Abstract
Objective
GH insensitivity (GHI) is caused in the majority of cases by impaired function of the GH receptor (GHR). All but one known GHR mutation are in the coding sequence or the exon/intron boundaries. We identified and characterised the first intronic defect occurring in the polypyrimidine tract of the GHR in a patient with severe GHI.
Design
We investigated the effect of the novel defect on mRNA splicing using an in vitro splicing assay and a cell transfection system.
Methods
GHR was analysed by direct sequencing. To assess the effect of the novel defect, two heterologous minigenes (wild-type and mutant L1-GHR8-L2) were generated by inserting GHR exon 8 and its flanking wild-type or mutant intronic sequences into a well-characterised splicing reporter (Adml-par L1–L2). 32P-labelled pre-mRNA was generated from the two constructs and incubated in HeLa nuclear extracts or HEK293 cells.
Results
Sequencing of the GHR revealed a novel homozygous defect in the polypyrimidine tract of intron 7 (IVS7-6T>A). This base change does not involve the highly conserved splice site sequences, and is not predicted in silico to affect GHR mRNA splicing. Nevertheless, skipping of exon 8 from the mutant L1-GHR8-L2 mRNA was clearly demonstrated in the in vitro splicing assay and in transfected HEK293 cells.
Conclusion
Disruption of the GHR polypyrimidine tract causes aberrant mRNA splicing leading to a mutant GHR protein. This is predicted to lack its transmembrane and intracellular domains and, thus, be incapable of transducing a GH signal.
Introduction
Primary GH insensitivity (GHI) is a rare inherited disorder characterised by severe postnatal growth failure, normal or increased GH secretion and insulin-like growth factor 1 (IGF1) deficiency. In the majority of GHI patients, a genetic defect in the GH receptor (GHR) gene leading to a functionless receptor is present (1). Approximately 20% of these defects alter the mechanism by which GHR coding exons are defined and correctly assembled to form the mature mRNA (2), a process known as mRNA splicing (3).
Defects affecting the efficiency of mRNA splicing comprise one-half of DNA point mutations responsible for human genetic disease (3, 4). The majority of splice mutations disrupt the native splice site through a base change within the invariant donor or acceptor dinucleotides (5). Mutations within other splice elements, such as the polypyrimidine tract and the branch point, may also cause genetic diseases through the exclusion of a constitutive exon from the mature mRNA (6), but these are less frequent.
The vast majority of GHR splice defects identified so far disrupt the invariant dinucleotide at the splice sites leading to aberrant mRNA splicing and a mutant GHR protein. We report the first mutation identified in the polypyrimidine tract of the GHR causing aberrant GHR mRNA splicing and GHI.
Materials and methods
Molecular analysis
A patient was referred for severe short stature. Informed consent for genetic analysis was obtained from his parents, and approval was obtained from the local ethics committee.
Genomic DNA was extracted from peripheral blood leucocytes. GHR coding exons, including the pseudoexon 6Ψ, and their intronic boundaries were amplified by PCR using specific primers (primer sequences available on request). PCR products were visualised on 1% agarose gel and were sequenced using the ABI Prism Big Dye Sequencing kit and the ABI 3700 automated DNA sequencer (Applied Biosystems, Warrington, UK) in accordance with the manufacturer's instructions.
Creation of minigenes
The wild-type minigene L1-GHR8-L2 was created by inserting the GHR exon 8 and its intronic boundaries between exons L1 and L2 of a well-characterised splice reporter Adml-par (7). The GHR exon 8 and 89 bp of its flanking introns were amplified from human genomic DNA, and exons L1 and L2 were amplified from Adml-par by PCR using specific primers (sequences available on request). The three exons were joined together by overlap extension PCR (8). Adml-par was amplified using primers T7-L1 and L2A (T7L1: 5′ TAATACGACTCACTATAGGGAGACCGGCAGATCAGCTT 3′, L2A: 5′ ATCCAAGAGTACTGGAAAGACCG 3′) and was used as a positive control for the splicing reaction. PCR products were run on a 1% agarose gel and those corresponding in size to the three-exon minigene were cut and purified by PCR gel extraction. The identity of the PCR product was confirmed by direct sequencing on the ABI 3700 sequencer. PCR products were cloned in the pGEM T-easy vector system (Promega), and the presence of the insert was assessed by direct sequencing of plasmid DNA. The mutant L1-GHR8-L2 minigene was obtained by site-directed mutagenesis using specific primers (sequences available on request) and the wild-type minigene as a template.
RNA preparation
Wild-type and mutant DNA minigenes and Adml-par were transcribed into RNA in the presence of [32P-α]GTP. Transcription reactions contained 200 ng DNA, 1 μl 10× transcription buffer (Ambion, Warrington, UK), 1 μl NTPs (5 mmol ATP, CTP and UTP), 10 μCi [32P-α]GTP (Perkin-Elmer, Massachusetts, USA), RNA CAP and 20 U/μl T7 RNA polymerase Plus (Ambion) in a final volume of 10 μl. Reactions mixtures were incubated for 1 h at 37 °C, run on a 4% polyacrylamide gel and gel purified. RNA was eluted from the gel (elution buffer: 0.5 M sodium acetate, pH 5.2, 1 mM EDTA and 0.2% SDS), ethanol precipitated and resuspended in RNAse-free H2O.
In vitro splicing assay
A splice reaction mixture containing 20 fmol RNA, 8 μl HeLa nuclear extracts (CilBiotech, s.a., Mons, Belgium), 1 μl 25× ATP/CP (12.5 mM ATP and 0.5 M creatine phosphate), 1 μl 80 mM MgCl2, 5 μl 13% polyvinyl alcohol, 1.25 μl 0.4 M Hepes-KOH (pH 7.3), 7 μl Buffer D (20 mM Hepes-KOH, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM phenylmethylsulphonyl fluoride and 1 mM dithiothreitol) in 25 μl final volume was incubated at 30 °C for 1 h. Control reaction mixtures were kept on ice for the same time. At the end of the incubation, reaction mixtures were deproteinised, precipitated and run on an 8% denaturing polyacrylamide gel before autoradiography. The bands of interest, corresponding in size to correctly spliced or aberrant products, were excised from the gel and retro-transcribed into cDNA. This was amplified by RT-PCR using primers T7L1 and L2A and products analysed by direct sequencing on the ABI 3700 DNA sequencer.
Cell transfection and RT-PCR
Wild-type and mutant minigenes L1-GHR8-L2 were subcloned from the bacterial pGEM T-easy vector into the mammalian pcDNA 3.1 vector. The identity and correct orientation of the pcDNA3.1 L1-GHR8-L2 insert were assessed by direct sequencing on the ABI 3700 sequencer. HEK293 cells were maintained in DMEM (Sigma–Aldrich) with 10% foetal bovine serum (Sigma–Aldrich) at 37 °C under 5% CO2 and split when confluent. Before transfection, HEK293 cells were seeded into a six-well plate. Cells were transiently transfected when ∼70% confluent with the wild-type or mutant pcDNA3.1 L1-GHR8-L2 plasmid (50 ng) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were harvested, and RNA was extracted and reverse transcribed into cDNA. cDNA was amplified in the presence of 0.5 μCi [32P-α] dCTP with primers T7L1 and L2A in a 12.5 μl PCR mixture. PCR products were electrophoresed on an 8% non-denaturating polyacrylamide gel prior to autoradiography and quantitation on a PhosphorImager (Molecular Dynamics Ltd, Chesham, Bucks, UK). The percentage of alternative splicing was calculated as the ratio of isoform/total of all isoforms, and results are presented as mean±s.d. of at least three separate experiments.
Hormone assay
Serum IGF1, IGF-binding protein 3 (IGFBP3), acid-labile subunit (ALS) and GH-binding protein (GHBP) were measured from venous blood samples using enzyme-linked immunosorbent assays (ELISA kit; Diagnostic System Laboratories, Inc., Webster, TX, USA). For IGF1, the assay sensitivity was 0.03 ng/ml. The intra- and inter-assay coefficients of variation (CV) were 8.6 and 6.8% for mean serum concentrations of 104 and 90 ng/ml respectively. For IGFBP3, assay sensitivity was 0.04 ng/ml. Mean intra- and inter-assay CV were 7.2 and 8.3% respectively. Serum GHBP was measured by the high pressure liquid chromatography–gel filtration method (9). GHBP is given as a percentage of specific binding, calculated as the difference between total binding and non-specific binding.
In silico analysis
The novel nucleotide change was studied using the Alex Dong Li's splice site finder (http://violin.genet.sickkids.on.ca/∼ali/splicesitefinder.html), which is an in silico prediction program that calculates the scores of donor, acceptor and branch sequences using an algorithm based on that created by Shapiro & Senapathy (10).
Results
Biochemical and auxological data
A 1.5-year-old boy of Bangladeshi origin from a consanguineous marriage was referred for severe short stature (−6.0 SDS for age and sex). He had facial features typical of Laron syndrome, with a prominent forehead and a depressed nasal bridge. Biochemical data showed low IGF1=30 ng/ml (n.r. 8–141 ng/ml) and IGFBP3=0.35 mg/l (n.r. 1.1–3.8 mg/l). GH levels were markedly elevated at baseline, which were equal to 1145 mUI/l and after stimulation test with glucagon were equal to 1195 mUI/l. Serum GHBP levels were 29% (normal range 15–20%). His parents' heights were −1.58 SDS (father) and −1.55 SDS (mother).
Genetic analysis
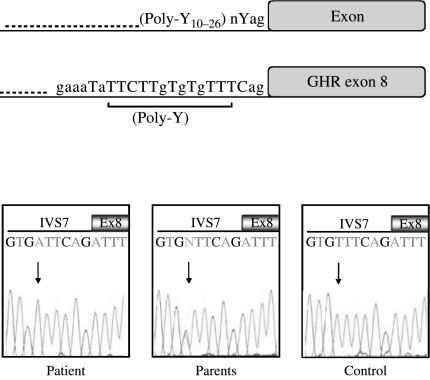
Sequencing of GHR revealed the presence of a homozygous T to A base change six bases upstream the acceptor splice site of intron 7 (IVS7-6T>A). In both parents, the same mutation was present in heterozygosity (Fig. 1). No other mutations were found in the GHR coding sequence, intronic boundaries and pseudoexon 6Ψ.
Figure 1.
GHR polypyrimidine tract. (A) Schematic representation of the typical eukaryotic intronic nucleotide sequence upstream the intron/exon splice junction (top) and the nucleotide composition of the intronic region upstream GHR exon 8 (bottom). Pyrimidines are shown in uppercase letters; position and length of the polypyrimidine tract (Poly-Y) are also indicated. (B) IVS7-6T>A. Chromatograms showing partial DNA sequences for the patient, his parents and a normal control are presented. The position of the mutation is indicated by the arrows. Y, pyrimidines; n, any nucleotide.
The T to A intronic defect does not abolish the GHR intron 7 acceptor splice site in silico
The thymine located six bases upstream the acceptor splice site of intron 7 is a conserved nucleotide (www.ensembl.org). The T to A base change was predicted, in silico, to cause a modest reduction of intron 7 acceptor splice site score (wild-type 85.57 versus mutant 82.80).
The T to A intronic defect causes GHR exon 8 skipping in vitro
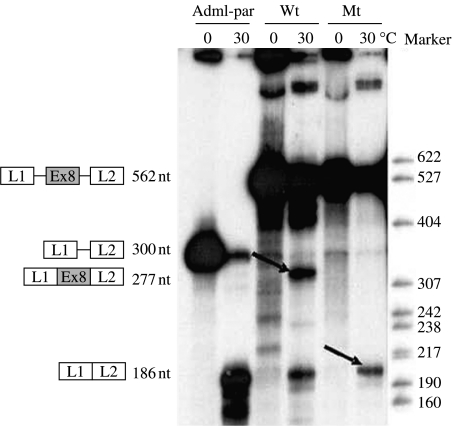
The wild-type minigene L1-GHRexon8-L2 and the corresponding mutant were created to study the nucleotide change IVS7-6T>A, which is located in the polypyrimidine tract before exon 8 and tested with the in vitro splicing assay. After 1 h under standard splicing conditions, the wild-type minigene produced a band of 277nt corresponding to the three-exon correctly spliced product L1-GHRexon8-L2 (54±2% of exon 8 inclusion) alongside a 186nt band corresponding to L1–L2 mRNA, the identity of which was confirmed by direct sequencing. The T>A mutation resulted in the complete skipping of GHR exon 8 and the appearance of the 186nt band, corresponding to exons L1 and L2 joined together, as confirmed by direct sequencing. No band of 277nt corresponding to L1-GHRexon8-L2 was present among the mutant minigene splice products (Fig. 2).
Figure 2.
Results for the in vitro splicing assay. The wild-type (Wt) and mutant (Mt) minigenes L1-GHRexon8-L2 and Adml-par were incubated in HeLa nuclear extracts for 60 min at 30 °C. Control reaction mixtures were incubated at 0 °C. The structures of the pre-mRNA and the mRNA splice products are indicated next to the autoradiogram. The expected size of the splice products is also indicated. Bands corresponding to correct and aberrant mRNA splice products are indicated by the arrows.
The T to A intronic defect causes GHR exon 8 skipping in HEK293 cells
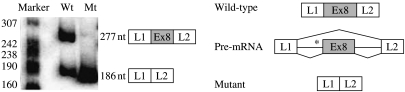
In order to confirm the effect of the T to A mutation on mRNA splicing, the wild-type minigene L1-GHRexon8-L2 and the corresponding mutant were subcloned in the mammalian pcDNA3.1 vector and transfected into HEK293 cells. After 48 h, the wild-type minigene produced a band of 277nt, corresponding to L1-GHRexon8-L2, alongside the 186nt band corresponding to L1–L2. The IVS7-6 T to A mutation resulted, instead, in the complete skipping of GHR exon 8 and the appearance of the 186nt band, corresponding to exons L1 and L2 joined together, as confirmed by direct sequencing (Fig. 3).
Figure 3.
Effects of the GHR T to A nucleotide change on mRNA splicing in HEK293 cells. (A) Radioactive RT-PCR analysis performed with mRNA from HEK293 cells transfected with the wild-type or mutant minigene L1-GHRexon8-L2. The structure of the correct (L1-Ex8-L2) and aberrant (L1–L2) splice products and their expected sizes are indicated. (B) Schematic representation of the splice events for the wild-type and mutant minigenes. Mutation location is indicated by an asterisk.
Discussion
This study describes the first mutation within the polypyrimidine tract of the GHR in a patient with GHI. This homozygous defect causes aberrant mRNA splicing, resulting in the skipping of GHR exon 8 and a prematurely truncated GHR protein.
Approximately 20% of GHR defects causing GHI are splice mutations (2). All defects disrupt the invariant splice site sequences, with rare exceptions involving the activation of cryptic intronic (11) or exonic splice sites (12). The splicing process involves the recognition of splice elements by a ribonucleoprotein complex called the spliceosome (13). The polypyrimidine tract is an intronic sequence rich in pyrimidines (14–16). It is located upstream of the acceptor splice site, and has been shown to be involved in the initial stages of spliceosome binding to the mRNA (17). Mutational studies have demonstrated that the length and location of the polypyrimidine sequence can affect spliceosome apposition and, thus, splicing efficiency (18).
The intronic base change identified and characterised in this study is located upstream of the acceptor splice site of GHR intron 7 in the polypyrimidine tract. Mutations within this splice element are known to contribute to genetic diseases by inducing the exclusion of a constitutive exon or inclusion of an alternative one in the mature mRNA (5, 19). Nevertheless, mutations in the polypyrimidine tract are a rare finding, possibly because of the difficulty in predicting their deleterious effect. In silico prediction programs are, in fact, fairly reliable when used to predict the effects of nucleotide changes occurring at the invariant donor and acceptor dinucleotide sequences, but become less efficient as nucleotide changes occur further away from the splice sites or in different splicing elements, such as the polypyrimidine tract (20).
The T>A nucleotide substitution identified in homozygosity in this GHI patient and in heterozygosity in his parents shortens the polypyrimidine tract of GHR intron 7. In silico analysis did not predict the abolition of the acceptor splice site, but only a modest reduction in its score, whose significance was unclear. As mRNA from the patient was not available, the effect of this novel GHR intronic nucleotide change was investigated by comparing mRNA splicing of the mutant and the wild-type minigenes in HeLa nuclear extract and in HEK293 cells. A clear GHR exon skipping was demonstrated for the mutant mRNA in both systems. The thymine substitution is likely to affect GHR splicing by interfering with the binding of the U2AF spliceosome element to the GHR mRNA. The resulting mutant GHR transcript will lack exon 8, with exon 7 splicing into exon 9, as was the case for the exon 8 splice site mutation described by Woods et al., which led to an exon 8-lacking mutant GHR (21). The skipping of exon 8 causes a frameshift and the appearance of a premature stop codon.
The presence of circulating GHBP in our patient is evidence that the mutant GHR is expressed. Moreover, elevated GHBP levels suggest that the aberrant splicing caused by the polypyrimidine tract defect and documented in the in vitro experiment also occurs in vivo. GHBP is the product of the cleavage of the GHR and corresponds to its extracellular domain (22). The prematurely truncated receptor lacks its transmembrane and intracellular domains and would be unable to signal or be anchored in the cell membrane, thus explaining the elevated GHBP levels seen in our patient, as well as in previously described cases of mutant GHR arising from exon 8 splice site defects (21, 23). The characteristics of the GHBP arising from this mutant GHR were described by Woods et al., who demonstrated a normal GH binding affinity and a molecular weight similar to that of normal controls (21). Analysis of GHBP characteristics in our patient would be expected to produce similar results and was, thus, not performed.
It is of interest that although the truncated mutant GHR is not able to signal, the patient had detectable IGF1 levels, albeit very low, especially in relation to the extremely elevated GH levels. The production of small amounts of IGF1 in patients with exon 8-lacking GHR has also been documented by other authors (22). In fact, splice defects may not always be 100% efficient in causing aberrant splicing (24–26), and very low amounts of wild-type GHR may be produced and could be responsible for the low IGF1 levels seen in our patient.
The patient's parents, heterozygous carriers of the polypyrimidine tract defect, are likely to express the mutant GHR. Although no mRNA was available from the parents of our patient, other authors have demonstrated the presence of mRNA for the exon 8-lacking mutant receptor in heterozygous carriers of exon 8 splice mutations (21). The polypyrimidine tract defect did not cause short stature or Laron features in heterozygosity, as was true for other exon 8-skipping mutations (23). However, elevated GHBP levels have been described in heterozygous carriers (23) and may be present in the parents of our patient, but could not be demonstrated as blood was not available.
GHR exon 8 skipping was observed in the presence of the wild-type polypyrimidine tract in both in vitro and cellular studies. It could be postulated that the GHR exon 8 splice site is constitutively weak, and that the shortening of its polypyrimidine tract, as in the presence of the novel T to A nucleotide change, favours its skipping.
In conclusion, we have described a novel rare cause of GHI resulting from the alteration of the GHR polypyrimidine tract. This nucleotide change, located outside the invariant splice site, was not predicted to abolish GHR splicing in silico, but was clearly shown in vitro to induce skipping of GHR exon 8. This strengthens the concept that the search for gene mutations should include careful analysis of non-coding sequences and, in particular, of intronic splice elements.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this scientific work.
Funding
This work has been supported by the Barts and the London Charitable Foundation (studentship to A David) and the Wellcome Trust (Research Grant 076430 to A J L Clark and a VIP award to A David).
Footnotes
(A J L Clark and L A Metherell contributed equally to this work)
References
- Burren CP, Woods KA, Rose SJ, Tauber M, Price DA, Heinrich U, Gilli G, Razzaghy-Azar M, Al-Ashwal A, Crock PA, Rochiccioli P, Yordam N, Ranke MB, Chatelain PG, Preece MA, Rosenfeld RG, Savage MO. Clinical and endocrine characteristics in atypical and classical growth hormone insensitivity syndrome. Hormone Research. 2001;55:125–130. doi: 10.1159/000049983. [DOI] [PubMed] [Google Scholar]
- David A, Metherell LA, Clark AJ, Camacho-Hubner C, Savage MO. Diagnostic and therapeutic advances in growth hormone insensitivity. Endocrinology and Metabolism Clinics of North America. 2005;34:581–595. doi: 10.1016/j.ecl.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Letters. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Human Genetics. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- De Klein A, Riegman PH, Bijlsma EK, Heldoorn A, Muijtjens M, Den Bakker MA, Avezaat CJ, Zwarthoff EC. A G→A transition creates a branch point sequence and activation of a cryptic exon, resulting in the hereditary disorder neurofibromatosis 2. Human Molecular Genetics. 1998;7:393–398. doi: 10.1093/hmg/7.3.393. [DOI] [PubMed] [Google Scholar]
- Anderson K, Moore MJ. Bimolecular ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Amselem S, Duquesnoy P, Attree O, Novelli G, Bousnina S, Postel-Vinay MC, Goossens M. Laron dwarfism and mutations of the growth hormone-receptor gene. New England Journal of Medicine. 1989;12:989–995. doi: 10.1056/NEJM198910123211501. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Research. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metherell LA, Akker SA, Munroe PB, Rose SJ, Caulfield M, Savage MO, Chew SL, Clark AJ. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. American Journal of Human Genetics. 2001;69:641–646. doi: 10.1086/323266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach L, Schiavi A, Bartlett R, Perera E, Day J, Brown MR, Stein S, Eidson M, Parks JS, Cleveland W. Clinical, biochemical, and molecular investigations of a genetic isolate of growth hormone insensitivity (Laron's syndrome) Journal of Clinical Endocrinology and Metabolism. 1997;82:444–451. doi: 10.1210/jcem.82.2.3784. [DOI] [PubMed] [Google Scholar]
- Wachtel C, Manley JL. Splicing of mRNA precursors: the role of RNAs and proteins in catalysis. Molecular BioSystems. 2009;54:311–316. doi: 10.1039/b820828j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends in Biochemical Sciences. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes and Development. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Hodges PE, Beggs JD. RNA splicing. U2 fulfils a commitment. Current Biology. 1994;1:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- Roscigno RF, Weiner M, Garcia-Blanco MA. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. Journal of Biological Chemistry. 1993;25:11222–11229. [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Reviews. Genetics. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Clark F. Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Research. 2001;29:2581–2593. doi: 10.1093/nar/29.12.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods KA, Fraser NC, Postel-Vinay MC, Savage MO, Clark AJ. A homozygous splice site mutation affecting the intracellular domain of the growth hormone (GH) receptor resulting in Laron syndrome with elevated GH-binding protein. Journal of Clinical Endocrinology and Metabolism. 1996;81:1686–1690. doi: 10.1210/jcem.81.5.8626815. [DOI] [PubMed] [Google Scholar]
- Conte F, Salles JP, Raynal P, Fernandez L, Molinas C, Tauber M, Bieth E. Identification of a region critical for proteolysis of the human growth hormone receptor. Biochemical and Biophysical Research Communications. 2002;290:851–857. doi: 10.1006/bbrc.2001.6261. [DOI] [PubMed] [Google Scholar]
- Silbergeld A, Dastot F, Klinger B, Kanety H, Eshet R, Amselem S, Laron Z. Intronic mutation in the growth hormone (GH) receptor gene from a girl with Laron syndrome and extremely high serum GH binding protein: extended phenotypic study in a very large pedigree. Journal of Pediatric Endocrinology and Metabolism. 1997;10:265–274. doi: 10.1515/jpem.1997.10.3.265. [DOI] [PubMed] [Google Scholar]
- Lemahieu V, Gastier JM, Francke U. Novel mutations in the Wiskott–Aldrich syndrome protein gene and their effects on transcriptional, translational, and clinical phenotypes. Human Mutation. 1999;14:54–66. doi: 10.1002/(SICI)1098-1004(1999)14:1<54::AID-HUMU7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Watanabe C, Liu T, Hollenbaugh D, Blaese RM, Kanner SB, Aruffo A, Ochs HD. Wiskott–Aldrich syndrome/X-linked thrombocytopenia: WASP gene mutations, protein expression, and phenotype. Blood. 1997;90:2680–2689. [PubMed] [Google Scholar]
- David A, Camacho-Hübner C, Bhangoo A, Rose SJ, Miraki-Moud F, Akker SA, Butler GE, Ten S, Clayton PE, Clark AJ, Savage MO, Metherell LA. An intronic growth hormone receptor mutation causing activation of a pseudoexon is associated with a broad spectrum of growth hormone insensitivity phenotypes. Journal of Clinical Endocrinology and Metabolism. 2007;92:655–659. doi: 10.1210/jc.2006-1527. [DOI] [PubMed] [Google Scholar]