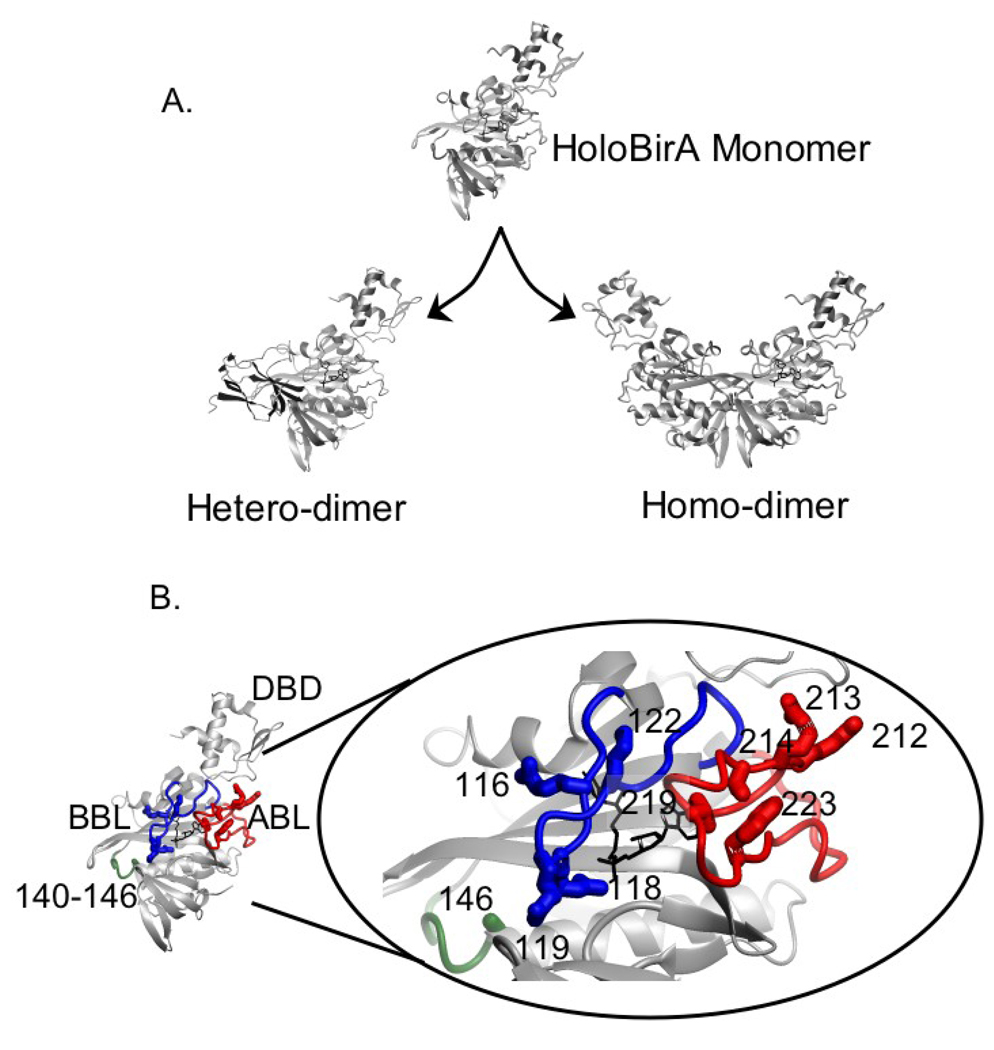
The ability of BirA to carry out distinct biological functions reflects the formation of interactions with distinct protein partners. Moreover, on a structural level the two interactions use a single BirA surface. This relatively simple system, thus, provides an opportunity to elucidate the rules governing bispecificity in protein-protein interactions. As indicated in Figure 1A, the active species in biotin transfer and transcription repression is not apoBirA, but the bio-5’-AMP bound holoBirA. The adenylated biotin, bio-5’-AMP, functions both as the activated intermediate in the biotin transfer reaction and as a corepressor in assembly of the transcription repression complex. The latter function reflects the ability of the adenylate to promote BirA homo-dimerization(11,12). The BirA monomer structure shown in Figure 1B is characterized by an N-terminal winged helix-turn-helix DNA binding domain (DBD), a central domain consisting of a core β-sheet sandwiched between α-helices that resembles an SH2 domain, and an all β-sheet C-terminal domain that is structurally similar to an SH3 domain(13,14).
Figure 1.
A. Switching of holoBirA between its interactions with apoBCCP and another holoBirA monomer required for biotin transfer and transcription repression, respectively. The models were constructed in MolMol(36) with PDB files 2EWN for the homo-dimer and a file provided by Dr. Zachary Wood (University of Georgia) for the hetero-dimer as input. B. The holoBirA monomer with amino acid residues relevant to these studies highlighted in the oval. The biotin binding loop (BBL) shown in blue, the adenylate binding loop (ABL) in red and the 140–146 loop in green feature amino acid residues relevant to these studies. The holoBirA monomer models in A and B were obtained from the dimer model by deletion of the second monomer.
One distinguishing feature of BirA is the presence of several loops, three of which are highlighted in Fig. 1B. In the apo- or unliganded monomer these three loops as well as a fourth composed of residues 193–199 are partially disordered.(13) Structures of two dimeric forms of BirA in which the protein is complexed to biotin or biotinol-5’-AMP (btnOH-AMP), an analog of bio-5’-AMP, have been determined(15,16). Both complexes form homo-dimers by side-by-side anti-parallel alignment of the central β-sheets of the two monomers. In addition, three loops that are flexible in apoBirA are fully ordered and located in the dimer interface. One of these loops, the biotin binding loop or BBL, is folded over biotin in the two structures. In the adenylate-bound structure, the fourth or adenylate binding loop (ABL) consisting of residues 212–233, folds around the adenine ring of btnOH-AMP. Studies of repressors with single amino acid replacements in the four loops indicate that they are functionally important for homo-dimerization.(17) In addition to the experimentally determined structures, a model of the complex of holoBirA bound to apoBCCP87, the domain of BCCP that is biotinylated, has been constructed ((18), Fig. 1A). The hetero-dimer, like the homo-dimer, is formed by extension of the central β-sheet of holoBirA, albeit in a parallel interaction. In addition, the loops on BirA that participate in the homo-dimer interface are also located in the hetero-dimer interface. However, nothing is known about the energetic contribution of these loop segments to apoBCCP binding. The basic features of the structural model are confirmed in an experimentally determined structure of the complex of the Pyrococcus horikoshii BirA paralog bound to a biotin acceptor domain.(19)
Measurements of homo and hetero-dimerization equilibria indicate that the two processes are roughly isoenergetic(20,21). HoloBirA homo-dimerization is characterized by an equilibrium dissociation constant of approximately 10 µM (200 mM KCl, 10 mM Tris HCl, pH 7.5, 20°C)(12). In the same conditions the hetero-dimerization reaction is governed by an equilibrium constant of 2–3 µM. Thus, despite the chemically distinct natures of the apoBCCP and holoBirA surfaces the two partners bind to a holoBirA monomer with similar energetics.
Although the free energies of homo- and hetero-dimerization are similar in magnitude, preliminary information indicates that distinct detailed chemistries underlie this superficial similarity. First, binding of bio-5’-AMP analogs to BirA produces different effects on the two protein-protein interactions. Although binding of the analogs biotinol-5’-AMP and biotin sulfamoyl 5’-AMP enhance BirA homo-dimerization by −3.0 and −1.0 kcal/mole, respectively, apoBCCP shows no tendency to interact with complexes of the protein bound to either ligand ((22)Streaker & Beckett, unpublished observations). Second, preliminary measurements suggest that the enthalpic and entropic driving forces for the two interactions are very different.(23) Finally, inspection of the structures of the two dimers indicates distinct chemistries with the homo-dimer characterized by more electrostatic interactions than the hetero-dimer.
In this work the structural basis of bispecificity in interaction of holoBirA with its two partners has been probed using combined thermodynamic analysis and comparison of the effects of single amino acid replacements in BirA on the alternative dimerization reactions. Measurements of the temperature and salt dependencies of hetero-dimerization using competition sedimentation equilibrium indicate modest or no dependencies of the process on these solution variables. Thus, in contrast to the very large positive enthalpy that opposes homo-dimerization, hetero-dimerization is characterized by a modest unfavorable enthalpy. Measurements of the function of BirA variants with amino acid replacements in three of the loops that are partially disordered in the apo-structure in homo- and hetero-dimerization, performed using combined sedimentation equilibrium and inhibition DNaseI footprint titrations, indicate considerable overlap in the structural requirements on BirA for both interactions. However, against the backdrop of this overlap, hotspots specific for one but not the other dimerization reaction were identified. Finally, the critical roles played by residues in the surface loops in both interactions support the importance of this structural feature in evolution of bi-specificity in BirA.
RESULTS
Distinct Driving Forces for Homo- and Hetero-dimerization of HoloBirA
Previous analysis of holoBirA homo-dimerization indicates that it is characterized by a large unfavorable enthalpy and large favorable entropy and a modest dependence on salt concentration(23). In order to assess the driving forces governing hetero-dimerization between holoBirA and apoBCCP87, the temperature and salt dependencies of the process were measured.
Binding of holoBirA to apoBCCP results in chemical transfer of biotin to form holoBCCP and apoBirA, thus prohibiting direct measurement of the binding interaction. An alternative method for measurement of the process has been developed that takes advantage of the positive linkage of bio-5’-AMP binding and homo-dimerization(21). This is an indirect method in which hetero-dimerization is monitored by its effect on holoBirA homo-dimerization. In the method, apoBCCP87, a C-terminal fragment of BCCP, is combined with an equilibrium mixture of holoBirA monomer and dimer. ApoBCCP87 hetero-dimerizes with holoBirA in a concentration-dependent manner, with resulting chemistry and conversion of holoBirA to the dimerization-incompetent apoBirA. The extent to which the homo-dimerization is perturbed provides a measure of the extent of the hetero-dimerization reaction. Previously this competition measurement was performed using sedimentation velocity(21). In this work, in order to conserve material, sedimentation equilibrium was employed.
In each measurement a 1:1 mixture of apoBirA and bio-5’-AMP was prepared in the absence and presence of two different apoBCCP87 concentrations and subjected to centrifugation. The resulting scans were analyzed for the effect of apoBCCP87 on the equilibrium constant for homo-dimerization. The use of equimolar amounts of bio-5’-AMP and apoBirA in sample preparation ensures that the perturbation associated with hetero-dimerization reflects a single interaction of a holoBirA monomer with apoBCCP87 and no BirA turnover occurs. In addition, because BCCP87 and bio-5’-AMP do not absorb at the 295 nm wavelength employed for the scans, the BirA signal can be monitored independent of these other species.
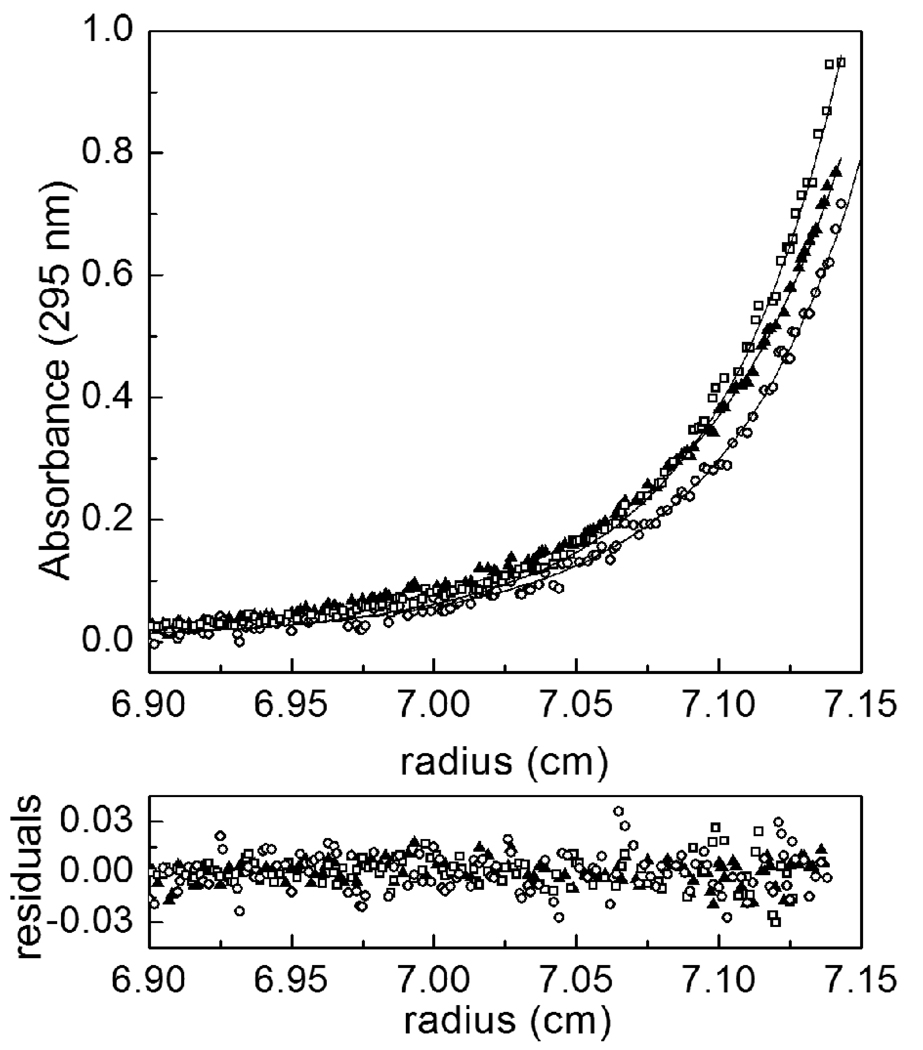
Addition of apoBCCP87 to holoBirA alters the sedimentation equilibrium profile (Fig. 2) with the scan of holoBirA alone significantly steeper than those obtained in the presence of apoBCCP87. This difference in steepness is consistent with a larger dimer population in the absence of the acceptor protein. Furthermore, the perturbation associated with the addition of the acceptor protein at10 µM is greater than that observed at 5µM. Two scans, obtained at speeds of 18K and 22K rpm, for each sample were subjected to global analysis using a monomer-dimer model to obtain the observed equilibrium dimerization constant for homo-dimerization. This observed equilibrium constant was then used to calculate the inhibition constant for hetero-dimerization using Equation 5 and Equation 6 (Materials & Methods). The value obtained at 20°C of 4(±2) ×10−6M is in excellent agreement with that obtained using sedimentation velocity or inhibition DNaseI footprint titrations(20,21).
Figure 2.
Results of competition sedimentation equilibrium measurements obtained at 25 °C in the absence and presence of apoBCCP87. The data scans were obtained from samples prepared at 20 µM holoBirA and apoBCCP87 at 0 (open squares), 5 (filled triangles) or 10 µM (open circles). The lines represent the best-fit curves obtained from global analysis of data in each sample to a monomer-dimer model performed as described in the Materials & Methods. The residuals of the fits are shown in the bottom panel.
Temperature Dependence of Hetero-dimerization
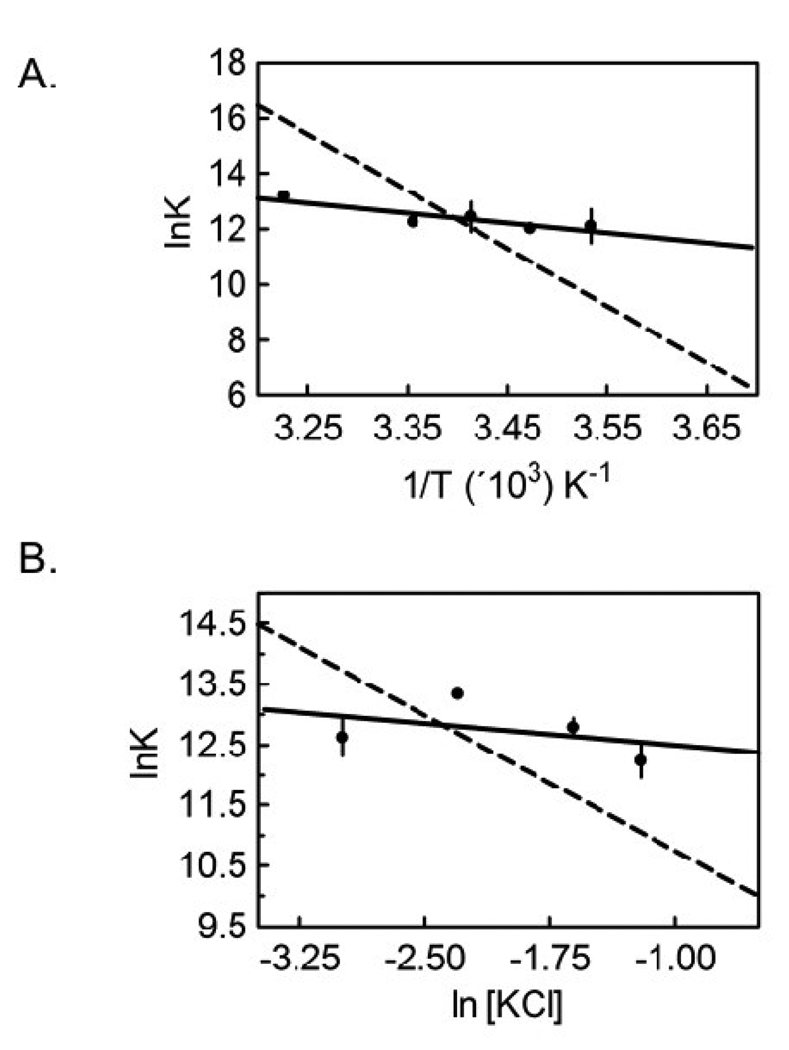
Measurements of the temperature dependence of hetero-dimerization from 10 °C to 25 °C reveals a modest change in the inhibition constant governing the reaction, KI, over the range employed (Table 1). The data were subjected to van’t Hoff analysis to obtain the enthalpy change associated with hetero-dimerization. The KI value at 37°C obtained using the inhibition DNaseI titration method was also included in the analysis(21). A linear dependence of the lnKa,I on 1/T was observed with a resolved enthalpy change from linear least-squares analysis of ΔHI = 7 ± 2 kcal/mol (Figure 3A, Table 3). Using the Gibbs Equation, ΔGI =ΔHI − TΔSI and this enthalpy change the entropic contribution to hetero-dimerization free energy (−TΔSI) at 20°C was calculated to be −14 ± 2 kcal/mole. Additionally, the linearity of the Van’t Hoff plot is consistent with the absence of a heat capacity change upon hetero-dimer formation over the temperature range studied. Finally, the unfavorable enthalpy associated with hetero-dimerization is much lower in magnitude than that associated with homo-dimerization (Figure 3A, Table 3).
Table 1.
Temperature-dependence of holoBirA hetero-dimerization
| T (°C) | KI (M−1) | ΔG°I (kcal/mol) |
|---|---|---|
| 10 | 6 (± 3) × 10−6 | −6.8 ± 0.3 |
| 15 | 6.1 (± 0.9) × 10−6 | −6.86 ± 0.08 |
| 20 | 4 (±2) × 10−6 | −7.2 ± 0.3 |
| 25 | 4.8 (± 0.8) × 10−6 | −7.2 ± 0.1 |
| 37* | 1.9 (± 0.1) × 10−6 | −8.09 ± 0.03 |
Measurements were performed in Standard Buffer (10 mM TrisHCl, 200 mM KCl, 2.5 mM MgCl2 adjusted to pH 7.50 ± 0.02 at the working temperature. The reported uncertainties represent the standard error associated with at least two independent measurements.
Measured using inhibition DNaseI footprinting method(21).
Figure 3.
(A) Van’t Hoff analysis of the temperature dependence of the inhibition equilibrium constant for hetero-dimerization between holoBirA and apoBCCP87. The best-fit line was obtained from linear regression of values obtained from averaging 2–4 independent measurements of hetero-dimerization at each temperature. The value at 37°C was obtained from inhibition DNaseI footprinting measurement(21). (B) Dependence of hetero-dimerization on [KCl]. The continuous line represents the results of linear regression of the lnK versus ln[KCl] data with the slope of the line yielding the number of ions taken up or released in the binding process. The plotted data points are averaged from 2–4 independent measurements at each salt concentration. The dashed lines in both plots represent the best-fit of analogous data obtained for holoBirA homo-dimerization.(23)
Table 3.
Physical-chemical properties of the homo- and hetero-dimerization processes
| Homo-dimerization | Hetero-dimerization | |
|---|---|---|
| ΔG° (kcal/mole)a | −7.0 ± 0.3 | −7.4 ± 0.2 |
| ΔH° (kcal/mole) | 41 ± 3 | 7 ± 2 |
| −TΔS° (kcal/mole)a | −48 ± 3 | −14 ± 2 |
| nion | −1.5 ± 0.1 | 0 |
Values reported were obtained from measurements of the dependence of homo-dimerization(23) and hetero-dimerization (this work) on temperature and salt concentration (KCl).
The reported values of these parameters are relevant to 20°C, 200 mM KCl.
Salt Dependence of Hetero-dimerization
The homo-dimerization reaction is characterized by a small dependence on salt concentration, consistent with a modest electrostatic contribution to this protein-protein interaction(23). In order to explore the contribution of electrostatic forces to hetero-dimerization, the process was measured by inhibition sedimentation equilibrium in Standard Buffer containing 50 – 300 mM KCl. The KI’s obtained at four salt concentrations are similar in magnitude (Table 2) and a plot of resolved ln Ka,I vs ln [KCl] (Figure 3B) shows a slope, within error, of zero, indicating that net ion release or uptake does not contribute to hetero-dimerization. By contrast, the salt-dependence of homo-dimerization is small but significant (Figure 3B, Table 3).
Table 2.
[KCl]-dependence of hetero-dimerization.
| [KCl] (mM) | KI (M) | ΔG°I (kcal/mol) |
|---|---|---|
| 50 | 3.3 (±0.9) × 10−6 | −7.3 ± 0.2 |
| 100 | 1.6 (± 0.2) × 10−6 | −7.8 ± 0.1 |
| 200 | 2.8 (±0.4) × 106 | −7.4 ± 0.1 |
| 300 | 4 (± 1) × 106 | −7.3 ± 0.3 |
Measurements were performed in Standard Buffer (10 mM TrisHCl, 50–300 mM KCl, 2.5 mM MgCl2, pH 7.50 ± 0.02 at 20.0 ± 0.1 °C). The reported uncertainties represent the standard error associated with at least two independent measurements.
Structural Requirements on BirA for Homo- and Heterodimerization
BirA variants that are Defective in Homo-dimerization
Although the thermodynamic measurements described above provide information about the overall driving forces for homo- and hetero-dimerization of holoBirA, they yield no information about the importance of specific structural elements on either protein for the two interactions. In order to investigate the sequence requirements of the interactions, the function of single-residue variants of BirA in homo- and hetero-dimerization were investigated. Previous analysis of the self-assembly properties of BirA variants has revealed two classes of defects including those that result from direct perturbation of the protein-protein interface and those that disrupt the coupling between bio-5’-AMP binding and dimerization. Dimer interface variants include R118G, R119W and A146Δ(17) (Figure 1B). Sedimentation equilibrium measurements performed on the three proteins at saturating bio-5’-AMP concentrations revealed that they are characterized by dimerization energetics too modest for quantitation by this technique. Indeed, at concentrations at which wild-type holoBirA is predominantly dimer, the three proteins are 100% monomeric. However, measurements of total assembly, which includes dimerization and bioO binding by the dimer, of these proteins on bioO by DNase I footprint titrations allows estimation of their dimerization properties(17). Assuming that for these three variants the energetic penalties associated with the total assembly reflect only decreased dimerization energetics, the dimerization free energies for R118G, R119W and A146Δ are estimated at −3.4, −2.2 and −5.1 kcal/mol, respectively (Table 4, Figure 5A). This assumption is based on the observed thermodynamic decoupling of dimerization and DNA binding with respect to the effect of bio-5’-AMP on the wild type repressor(24) and the fact that these three amino acids are located in the homo-dimer interface.(15,16) Single amino acid replacements in the adenylate binding loop, which is distal to the dimerization interface, disrupt the coupling between bio-5’-AMP binding and dimerization. These substitutions, which include V214A, V219A and W223A, are of side chains that form a hydrophobic cluster around the adenine ring of the adenylate ligand(25)(Figure 1B). Energetic penalties for homo-dimerization (ΔΔG°dim) of + 1 to +1.5 kcal/mol relative to wild-type BirA were measured for these proteins (Table 4, Figure 5A).
Table 4.
Homo-dimerization Properties of Wild Type and Variant BirA Proteins
| BirA Variant | Kdim (M) | ΔG°dim (kcal/mol) |
|---|---|---|
| WT | 6 (± 2) × 10−6 | −7.0 ± 0.3 |
| R116A | 5 (± 2) × 10−6 | −7.1 ± 0.3 |
| R118G † | 2.9 × 10−3 | −3.4 |
| R119W † | 2.2 × 10−2 | −2.2 |
| K122A | 2.1 (± 0.6) × 10−5 | −6.2 ± 0.2 |
| A146Δ† | 1.6 × 10−4 | −5.1 |
| R212A | 1.5 (± 0.6) × 10−5 | −6.4 ± 0.3 |
| R213A | 7 (± 4) × 10−6 | −6.9 ± 0.3 |
| V214A * | 3 (± 1) × 10−5 | −6.1 ± 0.2 |
| V219A * | 8 (± 1) × 10−5 | −5.5 ± 0.2 |
| W223A * | 3 (± 1) × 10−5 | −6.1 ± 0.2 |
All measurements were performed in Standard Buffer (10 mM Tris-HCl pH 7.50 ± 0.02 at 20.0 ± 0.1 °C. 200 mM KCl, 2.5 mM MgCl2) as described in Materials & Methods. The reported values represent the average of at least two independent sedimentation equilibrium measurements.
Self-association properties for these variants are from Kwon et al. and were estimated from energetics of assembly of the BirA-bioO complex from 2 holoBirA monomers and the free DNA and, therefore, have no errors are reported.(17)
Dimerization properties for these variants are from Naganathan & Beckett.(25)
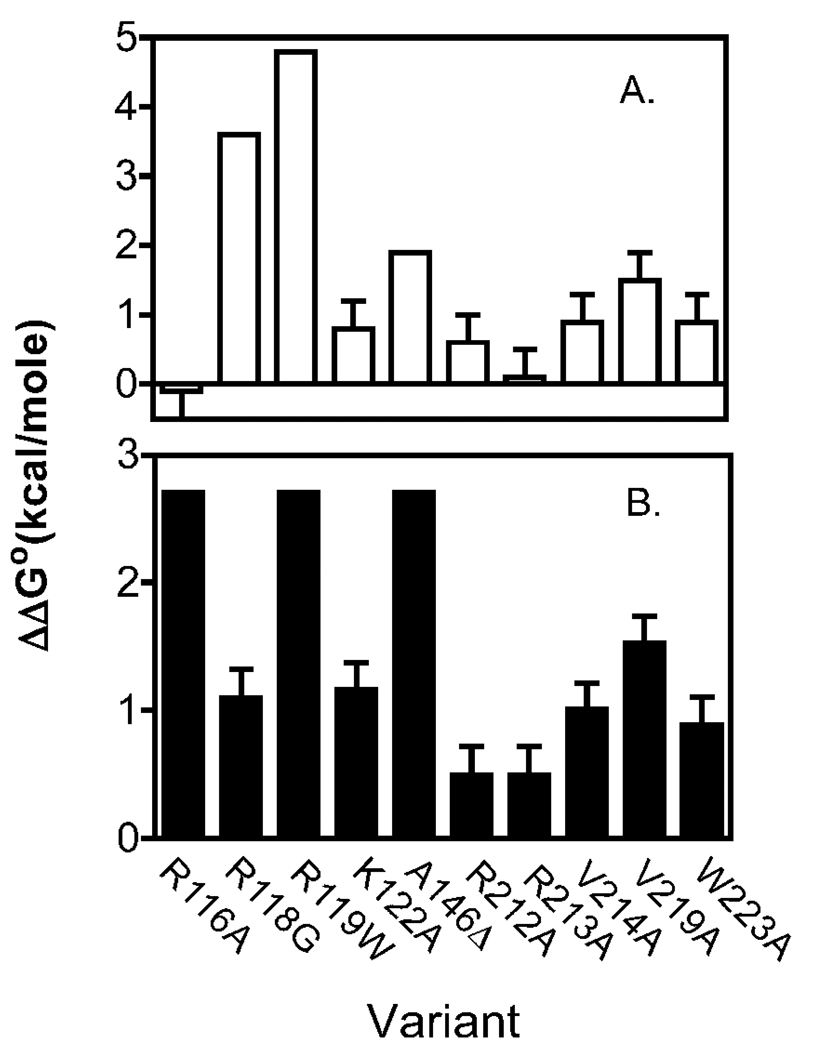
Figure 5.
Energetic perturbations (ΔΔG°) to homo-(Top panel) and hetero-dimerization (Bottom panel) resulting from single amino acid replacements in BirA calculated using the equation, ΔΔG° = ΔG°Variant − ΔG°WT. The reported values represent the average of at least two independent measurements and errors represent 68% confidence intervals. Values for homo-dimerization for were either estimated from previously reported measurements of bioO binding (R118G, R119W, A146Δ), previously measured by sedimentation equilibrium (V214A, V219A, W223A) or measured as described in the text. ΔΔGo° values for hetero-dimerization without error bars represent lower limits.
In this work sedimentation equilibrium measurements of homo-dimerization were performed on four additional BirA variants, two of which have amino acid replacements in the biotin binding loop (BBL) and two in the adenylate binding loop (ABL)(Figure 1B). Although these proteins were originally designed for an unrelated study, they proved useful for this investigation. In the homo-dimer residues R116 and K122 of the BBL are located in close proximity to and R212 and R213 of the ABL are distal to the interface (Figure 1A&B). These proteins combined with saturating bio-5’-AMP were subjected to sedimentation equilibrium measurements at three loading concentrations and two rotor speeds(26). For all variants analysis of the data using a single species model indicated molecular weights consistent with protein association. The data were then subjected to global analysis using a monomer-dimer association model to resolve the dimerization constants and the quality of each fit was assessed from the magnitude of the square root of the variance of the fit and the distributions of the residuals. The resolved equilibrium dissociation constants indicate that BirA variants R116A and R213A possess dimerization properties identical to those of the wild type protein (Table 4). By contrast, BirA variants K122A and R212A demonstrate modest but significant energetic defects (ΔΔG°dim of +0.6 to +0.8kcal/mol, Table 4, Figure 5A) in homo-dimerization.
Hetero-dimerization Properties of the Single Amino Acid Variants of BirA
Hetero-dimerization of each BirA variant with apoBCCP87 was measured using inhibition DNase I footprint titrations. Addition of apoBCCP87 to a mixture of holoBirA and bioO results in perturbation of holoBirA homo-dimerization that is required for bioO occupancy(20). As the apoBCCP87concentration is increased, the equilibrium holoBirA dimer concentration decreases, thereby decreasing the fractional occupancy of bioO. Inhibition data obtained over a range of apoBCCP87 concentrations yield an inhibition curve that can be analyzed to extract an “inhibition constant.” This inhibition constant, KI, provides a measure of the energetics of the holoBirA:apoBCCP87 interaction.
Design and analysis of the inhibition footprint titrations required initial assessment of bioO binding by all mutants using direct DNase I footprinting. Due to defects in adenylate synthesis associated with some BirA variants (data not shown), all footprint titrations, both direct and indirect, were performed with preformed complexes of each mutant protein bound to chemically synthesized bio-5’-AMP. Direct DNase I footprint titrations of bioO provide information to set the holoBirA concentration used in the inhibition titrations. The optimal concentration, which corresponds to a value just below 100% saturation of bioO, renders the system poised for perturbation by apoBCCP87 in the inhibition measurements. Analysis of the direct footprints also yielded the total equilibrium constant, KTOT, for each variant required for analysis of the inhibition footprint titrations.
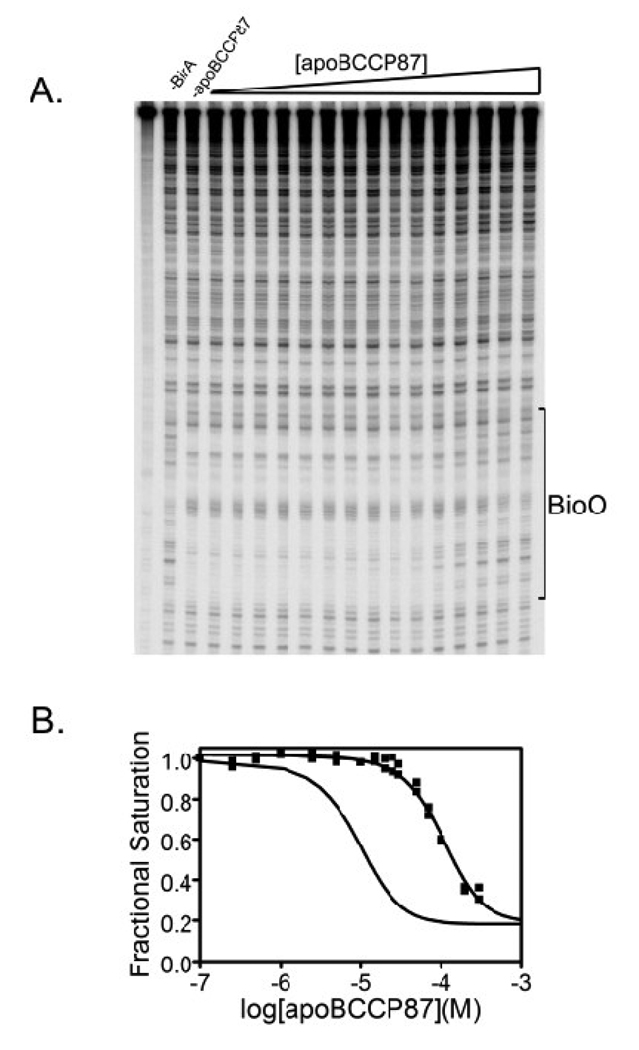
The inhibition footprint obtained for the K122A variant is shown in Figure 4A. Inspection of the gel image reveals a loss of the footprint with increasing apoBCCP87 concentration. Quantification of the data yields an isotherm in which the fractional occupancy of bioO decreases with increasing apoBCCP87 concentration (Figure 4B). For comparison, the best-fit curve for the inhibition footprint isotherm obtained with wild type holoBirA is also shown. The data obtained for the K122A variant were subjected to nonlinear least squares analysis to determine the KI, the equilibrium inhibition constant governing the hetero-dimerization reaction using Equation 7 (Materials & Methods). The results indicate a defect in hetero-dimerization for this variant of 7 to 8-fold (Table 5, Figure 5B).
Figure 4.
Results of inhibition DNaseI footprinting obtained with the K122A BirA variant (A) Representative image of a gel obtained for inhibition DNase I footprint titration of bioO with BirA K122A as a function of apoBCCP87 concentration. (B) Inhibition isotherms obtained from quantification of footprinting data obtained with K122A (■). Results obtained wild-type holoBirA are shown for comparison (solid line without data points).
Table 5.
Energetics of Hetero-dimerization of BirA variants with apoBCCP87 determined by inhibition DNaseI footprinting.
| BirA Variant | Ki (M)a | ΔG° (kcal/mol) |
|---|---|---|
| WT | 3 (± 1)×10−6 | −7.4 ± 0.2 |
| R116A | > 3×10−4 | >−4.7 |
| R118G | 20 (± 0.4)×10−6 | −6.3 ± 0.1 |
| R119W | > 3×10−4 | > −4.7 |
| K122A | 23 (± 3)×10−6 | −6.23 ± 0.01 |
| A146Δ | >3 ×10−4 | > −4.7 |
| R212A | 7 (± 1)×10−6 | −6.9 ± 0.1 |
| R213A | 7 (± 2)×10−6 | −6.9 ± 0.2 |
| V214A | 17 (± 2)×10−6 | −6.39 ± 0.07 |
| V219A | 42 (± 5)×10−6 | −5.87 ± 0.07 |
| W223A | 14 (± 2)×10−6 | −6.51 ± 0.08 |
All measurements were performed in Standard Buffer (10 mM Tris-HCl pH 7.50 ± 0.02 at 20.0 ± 0.1 °C. 200 mM KCl, 2.5 mM MgCl2) as described in the Materials and Methods, with the exception of that obtained for variant R119W, which was measured in 50 mM KCl. (a.) Inhibition footprinting isotherms were fit to equation (7) to determine Ka,I, the inverse of KI. In the analysis the parameter, KTOT was fixed at the value obtained from direct DNase I footprinting data. All reported values represent the average of two independent measurements and errors represent 68% confidence intervals. Equilibrium constants for apoBCCP87 hetero-dimerization with R119W, R116A, and A146Δ are lower limits and no errors are reported for them because they were beyond the concentration regime that could be used in the measurements.
Inhibition DNaseI footprint titrations performed on all of the variant repressors yielded a range of equilibrium constants governing their interaction apoBCCP87 (Table 4, Figure 5B). The KI values range from 3 – 200 fold weaker than that measured for the wild-type protein. The largest penalties to hetero-dimerization were obtained with the BirA variants R116A, R119W and A146Δ for which apoBCCP87 concentrations as high as 300 µM had no effect on bioO occupancy. Therefore, the KI values reported for these variants are lower limits.
Discussion
In this work the physical-chemical basis of use of a single surface on holoBirA to form alternative protein-protein interactions with similar total free energies was investigated. Measurements of the dependence of the two interactions on solution variables revealed the underlying thermodynamic driving forces for the two reactions. Comparison of the consequences of single amino acid replacements in BirA for the two protein-protein interactions provided insight into how specific structural features on repressor are utilized for the two processes. The results indicate distinct chemical driving forces for the two reactions. In addition, although some structural information on BirA is critical for both interactions, hotspots selective for one but not the other exist.
Homo- and hetero-dimerization are driven by distinct physical-chemical driving forces
The temperature and salt dependencies of the homo- and hetero-dimerization indicate distinct overall driving forces for the two protein-protein interactions. The dependencies of homo-dimerization on these two variables have previously been reported(23). In this work the temperature and salt dependencies of hetero-dimerization were measured using competition sedimentation equilibrium. The method takes advantage of the fact that interaction of the acceptor protein, apoBCCP87, with the holoBirA monomer results in biotin transfer with conversion of holoBirA to the apo species, with concomitant inhibition of homo-dimerization. The value of KI, the equilibrium inhibition constant, obtained from analysis of data acquired at multiple apoBCCP87 concentrations and multiple rotor speeds is 4 ± 2 micromolar, which is in excellent agreement with the inhibition constant previously determined using either competition sedimentation velocity or the inhibition DNaseI footprint titration method(20,21).
Van’t Hoff analysis of the temperature dependence of the KI indicates an enthalpy for the process of 7 ± 2 kcal/mole, a value in striking contrast to that determined for homo-dimerization of 41 ± 3 kcal/mole. Thus, although both reactions are characterized by unfavorable enthalpies, the magnitude of the value for the homo-dimerization reaction is much greater than that for hetero-dimerization. In addition, the calculated entropic contributions (−TΔS°) to the free energies of hetero- and homo-dimerization at 20°C are −14 kcal/mole and −47 kcal/mole, respectively. Again, although the entropic driving forces for both dimerization reactions are favorable, the magnitude of the value for homo-dimerization is significantly greater than that for hetero-dimerization. The very large enthalpic penalty and entropic driving force associated with homo-dimerization has previously been interpreted as indicative of solvent release upon formation of the protein-protein interactions(23). Structurally, side chains at both the homo- and hetero-dimeric protein-protein interfaces are rich in charged and polar groups. Assuming that these groups must be desolvated in forming either dimer interface, the observed enthalpic penalties and entropic gains associated with the two reactions are reasonable. However, the more modest enthalpic and entropic terms suggest a smaller contribution of solvent release to hetero-dimerization than to homo-dimerization.
The salt dependencies of the two dimerization reactions are also distinct, with homo-dimerization exhibiting a greater sensitivity to increasing monovalent salt concentration than hetero-dimerization. Superficially, the modest salt dependence runs counter to the known structural features of the two interfaces, which are rich in charged side chains. However, the studies presented in this work are limited in scope and a more extensive survey of the cation and anion dependence of the two dimerization reactions is required before any conclusion about the electrostatic driving forces for the two processes can be reached.
Roles of structural elements on BirA in the two protein-protein interactions
Initial assessment of the structural determinants of the alternative dimerization reactions was accomplished by comparing the homo- and hetero-dimerization of BirA variants. In all cases the holoBirA species was employed in these measurements. While several of these mutants have previously been assessed for their homo-dimerization properties, four additional mutants were analyzed in this work by sedimentation equilibrium. These include K122A, R116A in the biotin binding loop and R212A and R213A in the adenylate binding loop. Of the four only the K122A and R212A variants exhibit modest perturbations of +0.6 and 0.8 kcal/mole. The single amino acid variants were also subjected to analysis of their hetero-dimerization properties using the inhibition DNaseI footprint titration method. Results of the measurements indicate that all amino acid replacements are accompanied by energetic penalties in hetero-dimerization ranging from modest, approximately 1 kcal/mole, to greater than 3 kcal/mole (Table 5, Figure 5B). In general hetero-dimerization is more sensitive to amino acid substitutions than is homo-dimerization. In discussing the implications of the results of the measurements the variants are grouped into two classes including those with amino acid replacements in the ABL and those that have substitutions that directly perturb the dimer interfaces, including variants with single amino acid replacements in the BBL and the A146Δ variant.
Hetero-dimerization requires bio-5’-AMP induced folding of the ABL
The results obtained from the ABL variants indicate that structural determinants required for linkage of bio-5’-AMP to homo-dimerization are also important for hetero-dimerization. Replacement of side chains of residues V214, V219 or W223 with alanine results in energetic penalties to homo-dimerization ranging from +1.0 to +1.5 kcal/mole.(25) Since these three residues do not directly participate in the interface but do play a role in corepressor-induced ABL folding that is required for homo-dimerization (Figure 6A and 6C), the energetic penalties are interpreted as defects in coupling of bio-5’-AMP binding to dimerization. These same variants exhibit defects in hetero-dimerization of 0.8–1.5 kcal/mole, similar in magnitude to the perturbations to homo-dimerization. Therefore, like homo-dimerization, hetero-dimerization is positively linked to adenyate binding. In support of this conclusion, attempts to measure any interaction of apoBCCP87 with apoBirA have indicated no detectable interaction between the proteins (Nenortas & Beckett, unpublished results). In the context of BirA’s function in biotin transfer to apoBCCP, positive coupling of bio-5’-AMP binding and hetero-dimerization is reasonable because it ensures that acceptor protein binding does not occur until the activated intermediate is synthesized. The R212A and R213A variants exhibit modest or no effect on homo-dimerization. By contrast, the perturbation to hetero-dimerization associated with the two variants is on the order to 1 kcal/mole. Since both the R212 and R213 side chains project into solvent (Figure 1B), this result suggests that hetero-dimerization is more sensitive to the precise conformation of the ABL than is homo-dimerization.
Figure 6.
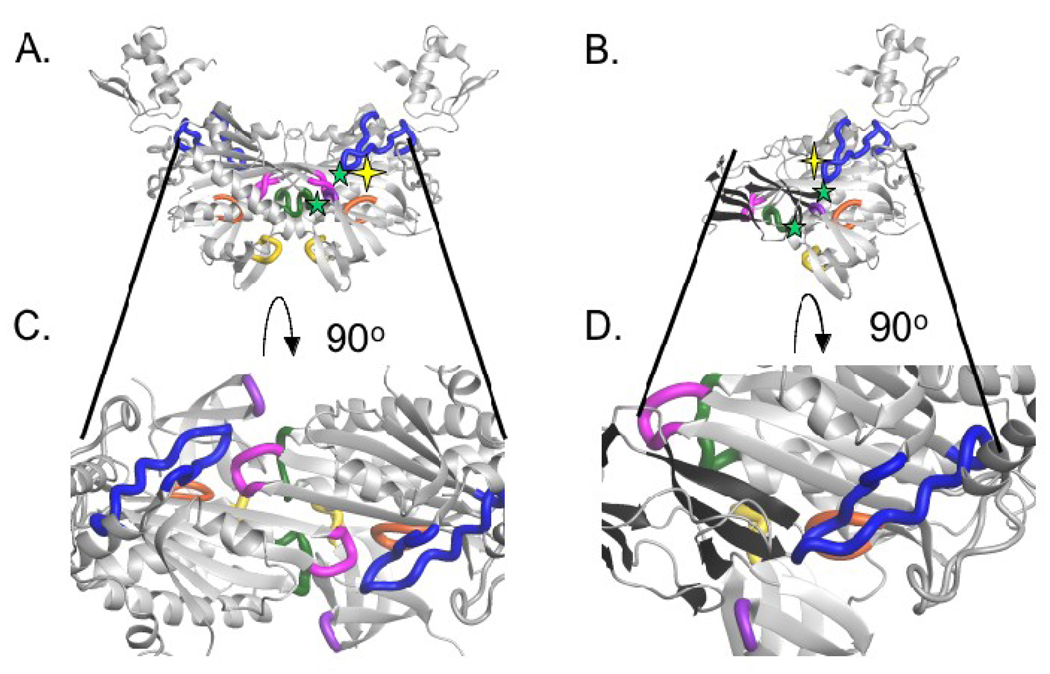
Interfacial residues on holoBirA that are hotspots for the homo- and hetero-dimerization processes highlighted on the dimer structures. (A and B) Replacement of interfacial side chains marked with yellow stars yields a large effect on one but not the other dimerization process. Replacement of those marked with green symbols has large consequences for both interactions (C and D). Cross sections of the homo- and hetero-dimer interfaces with interfacial loops on BirA highlighted. Loop 116–124 (blue), loop 140–146 (green), loop 170–175 (orange), loop 193–199 (pink), loop 280–286 (purple), loop 290–295 (yellow). All models were created using MolMol(36) with input file 2EWN for the homo-dimer and a file generously provided by Dr. Zachary Wood (University of Georgia) for the hetero-dimer.
Structural determinants of the homo- and hetero-dimer energetics
Homo- and hetero-dimerization properties of BirA variants with single amino acid replacements at positions in the homo-dimer interface reveal a complex relationship between structure and function in the two processes. Interface variants that have been subjected to measurements of the two reactions include R116A, R118G, R119W and A146Δ. The latter three variants were identified in genetic screens for defects in enzymatic and/or repression function and are all defective in repression(4,5). This phenotype is consistent with their defects in homo-dimerization measured in this laboratory. The penalties for homo-dimerization associated with the three variants, which were estimated from DNaseI footprinting titrations, are approximately 3.5, 4.8 and 2.0 kcal/mole for R118G, R119W and A146Δ, respectively(17). By contrast, although the R119W and A146Δ variants also exhibit large energetic penalties in the hetero-dimerization reaction, the penalty associated with the R118G mutant is relatively modest. Replacement of R116 with alanine also leads to different behaviors in the two reactions, with no effect on homo-dimerization but a dramatic effect on hetero-dimerization (Figure 5). The remaining BBL variant, K122A, exhibits a modest, but significant, effect on both homo- and hetero-dimerization (Figure 5A and B). This survey of single amino acid replacements in BirA indicates that although significant overlap exists in the structural requirements for the two dimerization reactions, R116 and R118, each represent hotspots for one but not the other reaction.
Function of loops in homo- and hetero-dimerization
The homo- and hetero-dimer structures provide a context for interpretation of the results obtained with the interface variants. Cross sections of the two interfaces are shown in Figures 6C and D. As described in the Introduction, the homo-dimer is formed by side-by-side antiparallel alignment of the central β-sheets of two monomers(15,16). In addition, several loops are located in the interface including those composed of residues 116–124 (BBL), 140–146, 172–175, 193–199 and 280–286, 290–295. In the BirA variants studied in this work the amino acid replacements are located in the 116–124 (blue) and 140–146 (green) loops. In the homo-dimer interface the side chains of R118 and R119 form electrostatic interactions with residues D176 and D197, respectively, of loop 193–199 (pink) on the second monomer. In the 140–146 loop, side chains of residues E140 and Q141 appear to contact the side chain of E283 and the backbone of I280, respectively, in loop 280–283 (purple) on a second monomer. These interactions may be disrupted by the shortening of the 140–146 loop that accompanies deletion of residue A146. In addition, due to the two-fold symmetry that characterizes the homo-dimer, each noncovalent interaction cited above occurs twice (Figure 6B). Thus the large perturbations to homo-dimerization reflect the loss of at least two interfacial contacts (Figure 6B). In the hetero-dimer model, which is formed by side-by-side parallel alignment of two β-sheets(18), the same loops on BirA that function in the homo-dimer interface also interact with apoBCCP. For example, the large defect in hetero-dimerization associated with the R119W variant reflects, in part, the loss of an electrostatic interaction between R119 on BirA and D119 on BCCP. The defect associated with the R116 mutation may be caused by the loss of an interaction with D98 on BCCP. The only large perturbation to hetero-dimerization for which the structural basis is not immediately obvious is that associated with A146Δ. Taken together the structural data and the results obtained with the interface variants indicate important roles for loops on BirA in both dimerization interactions. Furthermore, although there is considerable overlap in use of sequence for both interactions, residues specific to each exist. Establishment of the generality of this conclusion requires measurements of the properties of additional BirA variants with single amino acid replacements in all of the interfacial loops.
Surface loops and the evolution of protein-protein interactions and new function
In general, surface loops can contribute significantly to the ability of a protein to interact with other proteins and, thereby influence its biological function. Many protein-protein interactions are mediated at the domain level and, in the context of the modular structures of proteins, specific domains are frequently utilized for these interactions. Structure based sequence alignments of common domains indicate that surface loops dictate both the ability to interact as well as the mode of interaction(27). Indeed, the alignments reveal that loops can either promote or prevent protein self-association, and thus can be considered either enabling or disabling, respectively. The presence of one or more enabling loops in a domain structure promotes oligomerization. By contrast, the presence of a disabling loop prevents oligomerization.
The structural features of the homo- and hetero-dimeric interactions formed by BirA provide an illustration of this strategy of mediation of protein-protein interactions at the domain level by surface loops. The central and C-terminal domains of BirA are characterized by SH2 and SH3-like topologies, respectively(14). These two domains provide the interacting surfaces for both the hetero-dimeric and homo-dimeric protein interactions formed by holoBirA, albeit in mechanisms distinct from those associated with SH2 and SH3 domains involved in signal transduction. This distinct mode of interaction reflects, in part, the critical roles of the surface loops on BirA (Figure 6). In addition to mediating the two distinct protein-protein interactions the loops in BirA regulate the interactions. In apoBirA the flexibility associated with the BBL and ABL renders them disabling for both homo- and hetero-dimerization. In holoBirA, as a consequence bio-5’-AMP-induced folding, these same loops enable the interactions.
In BirA the participation of multiple loops, some of them partially disordered, was critical to evolution of its ability to bind to distinct protein partners in a regulated manner. The bifunctionality exhibited by BirA is a very ancient feature of many bacterial biotin holoenzyme ligases(28). Although the ligase function is required for viability, the transcriptional regulatory function is not. The use of surface loops in BirA for the alternative interactions provided a means of evolving a new function in transcription repression without compromising the enzymatic function that is essential for viability(5).
MATERIALS AND METHODS
Chemicals and Biochemicals
All chemicals used in buffer preparation were at least reagent grade. DNase I was purchased from Sigma. The Klenow fragment of DNA Polymerase I and restriction enzymes, HindIII and PstI, were purchased from Promega. The α32P dATP and dGTP used for labeling of DNA for footprinting was obtained from Perkin Elmer. The bio-5'-AMP was synthesized and purified in the laboratory using a published procedure(7). Truncated biotin carboxyl carrier protein (apoBCCP87) was purified as described in Nenortas and Beckett(29), with the exception of an additional chromatography step on a Hydroxyapatite ULTROL (PALL) to remove trace quantities of the DNase I that were introduced during one of the initial purification steps. This fragment has been shown to function identically to the intact protein in biotin transfer.(29)
Expression and Purification of BirA and its variants
Wild type BirA was purified as previously described(22). All BirA variants were expressed in the E. coli strain JM109 transformed with the plasmid pBtac2 carrying the E. coli birA gene (wild type or mutant) under transcriptional control of the tac promoter. In order to ensure purification of each protein way from the chromosomally encoded wild type BirA the plasmid borne gene was altered to encode a C-terminal (His)6 tag, which does not interfere with the protein’s function(17,30). Purification protocols for R118G, R119W, V214A, V219A, W223A and A146Δ were previously described.(17,25) The R116A, K122A, R212A and R212A mutants were constructed and purified as described in Naganathan & Beckett(25). Each purified protein was exchanged into Storage Buffer (10 mM TrisHCl, pH 7.50 ± 0.02 at 4°C, 200 mM KCl, 0.1 mM dithiothreitol, 5% (v/v) glycerol) and stored at −70 °C. Protein concentrations were determined spectrophotometrically using a molar extinction coefficient at 280 nm calculated by the method by Gill and von Hippel.(31) Each preparation was >95% pure as judged by Coomassie brilliant blue staining of samples subjected SDS-PAGE on a 12.5 % gel. Activities of all proteins were >90% as determined by stoichiometric binding titrations with bio-5'-AMP monitored by fluorescence spectroscopy.(7)
Preparation of bioO DNA for DNaseI Footprint Titrations
The plasmid pBioZ was digested with HindIII and the purified linearized DNA was radiolabeled with 32P- dATP and dGTP using the Klenow Fragment of DNA Polymerase I(7). The resulting labeled DNA was separated from unincorporated nucleotides using an Elutip-D column (Scheicher & Schuell) and subjected to phenol-chloroform extraction. The DNA was concentrated by ethanol precipitation and then cleaved with a second restriction enzyme, PstI. The desired restriction fragment containing the bioO sequence was separated from the second fragment by electrophoresis on a 1% agarose gel and recovered by electroelution(32). Following ethanol precipitation the radiolabeled DNA was resuspended to a final concentration of approximately 20,000 cpm/µl in TE buffer and stored at 4 ° C.
Sedimentation Equilibrium Measurements
HoloBirA homo-dimerization and hetero-dimerization with apoBCCP87 were measured using sedimentation equilibrium. All experiments were performed using a Beckman Optima XL-I (Beckman-Coulter) analytical ultracentrifuge equipped with a four-hole An-60 rotor. Either standard 12 or 3 mm double-sectored cells with charcoal-filled Epon centerpieces and sapphire windows were used for the experiments. Sample volumes were 150 µL for the 12 mm cells and 50 µL for the 3 mm cells. Prior to centrifugation proteins were fully exchanged into the appropriate buffer using Biospin6 columns (Biorad) according to manufacturer’s instructions. Immediately before use bio-5’-AMP was prepared by diluting a concentrated stock prepared in water into the working buffer and then combining it with the apoBirA solution to make holoBirA.
For homo-dimerization measurements, the complex was prepared in Standard Buffer (10 mM Tris-HCl (pH 7.50 ± 0.02 at 20.0 ± 0.1 °C), 200 mM KCl, 2.5 mM MgCl2) at stoichiometric conditions using a ratio of 1:1.5 of protein to bio-5’-AMP. Samples prepared at three final protein concentrations ranging from 15 µM to 75 µM were centrifuged at 18K and 22K rpm for 12 hours each(26). All scans were acquired at 295 nm with 5 averages per radial position and a step size of 0.001 cm.
Hetero-dimerization was measured indirectly by monitoring perturbation of holoBirA dimerization by apoBCCP87. This method, which has previously been performed using sedimentation velocity, relies on the positive coupling of bio-5’-AMP binding to homo-dimerization and the chemical transfer of biotin that occurs upon binding of apoBCCP87 to holoBirA(21). Thus, combination of apoBCCP87 with a mixture of holoBirA monomer and dimer results in interaction of some fraction of the holoBirA with the acceptor protein, biotin transfer and formation of apoBirA, which does not dimerize in the conditions employed. The effect of apoBCCP87 on holoBirA dimerization is evidenced as an apparently weaker equilibrium constant for homo-dimerization and the magnitude of the perturbation can be used to quantitate the hetero-dimerization reaction. The temperature dependence of hetero-dimerization was measured in Standard Buffer the pH of which was adjusted at each working temperature (10–25 °C). Salt dependence was also measured in Standard Buffer prepared at final KCl concentrations ranging from 50 to 300 mM and adjusted to pH 7.50 ± 0.02 at 20.1 ± 0.1 °C. In each experiment performed in a single buffer condition three solutions were centrifuged. The first contained only BirA and bio-5’-AMP in a 1:1 molar ratio. The remaining two solutions contained the same holoBirA concentration as the first but two different apoBCCP87 concentrations. Thus different levels of perturbation of homo-dimerization are observed in the latter two samples. The particular holoBirA and apoBCCP87 concentrations used in a measurement varied according to the buffer conditions employed. Samples were centrifuged at rotor speeds of 18K and 22K rpm with a 12-hour interval between the two speeds. Step scans were performed at 295 nm with five measurements at each radial position and a 0.001 cm spacing between steps.
Inhibition DNase I Footprint Titrations
Hetero-dimerization of holoBirA (wild type and mutants) with apoBCCP87 was measured by monitoring the effect of acceptor protein on the fractional occupancy of bioO by holoBirA(20,33). In the titrations holoBirA concentration was held constant and apoBCCP87 concentration was varied. The holoBirA concentration employed, which was just sufficient to obtain full occupancy of bioO, renders the system poised for perturbation by apoBCCP87. All titrations, with the exception of those performed with the R119W mutant, were performed in buffer containing 10 mM Tris-HCl (pH 7.5 ± 0.02 at 20 ± 0.1 °C), 200 mM KCl, 2.5 mM MgCl2, 1.0 mM CaCl2, 100 µg/mL BSA, 2 µg/mL sonicated calf thymus DNA and saturating concentration of bio-5’-AMP for each variant protein. The weak binding of the R119W mutant necessitated performing the measurement in 50 mM KCl. Each reaction contained 12,000 cpm 32P-labeled (8 pM) bioO DNA in a total volume of 45 µL and a different apoBCCP87 concentration. The binding reactions were initiated by addition of 5 µL of holoBirA to bring both proteins to the desired final concentrations. After an 80 s incubation, 5 µL of a 0.0055 mg/mL DNaseI solution was added and digestion was allowed to proceed for 20 s. DNA cleavage was quenched by addition 33 µL of 50 mM Na2EDTA and the products were then precipitated with 700 µL of 0.4 M NH4OAc, 50 µg/mL yeast phenylalanyl tRNA in ethanol and incubation in a dry ice/ethanol bath for approximately 20 minutes.(8)Samples were centrifuged and DNA pellets were washed with 80% (v/v) ethanol in H2O and dried by lyophilization. After resuspension in 7 µL loading buffer (80% deionized formamide, 1 × TBE buffer, 0.02% bromophenol blue, and 0.02% xylene cyanol) the samples were heated to 90°C for 10 min and quick cooled on ice. The DNA fragments were resolved by electrophoresis on a 10% denaturing polyacrylamide gel (19:1 acrylamide/bisacrylamide in 8 M urea).
Data Analysis
Analysis of Sedimentation Equilibrium Data
All sedimentation equilibrium data were subjected to non-linear least squares analysis using the program MacNonLin or WinNonLin(34). The reduced molecular weight or σ-values were obtained for each scan individually and related to the molecular weight using the following equation:
| (1) |
in which M is the molecular weight, ν͞ is the partial specific volume of the protein, ρ is the density of the buffer, ω is the angular velocity of the rotor, R is the gas constant and T is the temperature in Kelvin. Data were also analyzed globally to a monomer-dimer association model(35) to obtain the monomer-dimer association constant, Ka using the following equation:
| (2) |
in which δc is the baseline offset, ct is the total concentration at each radial position, r, cm(ro) is the monomer concentration at ro, ro is the reference radial position (at the first data point), σmon is the reduced molecular weight of the monomer, and Ka is the equilibrium inhibition constant governing assembly of the dimer from the monomers. In the analysis, the reduced molecular weight of the dimer is assumed to be twice that of the monomer. The quality of each global analysis was evaluated by the magnitude of square root of variance of the fit and distribution of residuals.
Homo-dimerization measurements
In analyzing the data obtained from sedimentation equilibrium measurements of homodimerization of the BirA variants the σmon value for each BirA variant was obtained from sedimentation equilibrium measurements performed on the protein in its apo or unliganded state. Global analysis of scans acquired for samples prepared at three protein concentrations and centrifuged at two speeds yielded Ka, the association equilibrium constant for homo-dimerization of the adenylate-bound protein.
Hetero-dimerization measurements
For hetero-dimerization, global analysis of scans obtained for each cell at two speeds was performed using Equation (2) to yield the resolved Ka,OBS, the observed equilibrium constant for homo-dimerization. In the analysis the σmon was fixed at a value obtained from measurements performed at low protein concentration in the relevant buffer conditions. The observed equilibrium constant, Ka,OBS, was used to calculate the Ka,I, the association constant governing inhibition of homo-dimerization.
The relationship between the Ka,I and the Ka,OBS resolved from the data analysis can be understood in terms of the simultaneous equilibria that occur in solutions containing holoBirA and apoBCCP87. A reversible equilibrium between holoBirA monomer and dimer governed by an equilibrium constant Ka takes place along with reversible binding of the holoBirA monomer to apoBCCP87. This second binding reaction is followed by irreversible chemistry that produces holoBCCP87 and, more importantly, apoBirA, a species that is not competent to dimerize in the buffer conditions employed. Equations illustrating these two reactions are as follows:
| (3) |
| (4) |
The KA,OBS resolved from analysis of sedimentation equilibrium measurements performed in the presence of holoBirA and apoBCCP87 is related to the true equilibrium constant for dimerization, KDIM, and Ka,I, the constant governing the hetero-dimerization process, using the following two equations:
| (5) |
| (6) |
in which [holoBirA] is the concentration of holoBirA monomer and [BCCP87]total is the total concentration of BCCP87 in the reaction. In the analysis KDIM in each buffer condition was fixed at the value previously determined(23) and Ka,I was resolved from the analysis. In the Results section the inverse of this resolved parameter is presented as KI, the inhibition constant associated with apoBCCP87-mediated inhibition of homo-dimerization.
Quantification and Data Analysis of Inhibition DNaseI Footprint Titration
Gels were dried and exposed to phosphor screens for at least 40 hours prior to acquiring the image using a Storm phosphorimaging system (Perkin Elmer). The optical densities of bands representative of the bioO sequence were boxed and integrated at each apoBCCP87 concentration (ImageQuant) to produce inhibition binding isotherms.(33) The resulting isotherms were subjected to non-linear least squares analysis (GraphPad Prism 4.0) using the following
| (7) |
in which Ka,I is the inhibition association constant governing the hetero-dimeric interaction between holoBirA and apoBCCP. In the Results section the inverse of the parameter, KI, reported. Equation (7) is a modification of the expression that relates the fractional saturation, Y͞, of bioO to holoBirA concentration:
| (8) |
in which KTOT the equilibrium constant for total assembly of the holoBirA dimer:bioO complex, or the product of the equilibrium association constant for holoBirA dimerization and binding of the dimer to bioO, KDIMKbioO. In the analysis KTOT was fixed at the value obtained from direct DNaseI footprint titrations of bioO with each of the repressor proteins (wild type or variant).
Acknowledgments
This work was supported by NIH Grants R01-GM-46511 and S10-RR15899 to DB
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 3.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 4.Barker DF, Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981;146:469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- 5.Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981;146:451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- 6.Lane MD, Rominger KL, Young DL, Lynen F. The Enzymatic Synthesis of Holotranscarboxylase from Apotranscarboxylase and (+)-Biotin. Ii. Investigation of the Reaction Mechanism. J Biol Chem. 1964;239:2865–2871. [PubMed] [Google Scholar]
- 7.Abbott J, Beckett D. Cooperative binding of the Escherichia coli repressor of biotin biosynthesis to the biotin operator sequence. Biochemistry. 1993;32:9649–9656. doi: 10.1021/bi00088a017. [DOI] [PubMed] [Google Scholar]
- 8.Streaker ED, Beckett D. Coupling of protein assembly and DNA binding: biotin repressor dimerization precedes biotin operator binding. J Mol Biol. 2003;325:937–948. doi: 10.1016/s0022-2836(02)01308-6. [DOI] [PubMed] [Google Scholar]
- 9.Li SJ, Cronan JE., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cronan JE., Jr Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J Biol Chem. 1988;263:10332–10336. [PubMed] [Google Scholar]
- 11.Prakash O, Eisenberg MA. Biotinyl 5'- adenylate: corepressor role n the regulation of the biotin genes of Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5592–5595. doi: 10.1073/pnas.76.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenstein E, Beckett D. Dimerization of the Escherichia coli biotin repressor: corepressor function in protein assembly. Biochemistry. 1999;38:13077–13084. doi: 10.1021/bi991241q. [DOI] [PubMed] [Google Scholar]
- 13.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell RB, Barton GJ. An SH2-SH3 domain hybrid. Nature. 1993:364–765. doi: 10.1038/364765a0. [DOI] [PubMed] [Google Scholar]
- 15.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci U S A. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J Mol Biol. 2006;357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 17.Kwon K, Streaker ED, Ruparelia S, Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J Mol Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 18.Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J Biol Chem. 2008 doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- 20.Streaker ED, Beckett D. The biotin regulatory system: kinetic control of a transcriptional switch. Biochemistry. 2006;45:6417–6425. doi: 10.1021/bi052599r. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Beckett D. Kinetic partitioning between alternative protein-protein interactions controls a transcriptional switch. J Mol Biol. 2008;380:223–236. doi: 10.1016/j.jmb.2008.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown PH, Cronan JE, Grotli M, Beckett D. The biotin repressor: modulation of allostery by corepressor analogs. J Mol Biol. 2004;337:857–869. doi: 10.1016/j.jmb.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Streaker E, Pan W, Beckett D. Protein-protein interactions dominate the assembly thermodynamics of a transcription repression complex. Biochemistry. 2007;46:13667–13676. doi: 10.1021/bi7013097. [DOI] [PubMed] [Google Scholar]
- 24.Streaker ED, Gupta A, Beckett D. The biotin repressor: thermodynamic coupling of corepressor binding, protein assembly, and sequence-specific DNA binding. Biochemistry. 2002;41:14263–14271. doi: 10.1021/bi0203839. [DOI] [PubMed] [Google Scholar]
- 25.Naganathan S, Beckett D. Nucleation of an allosteric response via ligand-induced loop folding. J Mol Biol. 2007;373:96–111. doi: 10.1016/j.jmb.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roark DE. Sedimentation equilibrium techniques: multiple speed analyses and an overspeed procedure. Biophys Chem. 1976;5:185–196. doi: 10.1016/0301-4622(76)80034-8. [DOI] [PubMed] [Google Scholar]
- 27.Akiva E, Itzhaki Z, Margalit H. Built-in loops allow versatility in domain-domain interactions: lessons from self-interacting domains. Proc Natl Acad Sci U S A. 2008;105:13292–13297. doi: 10.1073/pnas.0801207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nenortas E, Beckett D. Purification and characterization of intact and truncated forms of the Escherichia coli biotin carboxyl carrier subunit of acetyl-CoA carboxylase. J Biol Chem. 1996;271:7559–7567. doi: 10.1074/jbc.271.13.7559. [DOI] [PubMed] [Google Scholar]
- 30.Kwon K, Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 33.Brenowitz M, Senear DF, Shea MA, Ackers GK. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 34.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laue TM. Sedimentation equilibrium as thermodynamic tool. Methods Enzymol. 1995;259:427–452. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- 36.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]