Abstract
The core complex in photosynthetic bacteria plays a central role in photosynthesis. This molecular assembly is composed of two protein complexes, viz., the light-harvesting complex I (LH1), which absorbs sunlight by means of the protein-bound bacteriochlorophylls, and the reaction center (RC), which uses the light-excitation energy absorbed by the LH complexes to produce a transmembrane (TM) charge gradient, subsequently employed for energy conversion. In Rhodobacter (Rba.) sphaeroides, the core complex contains, in addition, two copies of the single TM α-helix protein, PufX, and forms a (RC-LH1-PufX)2 dimer. To this date, no high-resolution structure has been reported for the entire core complex. In particular, the location of PufX within the (RC-LH1-PufX)2 dimer is still the subject of much debate. Here, one of the proposed locations for PufX, requiring its dimerization, is examined. The PufX-dimer model on the basis of the glycophorin A (GpA) dimer was constructed, and its robustness was probed through a series of molecular dynamics (MD) simulations. The free-energy change due to the replacement of Gly35 by valine was also determined to assess whether this mutation is responsible for distinct PufX oligomerization states in different Rba. species. The present study shows that PufX helices form a stable GpA-like dimer with a helix-helix crossing angle that could constitute the molecular basis of the reported highly bent and V-shaped structure of the Rba. sphaeroides core complex dimer.
The core complex in photosynthetic bacteria plays a central role in photosynthesis. This molecular assembly is composed of two protein complexes, viz., the light-harvesting complex I (LH1), which absorbs sunlight by means of the protein-bound bacteriochlorophylls, and the reaction center (RC), which uses the light-excitation energy absorbed by the LH complexes to produce a transmembrane (TM) charge gradient, subsequently employed for energy conversion. In arguably the most studied purple bacterium, Rhodobacter (Rba.) sphaeroides, the core complex contains, in addition, two copies of the single TM α-helix protein, PufX, and forms a (RC-LH1-PufX)2 dimer. 1 To this date, no high-resolution structure has been reported for the entire core complex. In particular, the location of PufX within the (RC-LH1-PufX)2 dimer is still the subject of much debate. Here, one of the proposed locations for PufX,2 requiring its dimerization, is examined. The PufX-dimer model suggested by Busselez et al. 3 on the basis of the glycophorin A (GpA) dimer4 was constructed, and its robustness was probed through a series of molecular dynamics (MD) simulations. The free-energy change due to the replacement of Gly35 by valine was also determined to assess whether this mutation is responsible for distinct PufX oligomerization states in different Rba. species, as proposed by Busselez et al. 3 The present study shows that PufX helices form a stable GpA-like dimer with a helix-helix crossing angle that could constitute the molecular basis of the reported 5 highly bent and V-shaped structure of the Rba. sphaeroides core complex dimer.
A number of intriguing aspects of the bacterial photosynthetic core complex are yet to be explained. 6,7 In different species, core complexes can either be ring-like monomers, i.e., (RC-LH1)1, or S-shaped dimers, i.e., (RC-LH1-PufX)2 found in Rba. sphaeroides. 1 Although PufX is also present in other Rba. bacteria, some species, e.g., Rba. velkampii, possess monomeric core complexes ((RC-LH1-PufX)1). In Rba. sphaeroides, deletion of PufX results in monomeric rings, suggesting that PufX is directly involved in the dimerization of the core complex. 8,9 A recent three-dimensional electron microscope (EM) single-particle analysis revealed that Rba. sphaeroides core complex dimers are highly bent. 5 Without a high-resolution structure, however, a major and fundamental hurdle still remains in the understanding of the molecular arrangement of the core complex and the precise location of PufX — for reviews see Cogdell et al. 6 and Holden-Dye et al. 7 One of the suggested locations of PufX is at the center of the core complex dimer. 2 For this conjecture to hold, the natural propensity of PufX to dimerize needs to be established. Yet, only monomeric PufX solution structures have been reported. 10,11 To address the possibility of PufX dimerization, Busselez et al. 3 hypothesized that a GxxxG motif 12 found in the Rba. sphaeroides PufX sequence is responsible for PufX dimerization. A series of MD simulations were carried out to examine the viability of such dimerization scheme.
To investigate the possibility of a PufX dimer, different MD simulations were performed (see Supporting Information for the full list of simulations). According to the two-stage model for membrane-protein folding, 13,14 oligomerization of integral, TM α-helices is preceded by their insertion in the membrane, each helical segment assumed to be independently stable. For this reason, the first set of simulations focused on the robustness of the PufX monomer in a membrane environment. The second set of simulations explored the structural characteristics of a PufX dimer model based on the analogous GpA scaffold. 4 In addition, free-energy calculations were carried out to probe the effect of a point mutation in the GxxxG motif, in particular whether or not replacement of a glycine by a large aliphatic residue would abolish dimerization and, thus, lead to the formation of monomeric PufX.
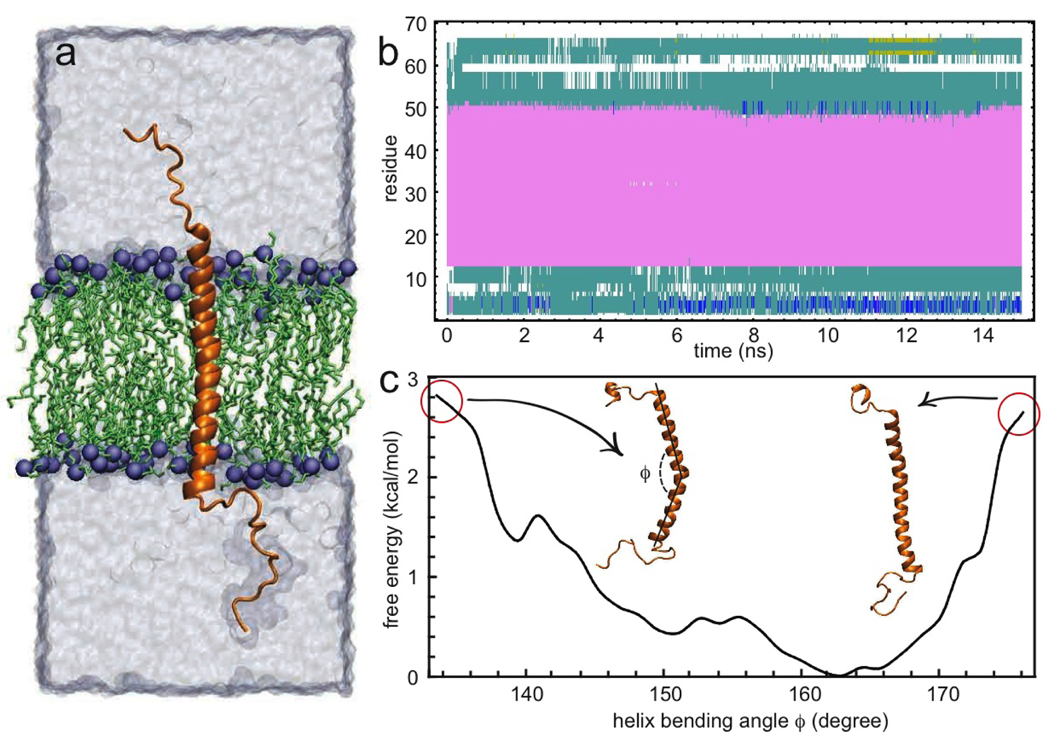
The initial conformation of the PufX monomer was taken from the solution structure of Wang et al.; 10 residues 1 to 69 were included in accordance with prior studies. 15 PufX was placed in an upright, TM orientation amidst a fully hydrated POPE membrane (Figure 1a) and simulated at thermodynamic equilibrium for 15 ns employing the molecular-dynamics package NAMD.16 During the simulation, the secondary structure of the PufX TM region fluctuated marginally, retaining the general conformation of an integral α-helix with flexible terminal loops (Figure 1b). Noteworthy changes in the PufX-membrane interactions were observed. In particular, the helical segment tilted in the membrane, and even bent spontaneously (Figure 1c; see also in Supporting Information MovieS1 depicting the simulation trajectory). The TM α-helix formed a bending angle about Gly31, and straightened back towards the end of the simulation. A glance at the bending free energy suggests that the helix is, in fact, naturally slightly bent, with an energetic cost for returning to an upright conformation. The free-energy penalty incurred to bend the helix by ± 10° is less than 2kB T (Figure 1c), reflecting the intrinsic flexibility of PufX. This flexibility resonates with the fact that two PufX solution structures were solved independently, one showing a seemingly straight helix, 10 while the other brought a prominent bend to light. 11 It is possible that the two different PufX solution structures can be reconciled on the basis of the intrinsic flexibility of PufX. Furthermore, the TM hydrophobic region of PufX is ca. 40 Å long, whereas the thickness of the POPE hydrophobic core is 35 Å. Such a “hydrophobic mismatch” is known to induce tilting and bending of TM helices, together with aggregation and oligomerization. 17,18
Figure 1.
MD simulation of the PufX monomer. (a) A single α-helix is immersed in a POPE bilayer. PufX is shown in orange, the water box in transparent gray, and POPE in green with phosphorus atoms shown as blue spheres. In all figures, PufX has a TM orientation, with its N-terminal end (cytoplasmic) pointing upwards. (b) Time evolution of the PufX secondary structure (pink: α-helix; cyan: turn; white: coil; red: π-helix). (c) Free-energy as a function of the bending angle, φ, of the helix. Insets: Snapshots showing a bent and a straight helix. Image rendered with VMD.19
Having established that the PufX monomer is structurally robust, a PufX dimer model was constructed, mimicking the GpA dimerization motif (PDB code 1AFO),4 as suggested in Busselez et al. 3 The conformation of the equilibrated PufX monomer was employed to build the dimer model. Interestingly enough, the PufX dimer model produced is similar to the solutions supplied by the HEX docking code. 20 A 50 ns simulation was carried out for the dimer-membrane system (Figure 2a), during which the secondary structure of both α-helices showed little fluctuation (Figure 2b), indicating that PufX in a dimeric state retains structural integrity. At the beginning, due to the slight curvature witnessed in the monomer simulation, the two α-helical segments formed a smaller crossing angle at the N–terminus than at the C–terminus. By the end of the simulation, however, both α-helices straightened and intersected with a consistent crossing-angle. The crossing angle for PufX dimer was determined to be ca. −38° (Figure 2c), the minus sign denoting right-handedness, comparable to the −40° angle measured for GpA. 4 In addition, spatial arrangement of the TM regions departs only moderately from the initial configuration (Supporting Information).
Figure 2.
MD simulation of the PufX dimer. (a) PufX dimer-membrane system. The coloring scheme is the same as in Figure 1. PufX1 is colored in orange and PufX2 in blue. Two snapshots of the TM helices are shown at the beginning and at the end of the simulation. (b) Time evolution of the secondary structure of the two α-helices. (c) Time evolution of their crossing angle, θ. (d) Time evolution of their buried molecular surface area. (e) Two-dimensional plot highlighting the key residues responsible for PufX dimerization. Insets: Two highly interacting pairs of residues, viz. Trp32-Gly35 and Trp32-Gly34.
To assess whether and how the α-helices interact throughout the simulation, the buried molecular surface area of the dimer was determined to provide an estimate for the surface area of interaction (Figure 2d). The two helical segments remained closely associated during the simulation, with the buried surface area increasing to ca. 650 Å2. For GpA, this quantity was measured to be 587 Å2. 21 The MD study also allowed the key residues participating in PufX dimer interaction to be identified. In Figure 2e, a heptad of residues, viz., M27, K28, W32, G34, G35, V36, and F38, is found to contribute predominantly to PufX dimerization, reminiscent of the dimerization scheme of GpA.4
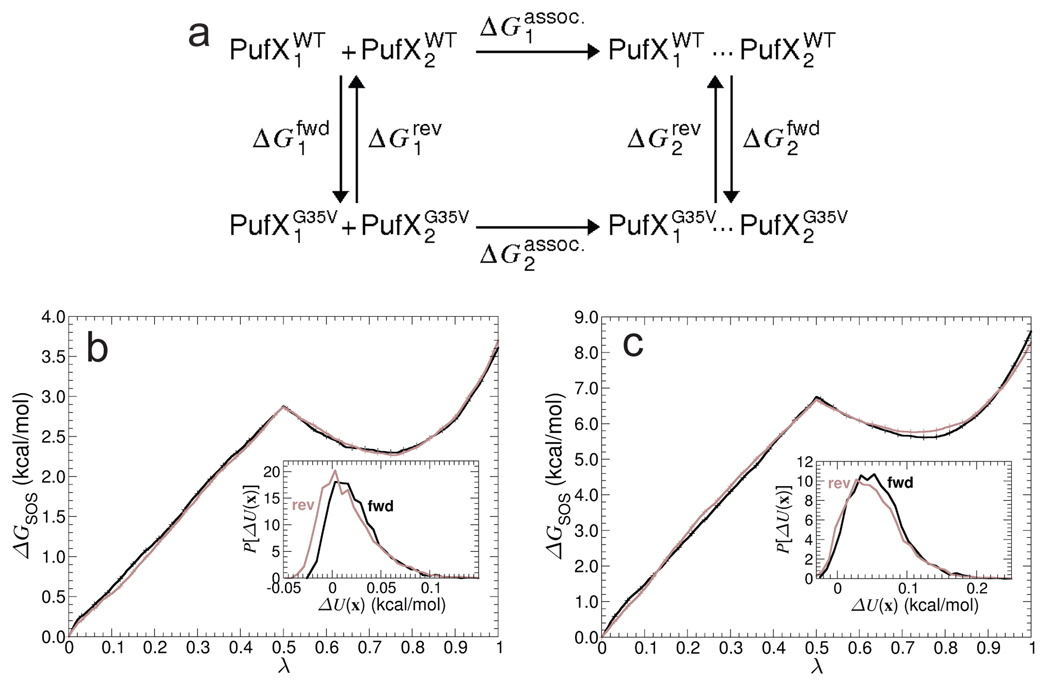
Whereas Rba. sphaeroides expresses dimeric core complexes, other Rba. species, e.g., Rba. velkampii, which also includes PufX, produce only monomeric core complexes. Busselez et al. 3 ascribed this difference in oligomerization states to differences in the PufX oligomerization states, which are rooted in sequence alterations. The Rba. sphaeroides PufX sequence contains a GxxxG motif. In Rba. velkampii, however, the corresponding sequence is GxxxV, and the Gly35 to Val35 mutation was hypothesized to abolish dimerization of PufX and lead to the formation of monomeric core complexes. 3 To examine the alleged3 disruptive effect of a G35V point mutation on PufX dimerization, free-energy perturbation (FEP) calculations were carried out 22,23 (Supporting Information). The present calculations follow the thermodynamic cycle shown in Figure 3a, where Gly35 is transformed quasi-statically into valine, in both the monomeric and the dimeric states, using a general-extent parameter, λ. 24 To improve the accuracy of the free-energy estimates, each calculation was repeated twice using different initial conditions, and the simple overlap sampling (SOS) scheme was employed to combine forward and reverse transformations. 25 Repeated simulations of the PufX monomer in both forward and reverse directions yielded a consistent free-energy change of ΔG1 = 3.7±0.3 kcal/mol, within chemical accuracy. Repeated simulations of the PufX dimer, also in both directions, yielded a free-energy change of ΔG2 = 8.5±0.3 kcal/mol, resulting in the net alchemical free-energy change of ΔΔG = 1.2±0.6 kcal/mol. Comparable site-directed mutagenesis experiments were conducted for GpA, replacing Gly83 with Val83, leading to a slightly higher apparent free-energy change, namely, on the order of 3–4 kcal/mol. 26 In view of the magnitude of the free-energy change measured here, the hypothesis of Busselez et al. 3 appears to be reasonable, yet somewhat incomplete. The free-energy penalty for the G35V mutation possibly reduces the probability of PufX dimer formation in Rba. velkampii. The low free-energy penalty, however, suggests that other factors may be at play for determining the actual oligomerization state of PufX.
Figure 3.
Free-energy calculation of the G35V point mutation. (a) Thermodynamic cycle delineating the G35V mutation. (b) and (c) Free energy as a function of the general extent parameter λ for both the forward (dark) and the reverse (light) transformations, in the case of the PufX monomer and the dimer mutation. Inset: Overlapping thermodynamic ensembles for the forward and the reverse transformations at λ = 0.5.
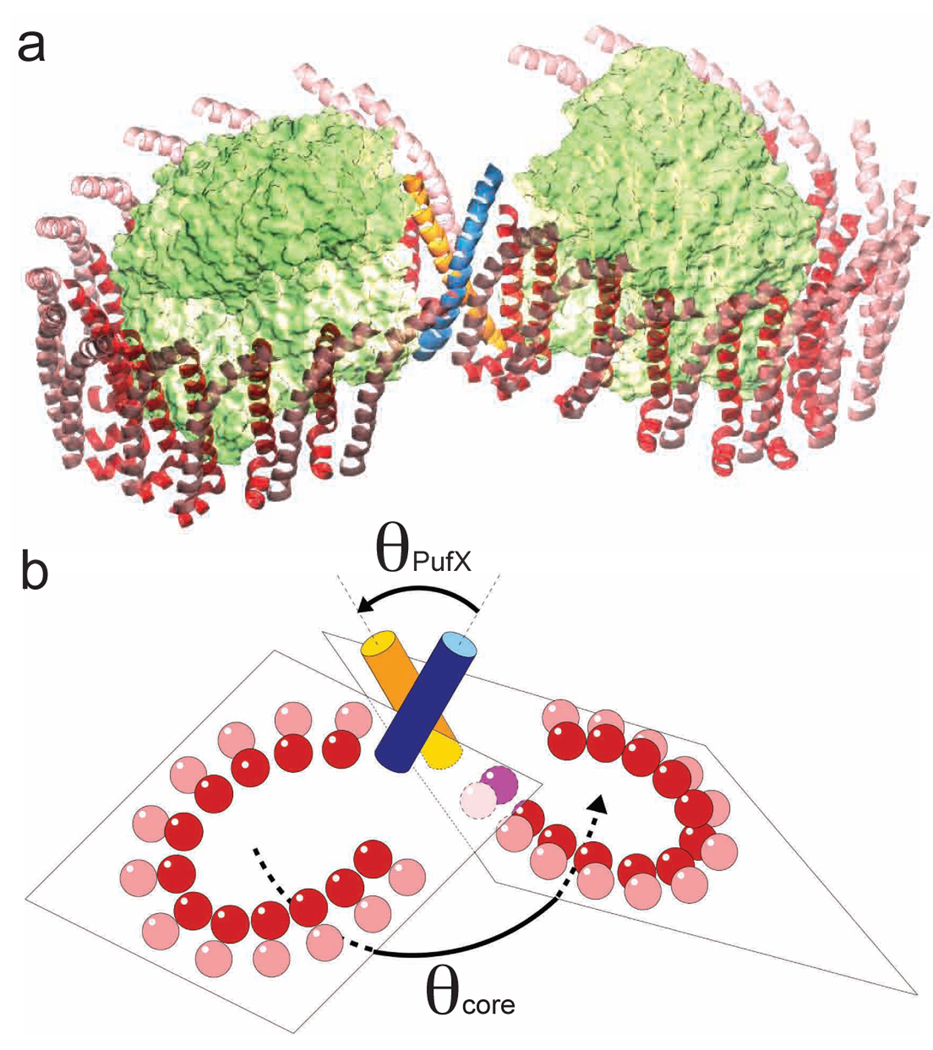
The present set of MD simulations reveals that a GpA-like dimerization construct for Rba. sphaeroides PufX is viable over the timescale explored. No hint of dissociation was observed. In contrast, PufX helix-helix association strengthened as the buried molecular surface increased. It is worth noting that earlier TOX-CAT27 measurement revealed significant self-association of the Rba. capsulatus PufX TM segment.28 Emergence of a dimeric state for PufX would suggest that the dimer is located at the center of the Rba. sphaeroides core complex, where it serves as a nucleation point for the self-assembly of the dimeric core complex. In this picture, PufX dimerization in the membrane initiates the assembly of the dimeric core complex, followed by docking of RC and LH1 subunits on both sides of the central PufX dimer29 (Figure 4a; see Supporting Information for an animation of the assembly process). In the resulting dimeric structure of the core complex, assuming that the LH1 helices remain parallel to that of PufX in each core complex monomer, the magnitude of the PufX helix-helix crossing angle (θPufX) can be related to the bending angle of the core-complex dimer (θcore) by means of an approximate geometric rule: θcore + θPufX ~ 180° (Figure 4b). In the present study, the equilibrium simulation of the PufX dimer yielded θPufX ~ 38°, and θcore was measured in single-particle EM study to be 146°. 5
Figure 4.
(a) Putative model of the Rba. sphaeroides core complex dimer, with a PufX dimer at the center. PufX: orange and blue; RC: green; LH1: red and pink. (b) The crossing angle of the PufX helices, θPufX, can be related to the bending angle of the core complex dimer, θcore, via an approximate geometric relation.
While MD simulations suggest that a GpA-like dimerization scheme is plausible for Rba. sphaeroides PufX, estimation of the free-energy cost incurred in the G35V mutation showed that the free-energy penalty, while disrupting dimer association, is not sufficiently high to completely abolish formation of PufX dimers. A GxxxG motif-based argument is, therefore, perhaps too rudimentary to rationalize in a nonambiguous fashion the different PufX oligomerization states in different Rba. species. A definitive answer would require further structural determination and biochemical mutation studies, which are expected to shed new light on the overall structure and assembly of photosynthetic core complexes.
Supplementary Material
Acknowledgement
This work was supported by grants NSF MCB-0744057 and NIH P41-RR005969. Computer time was provided by NCSA and TACC via Large Resources Allocation Committee grant MCA93S028, and resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, supported by the Office of Science of the U.S. DOE (contract DE-AC02-06CH11357). The authors thank James Sturgis, C. Neil Hunter, Simon Scheuring, and Christopher B. Harrison for insightful discussions and assistance.
Footnotes
Supporting Information Available: Movies and details on methods are available free of charge at http://pubs.acs.org.
References
- 1.Jungas C, Ranck J, Rigaud J, Joliot P, Verméglio A. EMBO J. 1999;18:534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheuring S, Francia F, Busselez J, Melandris BA, Rigaud J-L, Lévy D. J. Biol. Chem. 2004;279:3620–3626. doi: 10.1074/jbc.M310050200. [DOI] [PubMed] [Google Scholar]
- 3.Busselez J, Cottevielle M, Cuniasse P, Gubellini F, Boisset N, Lévy D. Structure. 2007;15:1674–1683. doi: 10.1016/j.str.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 4.MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 5.Qian P, Bullough PA, Hunter CN. J. Biol. Chem. 2008;283:14002–14011. doi: 10.1074/jbc.M800625200. [DOI] [PubMed] [Google Scholar]
- 6.Cogdell RJ, Gall A, Köhler J. Quart. Rev. Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 7.Holden-Dye K, Crouch LI, Jones MR. Biochim. Biophys. Acta. 2008;1777:613–630. doi: 10.1016/j.bbabio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Francia F, Wang J, Venturoli G, Melandri A, Barz W, Oesterhelt D. Biochem. 1999;38:6834–6845. doi: 10.1021/bi982891h. [DOI] [PubMed] [Google Scholar]
- 9.Frese R, Olsen J, Branvall R, Westerhuis W, Hunter C, van Grondelle R. Proc. Natl. Acad. Sci. USA. 2000;97:5197–5202. doi: 10.1073/pnas.090083797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z-Y, Suzuki H, Kobayashi M, Nozawa T. Biochemistry. 2007;46:3635–3642. doi: 10.1021/bi0618060. [DOI] [PubMed] [Google Scholar]
- 11.Tunnicliffe RB, Ratcliffe EC, Hunter CN, Williamson MP. FEBS Lett. 2006;580:6967–6971. doi: 10.1016/j.febslet.2006.11.065. [DOI] [PubMed] [Google Scholar]
- 12.Russ WP, Engelman DM. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 13.Popot J, Engelman D. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 14.Engelman DM, Chen Y, Chin C-N, Curran AR, Dixon AM, Dupuy AD, Lee AS, Lehnert U, Matthews EE, Reshetnyak YK, Senes A, Popot J-L. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 15.Parkes-Loach PS, Law CJ, Recchia PA, Kehoe J, Nehrlich S, Chen J, Loach PA. Biochemistry. 2001;40:5593–5601. doi: 10.1021/bi002580i. [DOI] [PubMed] [Google Scholar]
- 16.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Planque MRR, Killian JA. Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 18.Yeagle PL, Bennett M, Lemaitre V, Watts A. Biochim. Biophys. Acta Biomembr. 2007;1768:530–537. doi: 10.1016/j.bbamem.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 20.Ritchie DW. Proteins: Struct., Func., Gen. 2003;52:98–106. doi: 10.1002/prot.10379. [DOI] [PubMed] [Google Scholar]
- 21.Fleming KG, Ackerman AL, Engelman DM. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- 22.Zwanzig RW. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
- 23.Hénin J, Pohorille A, Chipot C. J. Am. Chem. Soc. 2005;127:8478–8484. doi: 10.1021/ja050581y. [DOI] [PubMed] [Google Scholar]
- 24.Chipot C, Pohorille A. Free energy calculations. Springer; 2007. [Google Scholar]
- 25.Lu N, Kofke DA, Woolf TB. J. Comp. Chem. 2004;25:28–39. doi: 10.1002/jcc.10369. [DOI] [PubMed] [Google Scholar]
- 26.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. J. Mol. Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russ WP, Engelman DM. Proc. Natl. Acad. Sci. USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aklujkar M, Beatty JT. Photosyn. Res. 2006;88:159–171. doi: 10.1007/s11120-006-9047-y. [DOI] [PubMed] [Google Scholar]
- 29.Pugh R, McGlynn P, Jones M, Hunter C. Biochim. Biophys. Acta. 1998;1366:301–316. doi: 10.1016/s0005-2728(98)00131-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.