Abstract
Here we describe an integrated software platform titled HD Desktop designed specifically to enhance the analysis of hydrogen/deuterium exchange (HDX) mass spectrometry data. HD Desktop integrates tools for data extraction with visualization components within a single web-based application. The interface design enables users to navigate from the peptide view to the sample and experiment levels, tracking all manipulations while updating the aggregate graphs in real time. HD Desktop is integrated with a relational database designed to provide performance enhancements, as well as a robust model for data storage and retrieval. Additional features of the software include retention time determination, which is achieved with the use of theoretical isotope fitting; here, we assume that the best theoretical fit will occur at the correct retention time for any given peptide. Peptide data consolidation for the rendering of data in 2D was realized by automating known and novel approaches. Designed to address broad needs of the HDX community, the platform presented here provides an efficient and manageable workflow for HDX data analysis and is freely available as a web tool at the project home page http://hdx.florida.scripps.edu.
Introduction
Hydrogen/deuterium exchange mass spectrometry (HDX MS) is a widely used method for the characterization of protein dynamics and protein-protein or protein-ligand interactions [1]. Despite advancements in instrumentation for the automated acquisition of HDX MS data, the subsequent analysis, statistical validation and presentation of the resultant exchange data remain a tedious process. Existing software platforms address a subset of these issues, but lack sufficient integration, functionality, and accessibility to meet the broader needs of the HDX community. For example, methods of determining deuterium content by deconvolution of spectral data have been described [2–4]. Software to simplify the determination of peptide ion centroid m/z value has been developed [5]. Command line tools have been made available which automate the extraction of deuterium content using approaches based on Fourier transform and natural isotopic abundance [6, 7]. Proprietary software has also been described that automates the extraction of deuterium content in standalone software applications such as DXMS [8] and HD Express (ExSAR Corp., NJ, unpublished). Unlike centroid based methods, which assume EX2 type exchange kinetics, other approaches have been described that measure the width of an isotopic distribution over the period of H/D exchange, thereby allowing for the characterization of EX1 type kinetics [9]. This approach has been offered as Microsoft Excel based tools [10]. A publicly available Windows based software package titled “TOF2H” has also been developed to automate the processing of LC-MALDI HDX data. This software automates several functions, including centroid calculations and deuterium uptake plots [11].
Recently, software titled “The Deuterator” [12] provided an automated platform for the calculation and validation of HDX MS data. The freely accessible web-based software was designed to accept common file format mzXML [13] data acquired with a variety of different mass spectrometers, and has been validated using low resolution MS data from quadrupole ion traps [14], intermediate resolution data acquired with an ESI quadrupole time-of-flight instrument (unpublished data provided by the Wysocki group) and high resolution MS data from both Orbitrap and 7 Tesla FT-ICR mass analyzers [15, 16]. In addition, a spectral range approach was described that enabled the software to disregard data from unrelated isotopic clusters resolved with high resolution mass spectrometers. The Deuterator software provided functionality so that HDX MS data can be extracted with either a “centroid” m/z approach or a “theoretical isotope fitting” model. Briefly, the “centroid” method relies on the calculation of the intensity weighted m/z average value of each isotope cluster. The shift in m/z of these average m/z values, with increasing deuterium incubation time, can be related to percent H/D exchange for a specific region of the protein. For the “theoretical isotope fitting” approach, a theoretical isotopic envelope is generated for the ion of interest and a chi-squared fit score obtained. The percent deuterium in the theoretical isotopic envelope is increased between zero and 100 percent deuterium (calculated from the number of exchangeable amide hydrogens within the sequence). The lowest chi-squared fit score provides the percent deuterium incorporation for that ion. This iterative fit model is similar to that initially described by Palmblad et al [6]. However, it should be noted that any peptide ion isotope clusters that cannot be resolved within the chromatography step or the mass analyzer, will not provide any meaningful HDX data.
While software such as The Deuterator greatly improves the throughput of HDX studies, the output from the software is limited to centroid m/z values and best theoretical fit percentages. No provision was made for data analysis (Microsoft Excel) visualization (Pymol and/or Jmol) or statistical analysis (GraphPad Prism). Even with the availability of several software tools for HDX data analysis, the task of assembling and visualizing the resulting data has remained a manual operation left to the end user. Not surprising, these processes remain very time consuming and are prone to user error. For laboratories that perform a large number of HDX studies, the use of multiple separate software tools for data analysis, visualization and statistical validation presents significant challenges to data workflow management.
To address the limitations described above we have developed “HD Desktop,” a fully integrated web-based application for the analysis, visualization and statistical validation of HDX MS data. HD Desktop provides a dramatic improvement in functionality and integration when compared to existing HDX software applications including The Deuterator. Similar to The Deuterator, HD Desktop accepts MS data in the common file format mzXML and supports HDX data acquired with low (< 2000), intermediate (~ 10,000) and high (> 60,000) resolution mass analyzers. Importantly, several new data analysis algorithms have been integrated. For instance, to reduce the requirement for manual correction of input retention time (RT) data, a retention time determination routine has been integrated into the preprocessing and synchronization modules of the application. This procedure operates in a similar fashion to the “best-spot determination” operation within the ToF2H program [11]. However, the RT best-fit in HD Desktop is determined by the best chi-squared fit score obtained across a small retention time window (described in detail below). If no retention time is provided, the software will determine the correct retention time from all the scans in the MS data set.
Although designed for use with HPLC ESI-MS data, HD Desktop can accommodate both MALDI MS and LC MALDI MS data. The “centroid” and “theoretical fit” methods for the determination of percent deuterium have been described previously [12]. The results from these methods for the determination of percent deuterium incorporation are analyzed and visualized using fully integrated tools within this new software application. The software can also accommodate off-exchange experiments if required.
Critically, the integration of a relational database within the software allows for the real time update of all data analysis/visualization steps following any manipulations that influence the percent deuterium calculation method. For example, changes in the “centroid” percent deuterium calculation may arise from a change in the retention time or m/z range in the centroid calculations. The new percent deuterium values are immediately reflected in all downstream calculation/visualization steps thus minimizing data curation by the end user. In addition, we have developed new tools for data analysis and statistical validation, and these have been integrated along with existing tools and are displayed within five new user interfaces. As expected, the software provides standard percent deuterium vs. time plots. However, HD Desktop provides the ability to plot HDX data (or differential HDX data) over a protein sequence or the three-dimensional protein structure, which is rendered by an integrated Jmol application. The end result is a streamlined HDX workflow that provides a significant reduction in the time required for the analysis, statistical validation and display of HDX MS data, thus significantly increasing the applicability and impact of HDX for the analysis of protein dynamics. The free availability of this software on-line and the inherent flexibility of the application will enable more researchers to take advantage of HDX technology.
Experimental
HD Desktop was designed as a web-based solution allowing the software to be scalable, and to run on multiple platforms/networks with minimal installation overhead. The existing code from the preceding software (The Deuterator) was leveraged whenever possible allowing the new software to build upon a number of established features. We have maintained the ability to process data from mass analyzers of differing resolving powers, and to work from peptide sets generated from a variety of sources. A schematic representation of the new software integrated within our HDX workflow is shown in Figure 1. There are three main inputs into the software; 1) the mzXML MS data files, 2) a list of peptide ion sequence, charge state and chromatographic retention time (this list is referred to as the “peptide set”), and 3) a text-based sequence file in fasta[17] format containing the target protein sequence. It should be noted that, as with our previous software, the methods for determining peptide ion identities and their acceptance criteria are left to the end user. Any attempt to integrate peptide identification and/or peak picking within a HDX application appears unnecessary given the vast efforts of the proteomics community and the rapid evolution being made in this field.
Figure 1. HDX Workflow Integration.
(a) The protein of interest is digested with an enzyme and the resulting peptides are analyzed by LC MS/MS. (b) Peptide identity is then established with database search tools such as Sequest or Mascot (note that the method for determining peptide identity is left to the user), and converted into PepXML and CSV formats to facilitate editing. (c) The user then selects the peptides of interest from the search results to create the peptide set. (d) The protein is incubated with D2O at multiple time points, digested with an enzyme and the resulting peptides are analyzed by LC MS. (e) The binary MS files, sequence file (in fasta format), and the peptide set are imported into the appropriate file store or LIMs. (f) The raw files are converted to mzXML, and HD Desktop pre-processing is conducted and the results are stored within a MySQL database. (g) Data are made available through interactive tools for HDX data curation and visualization components within the Grails framework.
Database integration
In contrast to our previous HDX software, this application was designed to operate in conjunction with a database. The first step in the preprocessing stage, described in more detail below and in Figure 1a–f, is to calculate the “reference data” from the peptide set. For each peptide of interest, the database is populated with the following information; monoisotopic mass, average mass, mz0, mz100, positional range, length, charge, score (optionally), number of prolines, and chemical formula. These data are calculated once from the peptide sequence and are subsequently referenced by many of the downstream algorithms. The subsequent step is to calculate the “result data”, which includes the following information: mzStart, mzEnd, retention time range, centroid, modified timestamp, and the “centroid” and “theoretical fit” percent deuterium values for a given peptide ion; %Dcentroid, and %Dtheoretical, respectively.
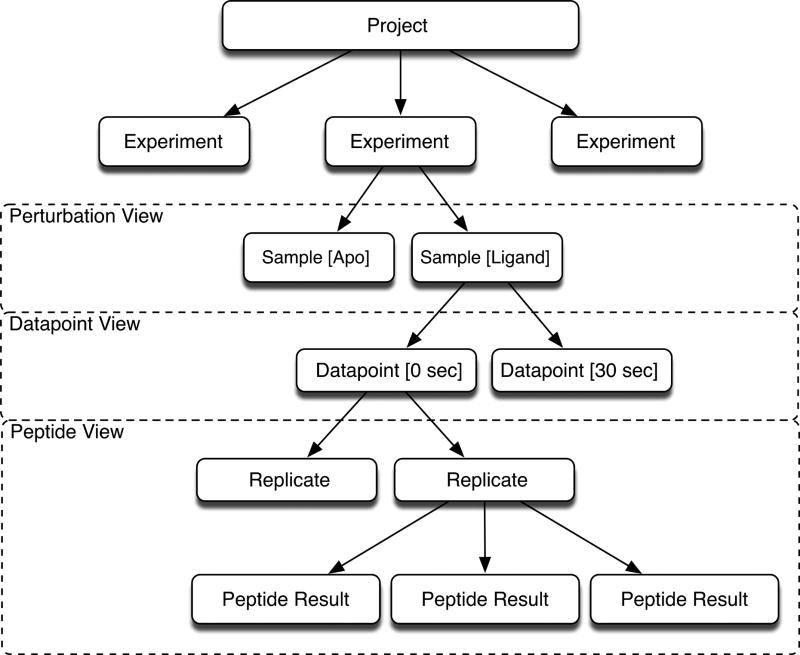
The database model (Figure S1) was designed and implemented to support the broad data management needs of the software. The database design reflects a typical HDX experiment, which often involves a comparison of two samples treated with different conditions such as, for example unbound protein versus ligand bound protein. Each sample is exposed to deuterium at varying time intervals and replicated either as reruns of the same sample (not advised) or as an independent replicate experiments, which is our preference[15]. Often it is desirable to run several experiments with similar samples, which falls under the category of a project; therefore each project may contain one or more experiments, and each experiment will contain one or more samples. Each sample has several datapoints, and each datapoint will have several replicates. The structural hierarchy of the software is shown in Figure 2.
Figure 2. The Data Hierarchy.
Hierarchical structure of the data outlines the relationships between data components. Each experiment contains one or more samples, and each sample has multiple datapoints. The boxes outline the relationships between the data structure and the corresponding software application. The “Perturbation view” application presents the comparison between samples and the “Datapoint view” presents the data from all datapoints for a given sample. The “Peptide view” allows users to view and manuipulate the data related to the peptide.
Technology
A synopsized overview of the technologies used within the application is presented in Table 1. A primary objective was to identify and integrate existing solutions where possible, and build new solutions only when necessary, effectively lowering development costs and expediting delivery times. With this in mind, several existing technologies were selected and integrated within the application. These decisions were based upon the technology having the required functionality and being freely available in a manner that would permit use in distributed applications, thus, paving the way for the eventual offering of HD Desktop as an open source solution. The use of flexible solutions was a priority to continue the effort to address the needs of the broader HDX community. Best of class technologies were also strongly considered, as this usually translates into a more vibrant community, better support, and simplified maintenance.
Table 1.
Software Components. Several technologies were integrated into the application. The selection criteria were based upon availability, functionality, flexibility, and support.
| Feature/Component | Project/Code base | Description |
|---|---|---|
| Application Framework | Grails [28] | Simplifies development of enterprise quality applications |
| Database | MySQL [29] | Robust freely available relational database |
| mzXML conversion | Sashimi [30] | Executables that convert native binary files into mzXML |
| Centroid m/z | Deuterator v1.0 [12] | Calculates the weighted m/z value for a peptide envelope within a user defined range |
| Theoretical fit %D | Deuterator v1.0 [12] | Calculates the best chi2 fit from all possible theoretical isotopic distributions at varying levels of deuterium incorporation |
| MS Data input | mzXML [13] | Common file format for representation of mass spectral data |
| 3D structure | Jmol [31], PyMol [32] | Open source molecule viewers |
| Plots | JFreeChart [33] | Open source Java chart library |
| Browser | Firefox [34] | Web browser with built in support for tabbed browsing |
| User Interface | Ext JS [35] | Javascript library for building rich applications |
| Database Persistence Layer | Hibernate [36] | Persistence service which facilitates class development for databases |
| Theoretical isotope calculation | Qmass [22] | Calculates theoretical isotopic distributions |
Retention Time Determination
To accommodate any experimental variation in chromatographic retention time (RT), we have incorporated a method to determine the correct retention time range for each peptide in all of the LC MS datasets. It should be noted that alignment of chromatographic retention time in HDX data must accommodate the increase in the m/z ratio of each peptide as %D increases. This change in the m/z ratio of each peptide precludes the use of a number of existing approaches for retention time correction [18, 19] that rely on finding specific m/z values across multiple LC MS data sets (feature detection).
Our approach employs a moving window of retention time around a user defined target RT value (from the peptide set) and is combined with our chi-squared fitting algorithm, as illustrated in Figure 3. When a target RT value is provided, the operational RT range becomes the target RT value ± 1 minute. Where a target RT value is available (from a database search, or previous HDX dataset for example) iteration over the entire retention time window becomes unnecessary and would contribute a significant computational expense. With a fixed window of 12 seconds (> the baseline width of typical chromatography peaks), the software calculates the co-added mass spectrum and determines the corresponding best-fit theoretical distribution (iterating from 0–100 %D). This process is repeated after moving the window in 0.05 digital minute increments. The retention time range with the lowest chi-squared-fit score is retained and can be assumed to represent the correct retention time for each peptide, regardless of %D incorporation. For the example shown in Figure 3, the input RT range value was 7.85 minutes. The software proceeded to iterate a moving window over the target RT value ± 1 min, determining the best chi-squared fit for each iteration. In this example the optimal RT range for this peptide was determined to be 7.05–7.25 minutes (Figure 3b). In situations where no target RT is provided, the moving window operates across the entire duration of LC MS dataset. These RT determination algorithms are integrated into the data preprocessing step and are available as an option within the synchronization tool.
Figure 3. Retention Time Determination.
To establish the optimal retention time (RT) for each peptide, a 0.05 digital minute RT window is combined with our chi-squared fitting algorithm and moved across the target RT value ± 1 minute. The RT that generates the theoretical plot with the lowest fit score is assumed to represent the correct retention time for each peptide. In this example, the target RT value was 7.85 minutes. In panels (a) and (c), the chi-squared scores were inferior to the score obtained from (b), which is a result of the difference between the observed co-added spectrum (illustrated in red)and the theoretical plot (shown in blue). The best score was found in panel (b) and the optimal RT range is therefore determined to be 7.05 – 7.25 minutes.
Data Extraction
The “centroid” approach has been previously described [12]. Briefly, the percent deuterium incorporation (%Dcentroid) for a given protein segment is determined from the intensity weighted average m/z ratio of the isotopic peaks with an approach similar to that described by Zhang and Smith 1993[20]:
| (1) |
Where m0%, and m100% are the neutral masses of the measured intensity weighted average m/z values of the undeuterated and fully deuterated controls. The neutral mass m is obtained from the experimentally measured intensity weighted average m/z value from the peptide ion of interest. If a non deuterated control (m0%) is not acquired, m0 becomes the theoretical intensity weighted average m/z value of the peptide. To accommodate situations where a fully deuterated control is not available m100 can be determined with equation (2):
| (2) |
where n = number of amino acids in the peptide, p = the number of prolines, and z = charge. The amino acid proline has no amide hydrogen and so they are subtracted from the equation. The subtraction of “2” within equation (2) is based on results from the work of Englander [21].
It is now possible for the user to edit the percentage value for the fully deuterated control in the present version of the software. For example, this can be changed to 50% if the use of a 50% back exchange control is preferred over a 100% control. This feature allows for greater flexibility in the experimental design.
The “Theoretical fit” approach to calculation of percent deuterium remains unchanged from the previous version of the software. A comprehensive description of the method has been published. Briefly, the software uses a modified version of Qmass [22] to generate 100 theoretical isotopic envelopes for the peptide ion of interest; one at every possible %D between 0% and 100%. A chi-squared fit score is obtained for each of these 100 isotopic distributions, and the lowest chi-squared fit score is assumed to be the correct percent deuterium incorporation for the peptide ion of interest.
The software provides two independent measures of percent deuterium incorporation into each of the peptides of interest. The output from each of the two approaches to percent deuterium calculations are treated equally for all downstream calculations. First, all of the %D values are corrected for the actual percent of D2O contained within the on-exchange buffer. This is a user-defined variable, which can be adjusted for each project. Our typical value is 80%, which corresponds to 4 μL protein (in H2O buffer) diluted to 20 μL with D2O buffer of an equivalent composition.
While HDX MS coupled with proteolysis has proven to be an effective approach in the determination of %D for specific regions of a protein, one inherent limitation of the technology is that the measured HDX rate represents the average value of all the exchangeable amides within a specific peptic peptide analyzed. Ideally it is desirable to obtain exchange data at the single amide level. In addition, the presentation of HDX data often requires the consolidation of overlapping peptide HDX data, which present time consuming data processing issues. For example, to represent HDX data over a three dimensional structure, overlapping peptides must be manually combined, or discarded, such that only a single measured HDX percentage remains for each amide. To address these issues, we have developed an automated method to consolidate and present HDX data with the greatest possible amide resolution.
While perfectly overlapping peptide regions that share both the start and the end residue (defined here as “twin” peptides) are simple to consolidate, the rendering of deuterium incorporation results from overlapping peptides of varying lengths is less straightforward, and specific rules need to be followed. The goal of our “Consolidation” approach was to render deuterium incorporation results with the highest amide resolution possible, while preserving measured data and accounting for error.
For overlapping peptide regions that share the same start or end position (referred to here as sibling peptides), the accepted approach [23, 24] is to assume that the non-overlapping segment would contain the difference between the two measured %D values. This approach will create theoretical peptides and make it possible to obtain more localized HDX data, sometimes resolved to a single amide. Taking this further, we have devised the first fully automated method for the determination of highly localized %D results.
First the software identifies “twin” peptides with the same start and end positions and generates a single consolidated peptide for the region. Each consolidated “twin” is comprised of the average %D values from all the contributors. At this stage in the process the first two residues are truncated [21]. Once this is accomplished, all peptides are examined for the presence of sibling peptides that share either start or end positions. For each pair of sibling peptides a theoretical peptide is generated representing the non-overlapping portion of the longer peptide. The %D value assigned to this theoretical segment becomes the difference between the two peptides.
Once all of the possible theoretical peptides have been generated and added to the consolidated peptide list, the software identifies any two sequential peptides that span the positional range of a single larger peptide. When this occurs, the larger peptide is discarded from the list. The next step is to iterate through the list to identify groups of overlapping peptides. For each group, the largest peptide is preserved, as well as any peptides from the group that do not overlap with this peptide. In cases where there are groups of peptides that do not overlap with the largest peptide, the largest from that group is preserved. By doing this, the smallest protein segments with known %D values are preserved, allowing for more localized HDX data (Figure 4).
Figure 4. HDX Data Consolidation.
To generate a condensed data set for display over a three dimensional structure we have automated the following process. 1) Peptides with shared start and end positions (twin peptides) are identified, averaged and consolidated (peptides A and B). 2) The remaining peptides are examined for “sibling peptides” which share either start or end position (peptides C and D), and a theoretical peptide is created from the non-overlapping portion (D–C). This theoretical peptide is assigned the difference between the %D values. 3) The process is repeated with all non consolidated peptides (real and theoretical). 4) In cases where the sum of two (or more) peptides equals another peptide, the larger peptide is removed from the list. 5) Peptides that do not meet the previous criteria “overlapping peptides” are either discarded (peptide E) or preserved if they do not overlap with the previously consolidated group (peptide F). 6) The “condensed data” illustrates the result of the process.
Since the standard deviations of all measured %D values are retained within the database, the standard deviation of the %D values within the theoretical peptides are calculated with the root sum square approach [25].
Data Display
The software contains five new web based user interfaces that mimic desktop applications. The reuse of common interface actions, such as the tree node selection, across each view maintains a consistent design. Three of these five interfaces contain rendering components to view the HDX data extracted from the underlying mass spectra. These three interfaces are the “Peptide view”, “Datapoint view” and “Perturbation view”. Aggregate statistics are calculated on the fly, ensuring the software presents the most current data.
The three primary interfaces share common tree and grid components to represent the data. Some of the components of the grid include the ability to sort, hide and move columns, as well as edit data directly within the cells. Another shared component is the tree view, which facilitates data navigation.
The calculated x-y spectral data obtained during the preprocessing operation are now stored within an indexed database (our previous software required that this x-y data be re-calculated during each display step). By removing the need for the calculation step when viewing the results we have significantly reduced the time required to render each isotopic cluster. This “load” time has dropped from approximately seven seconds to two seconds. Another benefit of the database backend is the ability to tightly integrate with outside systems. In our laboratory the integration with our Laboratory Information Management System (LIMs) would not have been possible in its absence.
HDX MS data
HDX MS data were acquired with a HTS PAL liquid handling robot (LEAP technologies, Carrboro, NC) interfaced with a linear ion trap mass spectrometer (Thermo Fisher, San Jose, CA). All experiments were performed as described previously [15, 26]. The test data set was acquired with a 10 μM stock solution of soluble recombinant human TNFα.
Results and Discussion
We have developed an interactive web based application for the analysis and display of HDX MS data. The four main user interfaces feature a collapsible bar that gives access to a tree view, which allows the users to easily drill down to items of interest. New features also include the ability to discard peptides, addition of a field containing the last modified date, chemical formula, calculated percent deuterium (from the “centroid” method) and theoretical fit percent deuterium. Each of these columns can be displayed, or hidden, from the view depending upon the user preference. Since these columns access data fields stored within the database, no data is lost by the removal of these columns from the user interface. We have evaluated the software with a test data set and 21 additional “live” projects. In total, these 22 projects contain over 1,100 LC MS datafiles and 35,000 data points. No problems with the stability of the software or the database have occurred.
Preprocessing
The software requires as input a list of candidate peptides, the associated protein sequence file in fasta format, and a series of MS data files in mzXML format. These files must be processed before the results can be presented to the user within the software interface. The test data set was comprised of two samples each containing 30 peptides of interest. Six on-exchange time points and the H2O control were acquired in triplicate for each sample, generating 42 LC MS files and 1440 data points. During the preprocessing steps, the database was populated with the “reference data” and “result data” described previously. To process our test data set and populate the database required approximately eight hours of processor time with a Sunfire v40z computer with four core 2.4 GHz processors and 16 GB RAM.
To achieve the greatest accuracy with each of the data extraction calculations (“centroid” and “theoretical fit”) the peptide ion isotopic distribution must be measured at the highest possible (S/N) ratio. This requires that the chromatography retention time limits are accurately defined. When a target RT value is provided within the peptide set the RT determination algorithm operates ± one minute from the target RT. The one minute value was determined with Mascot (Matrix Science, London, UK) search results from 15 liquid chromatography tandem mass spectrometry (LC-MS/MS) data sets containing over 800 peptide identifications with a peptide score > 30. In all cases the difference between the Mascot value for RT and the manually validated optimal RT value was < 1 min. In cases where the peptide ion was detected with good signal to noise ratio (~> 20), the RT determination was accurate 96% of the time with no additional curation required. While this increases the time in the preprocess phase, it dramatically reduces the time it takes the end user to curate the data.
Peptide view
The “Peptide view” provides a robust application to calculate and validate the “centroid” and “theoretical fit” percent deuterium values for a given peptide ion; %Dcent, and %Dtheo, respectively, and presents the reference and result data in a unified grid. Figure 5 shows a screen shot of the Peptide view, which covers much of the functionality present in the previous version of the software, and includes the ability to inspect and adjust the co-added m/z spectral data, and observe the determination of deuterium content by overlaying theoretical fit distributions onto observed data. A separate rendering of extracted ion chromatogram data is provided to ensure that the greatest possible (S/N) ratio for the isotopic distribution is achieved. New features include a dynamic grid component and significantly reduced loading times for the pre-calculated m/z and chromatographic data. Each isotope cluster and associated extracted ion chromatogram are now displayed in less than two seconds, down from approximately seven seconds.
Figure 5. Peptide View.
H/D exchange data are presented in the grid view, with each row representing a peptide. Selection of a peptide loads the associated data into the main spectral and extracted ion viewers. Tree navigation facilitates browsing to replicates; spectral and extracted ion data are presented in lower pane; measured isotopic envelope is shown in red. While the above example presents high-resolution spectral data, other resolutions are supported within the same interface. For high-resolution data, correct peak data is extracted from calculated sub-ranges only, displayed as yellow bars.
Datapoint view
The “Datapoint view”, illustrated in Figure 6 (left), presents one row for each peptide that is comprised of a series of columns containing the relevant %D values calculated from each of the datapoints. These columns contain the average %D for the peptides across all replicates. For each peptide, the mean %D value is color coded within a heat map grid, and selection of a peptide will result in a plotted deuterium uptake curve in the lower panel of the interface. The mean and standard deviation of each value are represented in these plots. The software allows for the automated rendering of data onto the three-dimensional protein structure, which is presented within a fully functional Jmol applet. It is often the case that the protein data bank structure file[27] (*.pdb) does not contain the identical protein sequence to that contained within the users fasta file. For this reason, the application provides an offset field that is accessible through the project setup pages. The offset provides for the manual alignment of the sequences within the fasta and pdb files.
Figure 6. Datapoint/Perturbation Views.
The Datapoint View (left), presents a heat map grid view with the %D results for all datapoints. Selection of a row in this view displays the deuterium uptake curves for the selected peptide for each datapoint. In the Perturbation View (right), for each peptide, perturbation data is presented in a color-coded grid. A number of automated rendering methods are available both of these views.
Perturbation view
The “Perturbation view” shown in Figure 6 (right) produces perturbation data in grid form and represents the difference in HDX data between two samples. The data are presented in a heat map grid and the selection of a peptide in this view produces a deuterium buildup graph for both samples that contributed to the change. The two-dimensional plot in the Perturbation view displays the positive and negative perturbation data in a bar chart format across the entire protein sequence with consolidated peptide data, presenting a simplified means for viewing peptide coverage and regions with significant exchange rate differences. As for the Datapoint view, the rendering of aggregate results onto the 3D protein structure has been automated.
Synchronization view
The synchronization interface has been enhanced from the previous version of the software. The synchronization tool will now only process data that have differences in the m/z and retention time ranges, making it much quicker to process especially in cases where most of the peptide data remains unchanged. The retention time determination functionality is also available through this interface. It is possible to use this tool across projects, provided the peptide set is the same. The navigation within this application is tree node driven, simplifying its use.
Project setup
The project setup interface provides a series of web pages that allow users to interact directly with the backend database. Projects can be created, deleted and altered from this portion of the application. Variables that need to be altered less frequently than those in the main interface are made available here. Examples of available variables experiment name, file paths and estimated rate of deuterium recovery during the LC MS experiment. Corrections for back exchange vary between experiments and labs due to factors such as sample temperature and pH variations for on-exchange stage of the experiment in samples. To account for this in the automated calculations, a value for estimated recovery value can be set through this interface. All calculations and interface components retrieve values from the database, in an effort to accommodate various users needs and approaches.
Conclusions
The means by which the candidate peptide list is determined is inconsequential to the software. In practice we have used various approaches such as combining multiple MS/MS search results to determine the peptide set. In our experience we have found that for the most part a single search result set is often sufficient. Regarding overlapping peptides; the existing spectral ranges approach described previously has been proven an effective method for the software to disregard non-peak data in situations when the instrument resolution is sufficiently to resolve the isotopic distributions. The ultimate goal of this software development program was twofold; 1) Automate all processes and use algorithms to reduce the need for manual data curation to less than 5% of all peptides present with (S/N) ratios greater than five, and 2) Provide an application flexible enough to accommodate different HDX MS experiments and data from different instrument manufacturers operating at varying resolving powers.
HD Desktop is a unique suite of software with direct relevance to the HDX community. The software provides a means of rapidly determining deuterium incorporation from HDX experiments, and aims to address all facets of the necessary analysis tasks. The combination of performance gained due to the database storage of pre-calculated m/z data, retention time determination in the preprocess step and automated data rendering, provides a robust application, with the ability to display correct HDX rate data with minimal manual intervention. The storage of all data within the database enables users to navigate between the data curation in the peptide view and the rendering components, accelerating the time it takes to get to the end results. Visualization tools provide users with additional options to inspect the data.
The benefits of our previous software, the Deuterator, have been leveraged and enhanced, and continue to address the needs of the broader community of users. The user base for the previous version of the software has grown substantially from the time of its inception, and we expect this to grow further as the current version is made available. HD Desktop is available as web based application for non-commercial purposes at http://hdx.florida.scripps.edu and will eventually be offered as an open source solution.
Supplementary Material
Figure S1. Database Schema - The database model was designed to accommodate a wide range of HDX experiments. Spectral plot and result data are stored in the database for enhanced performance.
Acknowledgments
This work was supported by The State of Florida. Recombinant human TNFα was provided by David Szymkowski (Xencor Corp.). The authors thank Jun Zhang and Scott Novick for the many hours spent testing the software.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Englander SW. Hydrogen Exchange Mass Spectrometry: A Historical Perspective. Journal of the American Society for Mass Spectrometry. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abzalimov RR, Kaltashov IA. Extraction of Local Hydrogen Exchange Data from HDX CAD MS Measurements by Deconvolution of Isotopic Distributions of Fragment Ions. Journal of the American Society for Mass Spectrometry. 2006;17(11):1543–1551. doi: 10.1016/j.jasms.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Analytical Chemistry. 2006;78(1):207–214. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 4.Dai SYaF, MC Accuracy of SUPREX (Stability of Unpurified Proteins from Rates of H/D Exchange) and MALDI Mass Spectrometry-Derived Protein Unfolding Free Energies Determined Under Non-EX2 Exchange Conditions. J Am Soc Mass Spectom. 2006;17(11):1535–1542. doi: 10.1016/j.jasms.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Zhang ZaMA. A Universal Algorithm for Fast Automated Charge State Deconvolution of Electrospray Mass-to-Charge Ratio Spectra. Journal of the American Society for Mass Spectrometry. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 6.Palmblad M, Buijs J, Hakansson P. Automatic Analysis of Hydrogen/Deuterium Exchange Mass Spectra of Peptides and Proteins Using Calculations of Isotopic Distributions. Journal of the American Society for Mass Spectrometry. 2001;12:1153–1162. doi: 10.1016/S1044-0305(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 7.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15(3):583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. Journal of Biomolecular Techniques: JBT. 2003;14(3):171–182. [PMC free article] [PubMed] [Google Scholar]
- 9.Weis DD, Hotchko M, Wales TE, Ten Eyck LF, Engen JR. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. Journal of the American Society for Mass Spectrometry. 2006;17(11):1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Weis DD, Engen JR, Kass IJ. Semi-Automated Data Processing of Hydrogen Exchange Mass Spectra Using HX-Express. Journal of the American Society for Mass Spectrometry. 2006;17(12):1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Nikamanon P, Pun E, Chou W, Koter MD, Gershon PD. “TOF2H”: a precision toolbox for rapid, high density/high coverage hydrogen-deuterium exchange mass spectrometry via an LC-MALDI approach, covering the data pipeline from spectral acquisition to HDX rate analysis. BMC Bioinformatics. 2008;9:387. doi: 10.1186/1471-2105-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nature Biotechnology. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 14.Quint P, Ayala I, Busby SA, Chalmers MJ, Griffin PR, Rocca J, Nick HS, Silverman DN. Structural mobility in Human manganese superoxide dismutase. Biochemistry. 2006;45:8209–8215. doi: 10.1021/bi0606288. [DOI] [PubMed] [Google Scholar]
- 15.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J Biomol Tech. 2007;18(4):194–204. [PMC free article] [PubMed] [Google Scholar]
- 16.Dai SY, Chalmers MJ, Bruning J, Bramlett KS, Osborne HE, Montrose-Rafizadeh C, Barr RJ, Wang Y, Wang M, Burris TP, et al. Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc Natl Acad Sci U S A. 2008;105(20):7171–7176. doi: 10.1073/pnas.0710802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasta Format.http://en.wikipedia.org/wiki/Fasta_format.
- 18.Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22(5):634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 19.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 20.Zhang ZaS, DL Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Science. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Milne JS, Englander SW. Primary Structure Effects on Peptide Group Hydrogen Exchange. Proteins: Struc Funct, Genet. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockwood AL, Haimi P. Efficient calculation of accurate masses of isotopic peaks. Journal of the American Society for Mass Spectrometry. 2006;17(3):415–419. doi: 10.1016/j.jasms.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL., Jr Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14(12):2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resing KA, Hoofnagle AN, Ahn NG. Modeling deuterium exchange behavior of ERK2 using pepsin mapping to probe secondary structure. J Am Soc Mass Spectrom. 1999;10(8):685–702. doi: 10.1016/S1044-0305(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 25.Sum Of Squares.http://en.wikipedia.org/wiki/Sum_of_squares.
- 26.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Analytical Chemistry. 2006;78(4):1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 27.The Protein Data Bankhttp://www.rcsb.org/pdb/home/home.do.
- 28.Grails.http://www.jmol.org/.
- 29.MySQLhttp://www.mysql.com/.
- 30.The Sashimi Project.[http://sashimi.sourceforge.net].
- 31.Jmol.http://www.jmol.org/.
- 32.Pymol.[http://pymol.sourceforge.net].
- 33.Gilbert D, Morgner T. The JFreeChart Library. 2005 [ http://www.jfree.org/jfreechart]
- 34.Firefox.[www.mozilla.com/firefox].
- 35.Ext JShttp://extjs.com/.
- 36.Hibernate.[http://www.hibernate.org].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Database Schema - The database model was designed to accommodate a wide range of HDX experiments. Spectral plot and result data are stored in the database for enhanced performance.