Abstract
Purpose
As a result of the questionable risk-to-benefit ratio of adjuvant therapies, stage II melanoma is currently managed by observation because available clinicopathologic parameters cannot identify the 20% to 60% of such patients likely to develop metastatic disease. Here, we propose a multimarker molecular prognostic assay that can help triage patients at increased risk of recurrence.
Methods
Protein expression for 38 candidates relevant to melanoma oncogenesis was evaluated using the automated quantitative analysis (AQUA) method for immunofluorescence-based immunohistochemistry in formalin-fixed, paraffin-embedded specimens from a cohort of 192 primary melanomas collected during 1959 to 1994. The prognostic assay was built using a genetic algorithm and validated on an independent cohort of 246 serial primary melanomas collected from 1997 to 2004.
Results
Multiple iterations of the genetic algorithm yielded a consistent five-marker solution. A favorable prognosis was predicted by ATF2 ln(non-nuclear/nuclear AQUA score ratio) of more than –0.052, p21WAF1 nuclear compartment AQUA score of more than 12.98, p16INK4A ln(non-nuclear/nuclear AQUA score ratio) of ≤ −0.083, β-catenin total AQUA score of more than 38.68, and fibronectin total AQUA score of ≤ 57.93. Primary tumors that met at least four of these five conditions were considered a low-risk group, and those that met three or fewer conditions formed a high-risk group (log-rank P < .0001). Multivariable proportional hazards analysis adjusting for clinicopathologic parameters shows that the high-risk group has significantly reduced survival on both the discovery (hazard ratio = 2.84; 95% CI, 1.46 to 5.49; P = .002) and validation (hazard ratio = 2.72; 95% CI, 1.12 to 6.58; P = .027) cohorts.
Conclusion
This multimarker prognostic assay, an independent determinant of melanoma survival, might be beneficial in improving the selection of stage II patients for adjuvant therapy.
INTRODUCTION
Adjuvant therapy is the standard of care for many low-stage cancers that can be completely resected with tumor-free margins. However, for some other cancers, the lack of effective and safe adjuvant therapy leads to an excess of mortality directly related to the development of metastatic disease in patients assumed to have undergone a complete resection of their malignancy. One important example is cutaneous malignant melanoma, the sixth most common cancer in the United States.1 Although more than 80% of new cases are still localized to the skin1 where a wide local excision should be curative in the setting of a negative sentinel lymph node biopsy, the unfavorable risk-to-benefit ratio of available adjuvant regimens advocates caution when administering such agents to individuals with stage I to IIA and even stage IIB or IIC disease, where high-dose interferon alfa-2b is currently approved by the US Food and Drug Administration in the adjuvant setting.2 Consequently, 20% of these patients will develop metastases and die of their disease within 10 years, with more than 30% 10-year mortality among patients with T3 and T4 tumors.3 Development of a prognostic tool that could selectively triage the subset of stage II patients at high risk of recurrence for adjuvant therapy could potentially lower the burden of untreatable metastatic cancer and enable us to selectively treat those patients who are more likely to develop distant metastatic disease.
Nine clinicopathologic prognostic markers have been identified and incorporated into clinically validated outcome risk stratification models.3,4 However, these do not account for all of the observed variability associated with melanoma-related survival. Immunohistochemistry (IHC) is a widely accepted and well-documented method for characterizing patterns of protein expression in formalin-fixed, paraffin-embedded samples while preserving tissue and cellular architecture.5 Although no IHC marker has become standard of care, new work may suggest the inclusion of Ki-67.6 Our recent systematic review of melanoma IHC data shows that individual contributions of IHC markers to overall prognosis are of narrow statistical significance and thus unlikely to demonstrate broad clinical utility7 or see wide adoption.
Here, we describe the generation of an independently significant, multimarker prognostic model for melanoma using genetic algorithms on a subset of 38 candidate proteins assessed on a cohort of 192 primary melanomas. Our model shows two prognostic groups (low risk and high risk), created from five markers, that were successfully validated as significant independent prognostic factors in a second cohort of 246 primary melanomas. These data demonstrate the potential for multimarker assays in improving melanoma prognostic assessment and warrant a prospective, randomized, controlled melanoma prognostic study. This test could be a valuable tool to help determine which sentinel node–negative stage II melanoma patients should seek adjuvant therapy or other aggressive management strategies.
METHODS
Patients and Tumor Samples
Seven hundred thirty-seven tumor samples from three nonoverlapping series of patients with cutaneous melanoma were analyzed for protein expression. The Yale Melanoma Discovery Cohort consisted of 192 white patients who underwent resection of a primary invasive cutaneous melanoma at Yale-New Haven Hospital during 1959 to 1994 for whom the surgical specimen was not exhausted during diagnosis and for which follow-up information is available. The Yale Melanoma Validation Cohort included 246 patients with serial Clark level III to V cutaneous melanoma who underwent sentinel lymph node biopsy by a single surgeon during 1997 to 2004.8 The Yale Metastatic Series includes 299 unique subcutaneous metastases, lymph node metastases, or visceral metastases occurring in patients previously diagnosed with cutaneous melanoma and surgically removed at Yale-New Haven Hospital during 1959 to 1994 (n = 198) or during 1995 to 2002 (n = 101). For the primary melanomas, clinical data describing patient demographics, date of diagnosis, clinical course, and follow-up through August 1, 2007 were obtained after a comprehensive review of the medical record, the archives of the Connecticut Tumor Registry, and, if applicable, the State of Connecticut Vital Records. This study was approved by the Yale Human Investigations Committee.
Tissue Microarray Construction, IHC, and Automated Image Acquisition and Analysis
Formalin-fixed, paraffin-embedded blocks were retrieved from the Yale Pathology Archives, and 0.6-mm tissue microarrays (TMAs) were constructed according to the published method.9 The discovery TMA included single cores from the 192 primary melanomas, the 299 metastases, and a series of controls. The validation TMA included two-fold redundant cores in separate blocks from the 246 patients plus a random selection of 60 individuals from the discovery series to facilitate normalization of the validation array. Fluorescence-based immunohistochemical staining was performed by using the automated quantitative analysis (AQUA) technology as previously described.10,11 Using this method, target antigen expression is automatically determined, blinded to any a priori clinical information, as the sum of intensities from the Cy5 channel in all pixels within a compartment defined by S100 staining divided by the number of pixels within that compartment (Appendix, online only, for details).
Statistical Analysis
Cores whose tumor mask covered less than 5% of the total histospot area were dropped from further analysis. For individuals represented by multiple cores on the TMA, AQUA scores were averaged before analysis. To normalize the AQUA scores between the discovery and validation cohorts, a regression equation was calculated for the set of 60 samples spotted on both arrays, and the mean values for the validation cohort were adjusted according to the regression equation.
To develop a multimarker prognostic model from the discovery cohort data, a genetic algorithm using standard methodology12,13 within the X-tile software suite14 was used (see Appendix). Bivariate and survival analyses were performed using SAS version 9.1.3 and Statview 5.0 (SAS Institute, Cary, NC), and adjustments for multiple comparisons were performed using the standard Bonferroni method.
RESULTS
Patient Characteristics
The distribution of demographic and clinicopathologic characteristics for both the discovery and validation cohorts is presented in Table 1. In addition to the longer follow-up time (P < .0001), the discovery cohort displayed overall thicker tumors (P = .01), a more balanced sex distribution (P = .04), a higher prevalence of ulcerated melanomas (P = .01), and fewer superficial spreading melanomas (P = .04) than the validation cohort.
Table 1.
Demographics and Clinical Characteristics of the Yale Melanoma Discovery and Validation Cohorts
| Characteristic | Discovery Cohort (n = 192) |
Validation Cohort (n = 246) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Follow-up time for censored individuals, years | < .0001* | ||||
| Mean | 9.50 | 4.05 | |||
| Standard deviation | 9.14 | 2.12 | |||
| Breslow thickness, mm | .01* | ||||
| Mean | 2.42 | 1.95 | |||
| Standard deviation | 2.01 | 1.78 | |||
| Age at diagnosis, years | .34 | ||||
| Mean | 57.77 | 59.28 | |||
| Standard deviation | 15.65 | 16.76 | |||
| Sex | .04* | ||||
| Male | 96 | 50.0 | 147 | 59.8 | |
| Female | 96 | 50.0 | 99 | 40.2 | |
| Stage at diagnosis | NA | ||||
| Localized | 160 | 84.2 | 246 | 100 | |
| Regional spread | 16 | 8.4 | 0 | 0 | |
| Distant metastases | 14 | 7.4 | 0 | 0 | |
| Ulceration | .01* | ||||
| Absent | 135 | 70.3 | 198 | 80.5 | |
| Present | 57 | 29.7 | 48 | 19.5 | |
| Tumor-infiltrating lymphocytes | .09 | ||||
| Nonbrisk | 150 | 78.5 | 208 | 84.9 | |
| Brisk | 41 | 21.5 | 37 | 15.1 | |
| Histologic subtype | .04* | ||||
| Superficial spreading | 127 | 66.1 | 132 | 73.7 | |
| Nodular | 30 | 15.6 | 24 | 13.4 | |
| Lentigo maligna | 8 | 4.2 | 4 | 2.2 | |
| Acral lentiginous | 11 | 5.7 | 1 | 0.6 | |
| Other | 16 | 8.3 | 18 | 10.1 | |
| Chronically sun-exposed anatomic site† | .14 | ||||
| No | 95 | 49.7 | 105 | 42.7 | |
| Yes | 96 | 50.3 | 141 | 57.3 | |
| Received any nonsurgical therapy | .37 | ||||
| No | 153 | 79.7 | 201 | 83.1 | |
| Yes | 39 | 20.3 | 41 | 16.9 | |
| Microsatellitosis | NA | ||||
| Absent | 149 | 77.6 | 0 | 0 | |
| Present | 43 | 22.4 | 0 | 0 | |
| Positive sentinel lymph node biopsy | NA | ||||
| No | 0 | 0 | 211 | 87.6 | |
| Yes | 0 | 0 | 30 | 12.4 | |
NOTE. Numbers may not sum to total because of missing values; percentages may not sum to 100% as a result of rounding.
Abbreviation: NA, not applicable.
Significant at P < .05.
Anatomic location was dichotomized as chronically sun exposed (face, scalp, neck, arms, legs, and nonacral lentiginous lesions of hands and feet) and non–chronically exposed (chest, back, abdomen, groin, and hand and foot acral lentiginous lesions).
Clinicopathologic Correlates of Candidate Marker Expression
Thirty-eight unique protein markers were assayed by AQUA on the discovery cohort (n = 192) and, for comparison, the metastatic series (n = 299). Exclusion of individual tumors as a result of random failure for individual histospots and attrition of samples as a result of exhaustion of the arrayed tumor core resulted in less than 100% of tumor samples available for analysis from each assay. Only the subset of 20 markers with missingness completely at random was included in subsequent analyses.
Associations between levels of protein expression and tumor progression were evaluated by the Mann-Whitney U test (Appendix Table A2, online only). After adjustment for multiple comparisons, levels of fibronectin, Ki-67, and p21WAF1 and the ratios for both ATF2 and p16INK4A were significantly elevated, whereas Hey1, HDM2, N-cadherin, nuclear p16INK4A, and non-nuclear ATF2 were significantly decreased among the metastases compared with the primary tumors (P ≤ .0025).
To determine the independent crude and adjusted effects of each marker on melanoma-specific mortality, the AQUA scores or calculated ratios were divided into quartiles, and the hazard ratios and the associated P values were calculated using Cox proportional hazards modeling (Appendix Table A4, online only). Using these cut points, five markers, one that increased risk with increasing value (p16INK4A ratio, P = .04) and four that decreased risk with increasing value (ATF2, P = .001; β-catenin, P = .04; N-cadherin, P = .001; p16INK4A, P = .047), were significant at P < .05 on univariate analysis, but only two markers, ATF2 and N-cadherin, remained significant after adjustment for multiple comparisons (P ≤ .0025).
Multivariable Cox proportional hazards modeling included adjustment for age at diagnosis, sex, Breslow thickness (millimeters), stage at diagnosis, presence of microsatellitosis, sun exposure to anatomic site, and receipt of systemic therapy. Two of the five markers significant on univariate analysis (β-catenin, P = .04; p16INK4A, P = .04) retained both their significance at P < .05 and directionality of effect after adjustment for clinicopathologic parameters. Three additional markers that were not significant on crude analysis became significant at P < .05 on multivariable analysis (α-catenin, p27/KIP1, and tenascin-C).
Constructing a Genetic Algorithm–Based Multimarker Prognostic Model
Because the power of multiplexed biomarker assays is thought to be greater than that obtainable with any single marker, we sought to identify a robust prognostic indicator by combining information from all 20 available markers, regardless of whether a significant independent association with progression or prognosis was obtained, using genetic algorithms. Our selected model, obtained in each of the five independent iterations, yielded a log-rank χ2 of 24.27 (P = 1.5 × 10−6) and consisted of the following five markers and associated cut points: ATF2 ratio more than –0.052, β-catenin more than 38.68, fibronectin ≤ 57.93, p16INK4A ratio ≤ –0.083, and p21WAF1 more than 12.98.
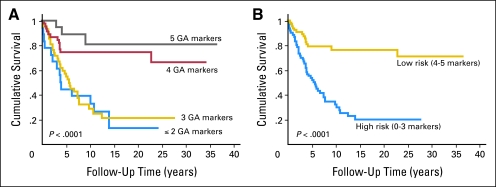
The Kaplan-Meier curves for the four classes obtained from the genetic algorithm are presented in Figure 1A. On the basis of the similar survival experiences of the groups with ≤ two or three conditions and those with four or five conditions, we further simplified our model to the following two groups: a low-risk group with four or five marker conditions being met and a high-risk group with less than four marker conditions being met (Fig 1B). Crude and multivariable survival estimates were calculated for the multimarker predictor and the clinicopathologic covariates using Cox proportional hazards modeling (Table 2). In our final multivariable model, the high-risk group demonstrated a nearly three-fold increased risk of mortality (P = .002) compared with the low-risk group. Other variables remaining significant in the multivariable model included stage at diagnosis and receipt of nonsurgical therapy (P ≤ .01), with Breslow thickness trending toward significance (P = .06).
Fig 1.
Kaplan-Meier estimates of melanoma-specific mortality among the 129 Yale Melanoma Discovery Cohort participants with complete data across the five markers comprising the genetic algorithm (GA) –based multimarker prognostic assay according to algorithm-derived prognostic score. (A) Survival curves drawn according to number of prognostic conditions met. (B) Survival curves for the dichotomized model describing low-risk (four to five conditions met) or high-risk (≤ three conditions met) groups.
Table 2.
Crude and Multivariable-Adjusted Melanoma-Specific Mortality Hazard Ratios for the Genetic Algorithm–Based Multimarker Predictor in the Yale Melanoma Discovery Cohort
| Parameter | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P* | Hazard Ratio | 95% CI | P | |
| Genetic algorithm-based predictor | < .0001† | .002† | ||||
| Low-risk group (4 or 5 conditions met) | 1.00 | 1.00 | ||||
| High-risk group (< 4 conditions met) | 3.88 | 2.16 to 6.94 | 2.84 | 1.46 to 5.49 | ||
| Breslow thickness, mm | 1.28 | 1.14 to 1.43 | < .0001† | 1.14 | 0.99 to 1.31 | .06 |
| Age at diagnosis, years | 1.01 | 0.99 to 1.03 | .41 | 1.01 | 0.99 to 1.03 | .39 |
| Sex | .14 | .14 | ||||
| Male | 1.00 | 1.00 | ||||
| Female | 0.68 | 0.41 to 1.14 | 0.66 | 0.38 to 1.14 | ||
| Stage at diagnosis | ||||||
| Localized | 1.00 | 1.00 | ||||
| Regional spread | 3.54 | 1.72 to 7.30 | .0006† | 4.67 | 2.08 to 10.47 | .0002† |
| Distant metastases | 5.05 | 2.35 to 10.94 | < .0001† | 3.32 | 1.31 to 8.39 | .01† |
| Chronically sun-exposed anatomic site | .03† | .24 | ||||
| No | 1.00 | 1.00 | ||||
| Yes | 0.56 | 0.33 to 0.95 | 0.70 | 0.39 to 1.26 | ||
| Microsatellitosis | .047† | .63 | ||||
| Absent | 1.00 | 1.00 | ||||
| Present | 1.73 | 1.01 to 2.96 | 1.16 | 0.64 to 2.11 | ||
| Receipt of nonsurgical therapy | .0005† | .008† | ||||
| No | 1.00 | 1.00 | ||||
| Yes | 2.54 | 1.50 to 4.30 | 2.31 | 1.25 to 4.26 | ||
P values were calculated according to the Wald method.
Significant at P ≤ .05.
Assessment of Multimarker Model Reproducibility in the Validation Cohort
To determine the prognostic breadth and strength of our genetic algorithm-based multimarker predictor, we performed the assay on the independent validation TMA, normalizing the two builds as described. Complete AQUA data were obtained for 226 of the 246 eligible individuals, with 76 individuals (33.6%) meeting criteria for the low-risk group and 150 (66.4%) belonging to the high-risk group. Notably, our predictor was independent of both Breslow thickness (P = .41) and sentinel lymph node status (P = .52; Table 3). Our predictor trended toward, but did not achieve, significance for melanoma-specific mortality in univariate analysis (Table 4). However, multivariable modeling that adjusted for Breslow thickness, age at diagnosis, anatomic site, sentinel lymph node biopsy status, and receipt of nonsurgical therapy revealed a significantly increased melanoma-specific mortality for the high-risk group (adjusted hazard ratio = 2.72; 95% CI, 1.12 to 6.58; P = .027; Table 4), consistent with the possibility of negative confounding by clinicopathologic parameters in the validation set. Our predictor is independent of sentinel lymph node status, and the interaction between multimarker assignment and sentinel lymph node status was not significant (P = .78).
Table 3.
Bivariate Associations Between the Genetic Algorithm–Derived Prognostic Indicator and Clinicopathologic Correlates of Melanoma Outcome for the Yale Melanoma Validation Cohort
| Parameter | Low-Risk Group (n = 76) |
High-Risk Group (n = 150) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Breslow thickness, mm | .41 | ||||
| Mean | 1.86 | 2.08 | |||
| Standard deviation | 1.73 | 1.89 | |||
| Age at diagnosis, years | .08 | ||||
| Mean | 57.17 | 61.14 | |||
| Standard deviation | 15.66 | 16.57 | |||
| Sex | .12 | ||||
| Male | 41 | 54.0 | 97 | 64.7 | |
| Female | 35 | 46.1 | 53 | 35.3 | |
| Ulceration | .39 | ||||
| Absent | 63 | 82.9 | 117 | 78.0 | |
| Present | 13 | 17.1 | 33 | 22.0 | |
| Tumor-infiltrating lymphocytes | .94 | ||||
| Nonbrisk | 64 | 84.2 | 126 | 84.6 | |
| Brisk | 12 | 15.8 | 23 | 15.4 | |
| Histologic subtype | .43 | ||||
| Superficial spreading | 45 | 76.3 | 76 | 72.4 | |
| Nodular | 7 | 11.9 | 16 | 15.2 | |
| Lentigo maligna | 2 | 3.4 | 1 | 1.0 | |
| Acral lentiginous | 1 | 1.7 | 0 | 0.0 | |
| Other | 4 | 6.8 | 12 | 11.4 | |
| Chronically sun-exposed anatomic site | .99 | ||||
| No | 33 | 43.4 | 65 | 43.3 | |
| Yes | 43 | 56.6 | 85 | 56.7 | |
| Received any nonsurgical therapy | .50 | ||||
| No | 60 | 80.0 | 123 | 83.7 | |
| Yes | 15 | 20.0 | 24 | 16.3 | |
| Sentinel lymph node biopsy | .52 | ||||
| Negative | 64 | 85.3 | 129 | 88.4 | |
| Positive | 11 | 14.7 | 17 | 11.6 | |
NOTE. Numbers may not sum to total because of missing values; percentages may not sum to 100% as a result of rounding.
Table 4.
Crude and Multivariable-Adjusted Melanoma-Specific Mortality Hazard Ratios for the Genetic Algorithm-Based Multimarker Predictor in the Yale Melanoma Validation Cohort
| Parameter | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P* | Hazard Ratio | 95% CI | P | |
| Genetic algorithm-based predictor | .14 | .027† | ||||
| Low-risk group (4 or 5 conditions met) | 1.00 | 1.00 | ||||
| High-risk group (< 4 conditions met) | 1.75 | 0.83 to 3.72 | 2.72 | 1.12 to 6.58 | ||
| Breslow thickness, mm | 1.20 | 1.11 to 1.31 | < .0001 | 1.14 | 1.01 to 1.29 | .029† |
| Age at diagnosis, years | 1.03 | 1.00 to 1.05 | .027 | 1.04 | 1.01 to 1.07 | .007† |
| Sex | .07 | .10 | ||||
| Male | 1.00 | 1.00 | ||||
| Female | 0.51 | 0.25 to 1.06 | 0.52 | 0.24 to 1.14 | ||
| Chronically sun-exposed anatomic site | .20 | .11 | ||||
| No | 1.00 | 1.00 | ||||
| Yes | 1.55 | 0.79 to 3.04 | 1.96 | 0.87 to 4.44 | ||
| Sentinel lymph node biopsy status | < .0001 | .017† | ||||
| Negative | 1.00 | 1.00 | ||||
| Positive | 4.41 | 2.23 to 8.71 | 2.78 | 1.20 to 6.47 | ||
| Receipt of nonsurgical therapy | < .0001 | .0001† | ||||
| No | 1.00 | 1.00 | ||||
| Yes | 7.09 | 3.60 to 13.96 | 4.65 | 2.11 to 10.24 | ||
P values were calculated according to the Wald method.
Significant at P ≤ .05.
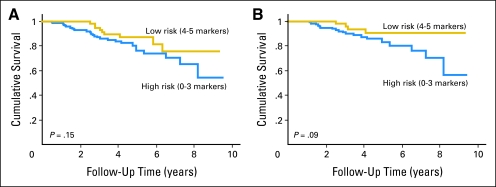
Although the multivariate analysis of the validation set is statistically significant (Table 4), the Kaplan-Meier analysis of the validation set is not (Fig 2A) most likely because of the confounding effect of nonuniform treatment. McShane et al,15 in the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines, point out the value of the multivariate analysis over the log-rank assessment performed on the Kaplan-Meier data. This work is an example of the multivariate analysis adjusting for confounding to show significance, as anticipated by the REMARK criteria. However, the Kaplan-Meier plot is shown to help convey the data in a more simple form related to the envisioned utility of the test in sentinel node–negative patients. In this population, the high-risk group has only a 60% 10-year survival rate compared with a 10-year survival rate of more than 90% in the low-risk group (Fig 2B, log-rank P = .09).
Fig 2.
Kaplan-Meier estimates of melanoma-specific mortality for the dichotomized model describing favorable or unfavorable profiles among (A) all 226 participants of the Yale Melanoma Validation Cohort scored completely for the multimarker prognostic assay, and (B) the 193 members of the Yale Melanoma Validation Cohort who are sentinel lymph node negative (stage II melanoma).
DISCUSSION
Over the last few years, multimarker molecular models have been constructed to supplement available clinicopathologic parameters for refining prognosis in some tumor types. Here, we report on a multimarker melanoma prognostic assay with potential for translation into the clinic that may be especially useful for identifying the subset of stage II melanoma patients most appropriate for supplemental therapy. Presently, up to 40% of patients with stage IIA or IIB melanoma will die of their disease within 10 years of diagnosis.3,16 Because of the poor risk-to-benefit ratio and toxicity of current adjuvant therapy regimens,2 these are not often administered in this population. We believe there is a significant clinical need to stratify this population at the time of diagnosis into a subset of stage II patients with the highest risk for recurrence and a lower risk group. The goal of this stratification, using the test described here, would be to offer adjuvant intervention or at least aggressive follow-up screening to high-risk stage II patients. We believe this would improve the overall survival of these vulnerable patients without exposing the remaining patients to the risk of excessive toxicity; thus, this test has the potential to alter the standard of care for management of melanoma. However, such an approach would require prospective validation in the target (sentinel node–negative) population.
To our knowledge, only one other prognostic multimarker molecular classifier for primary melanoma has been described specifying a 254-gene classifier obtained from differential mRNA expression profiling on a series of 83 snap-frozen samples.17 Although protein expression by IHC was confirmed for the 23-gene subset with commercially available antibodies, the authors only reported on their marginal univariate and multivariate prognostic relationships. Although this study is valuable, to date, the multimarker classifier has not been validated on a second population. Additional molecular classifiers of melanoma phenotype that integrate either somatic mutation18 or gene expression information19 have been reported but have not been evaluated for prognostic relevance. Efforts that use hierarchical clustering, which is valuable for classification, suffer from the inabilities to both calculate error associated with a clustering run and prospectively assign new patients to existing clusters without re-executing the clustering, which risks reorganizing cluster assignment. Assignment of new patients according to our genetic algorithm profile, as demonstrated in our validation strategy, only requires simultaneous AQUA analysis of selected reference standards.
Assignment to the low-risk group requires elevated levels of overall β-catenin and nuclear p21WAF1, decreased levels of fibronectin, and distributions that favor nuclear concentration for p16INK4A but cytoplasmic concentration for ATF2. Each of these assignments is consistent with the previous literature for melanoma. Our data, as well as data from others,20,21 support that increased nuclear p16INK4A expression significantly improves melanoma prognosis in multivariable modeling, consistent with its role in cell cycle inhibition.22 Although specific cytoplasmic p16INK4A expression has been confirmed by multiple high-resolution imaging technologies,23,24 little is known about its functional role or prognostic implications. Our data suggest that a ratio that favors nuclear abundance contributes to improved cell cycle control. A similar rationale can be suggested for elevated nuclear p21WAF1; however, neither we nor others have shown a significant effect for the marginal effects of nuclear p21WAF1 on univariate25,26 or multivariate20,27 analysis. The requirement for a higher proportion of cytoplasmic ATF2 is supported by the observation that although ATF2 possesses both nuclear export and nuclear localization signals and shuttles between both locations, nuclear heterodimerization with c-Jun and subsequent phosphorylation of both subunits by MAP kinases are required for transcriptional activation activity.28,29 Although we did not distinguish between membranous cadherin-associated and cytoplasmic/nuclear Wnt signaling–associated β-catenin, our association between improved prognosis and elevated β-catenin is consistent with others.30,31 Finally, our requirement for reduced fibronectin supports both tissue- and cell-based observations that increased tumor-derived expression facilitates melanoma cell invasion and metastasis.32–34
This work suffers from a number of limitations. Perhaps the most significant limitation is the relatively limited set of available markers eligible for our analysis. Unlike nucleic acid arrays where tens of thousands of genes can be interrogated in each experiment, we can only assess one gene product at a time (although we have the advantage of assessing hundreds of patients per experiment). Furthermore, more than half of the markers initially considered for this study were ultimately eliminated from our genetic algorithm as a result of preferential attrition of longer surviving (typically thinner) melanomas as a result of exhaustion of their tissue cores with higher cuts of the TMA. Future replication of these results on parallel blocks of the discovery TMA may both fill gaps and also provide useful information regarding heterogeneity of marker expression. Although we selected a broad range of candidate targets, the inherent limitation of the candidategene approach omitted sufficient markers from some cancer progression pathways such as evading apoptosis, sustained angiogenesis, orinsensitivity to antigrowth signals.35 Additionally, several proteins previously shown by others to have significant independent marginal associations with melanoma outcome, such as MMP-2,36,37 osteopontin,38 MCAM/MUC18,39,40 and AIB-1,41 were not assayed (in some cases because of antibody validation failure). Another theoretical weakness of this approach is that our genetic algorithm equally weighted each protein's individual contribution. This is in contrast to a commercially available breast cancer diagnostic (Oncotype DX; Genomic Health, Redwood City, CA) where individual marker contribution is weighted according to its relative marginal contribution to the overall model.42 The genetic algorithm approach risks bias in group assignment should the presence or absence of one specific marker disproportionately drive assignment into one of the algorithm states. However, as shown earlier, we found that this bias did not occur in our discovery phase.
Strengths of our approach include the use of equally large and robust, yet completely independent, training and validation study populations as well as choice of a computational method that supports the prospective evaluation of new patients according to its calculated criteria. Given that we were able to replicate a significant, independent association between our multimarker prognostic assay and melanoma-specific mortality after adjustment for relevant clinicopathologic covariates in our independently collected validation set, we believe these data could support the use of this test to assist management of patients with sentinel node–negative melanoma. For example, a high-risk test result in a sentinel node–negative patient might prompt that patient to choose adjuvant therapy. Although the data on the efficacy of adjuvant interferon are controversial,43 other adjuvant therapies such as ipilimumab and vaccine therapies are currently under investigation, and these studies typically include only stage III patients. However, high-risk stage II patients identified by improved prognostic assays such as this should also be considered for these studies. Prospective validation is planned in a broader geographic constituency to determine whether this method should become part of the routine work-up for patients with malignant melanoma.
Acknowledgment
We thank the Cell Culture Core Facility of the Yale Skin Disease Research Core Center (YSDRCC), supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant No. 5 P30 AR 041942-12 (Dr Robert Tigelaar, primary investigator), for providing normal human melanocytes and melanoma cells and Dr Ruth Halaban for providing cell line controls, supported by the Yale Specialized Program of Research Excellence for Skin Cancer (Grant No. P50 CA121974).
Glossary Terms
- Confounding:
Confounding variables are extraneous variables in a statistical model that are associated/correlated with both the independent and dependent variables but are not on the causal pathway between independent and dependent variables. When confounding variables are present, crude (unadjusted) statistical models describing the association between independent and dependent variables are biased (i.e., wrong) as the risk estimate includes the effect of the confounding variable as well (Type 1 error). As a result, to properly describe the relationship between independent and dependent variables, a multivariable model that includes both the independent variable and all relevant confounding variables as predictors must be executed.
- Immunofluorescence:
Refers to laboratory methods that combine the use of antibody reagents to detect the presence of specific biomolecule antigens in situ with a detection system that uses fluorescent molecules to visualize the localization of the target antigen/antibody complex.
- Proportional hazards:
Semiparametric approach to survival analysis developed by Cox in 1972. Unlike product-limit (Kaplan-Meier) survival analyses that are restricted to categorical predictor variables and do not produce a risk estimate, proportional hazards models can accommodate continuous and ordinal variables as well as allow for the inclusion of multiple predictor variables to compute adjusted risk estimates. Proportional hazards models are based on the fundamental premise that all individuals have the same baseline hazard that varies as a function of time [λ(t)] but that exposure to the independent variable changes the hazard by a fixed value [h(x)]. What is parameterized in the model is the value of this fixed effect per unit increase of the predictor variable whereas the value of λ(t) remains uncharacterized.
- Sentinel lymph node:
The lymph node that is anatomically located such that it is the first site of lymph drainage from the location of the primary tumor. It is suspected and assumed that if a malignancy is going to disseminate via the lymphatic system, metastases will first be evident in the sentinel lymph node. In this manner, this lymph node is said to stand guard or sentinel over the metastatic state of the tumor. For many cancers, the sentinel lymph node is biopsied as part of the staging process and presence of macro- or micrometastases in the sentinel lymph node is a negative prognostic factor.
- Genetic algorithm:
Genetic algorithms are a type of iterative mathematical modeling technique used to find the optimal combinatorial state given a set of parameters of interest. Usage of the term “genetic” refers to the mechanism of the algorithm where, through the process of iteration, individual models “evolve” over time and compete with each other in a Darwinian fashion where the fittest model emerges as the solution, similar to how chromosomes evolve to create speciation. Given a set of parameters of interest, a baseline model is fit from a subset of the eligible variables, each dichotomized about a random cut point. Then, through successive iterations, the model is altered by either swapping one of the included parameters (a crossover) or by changing the dichotomization cut point for an included parameter (a mutation) and the model's fitness is reassessed. After several million iterations, the model with the best goodness of fit is selected.
- Immunohistochemistry (IHC):
The application of antigen-antibody interactions to histochemical techniques. Typically, a tissue section is mounted on a slide and is incubated with antibodies (polyclonal or monoclonal) specific to the antigen (primary reaction). The antigen-antibody signal is then amplified using a second antibody conjugated to a complex of peroxidase-antiperoxidase (PAP), avidin-biotin-peroxidase (ABC) or avidin-biotin alkaline phosphatase. In the presence of substrate and chromogen, the enzyme forms a colored deposit at the sites of antibody-antigen binding. Immunofluorescence is an alternate approach to visualize antigens. In this technique, the primary antigen-antibody signal is amplified using a second antibody conjugated to a fluorochrome. On UV light absorption, the fluorochrome emits its own light at a longer wavelength (fluorescence), thus allowing localization of antibody-antigen complexes.
- Prognostic marker:
A marker that predicts the prognosis of a patient (eg, the likelihood of relapse, progression, and/or death) independent of future treatment effects. A factor can be both prognostic and predictive.
- Prognostic factor:
A measurable patient characteristic that is associated with the subsequent course of disease (whether or not therapy is administered). The identification of a prognostic factor does not necessarily imply a cause-and-effect relationship. However, within a suitable outcome model, the measurement of a prognostic factor contributes to an estimate of an outcome probability (eg, the probability of disease-free survival within a given time interval).
- Tissue microarray (TMA):
Used to analyze the expression of genes of interest simultaneously in multiple tissue samples, tissue microarrays consist of hundreds of individual tissue samples placed on slides ranging from 2 to 3 mm in diameter. Using conventional histochemical and molecular detection techniques, tissue microarrays are powerful tools to evaluate the expression of genes of interest in tissue samples. In cancer research, tissue microarrays are used to analyze the frequency of a molecular alteration in different tumor type, to evaluate prognostic markers, and to test potential diagnostic markers.
Appendix
Supplemental Methods
Patients and tumor samples.
Stage at diagnosis (localized, regional, and distant) and anatomic location were obtained from the surgical report. Receipt of nonsurgical therapy referred to administration of cytotoxic chemotherapy, immunomodulators, or radiotherapy either in the adjuvant setting or after clinical recurrence. For each cohort, a single investigator reviewed all slides to reconfirm the diagnosis of melanoma and to determine Breslow thickness, Clark level of invasion, histopathologic subtype, and the presence of ulceration, microsatellitosis, and tumor-infiltrating lymphocytes.
Tissue microarray construction, immunohistochemistry, and automated quantitative image analysis.
Internal positive and negative controls for both the discovery and validation tissue microarrays (TMAs) comprised single cores from formalin-fixed, paraffin-embedded preparations of 15 melanocytic cell lines for which protein expression is verified by Western blot (DiVito KA, Charette LA, Rimm DL, et al: Lab Invest 84:1071-1078, 2004). The cell lines included BHP 18-21, Mel 501, Mel 624, Mel 888, Mel 928, Mel 1241, Mel 1335, MM127, MNT-1, SK23, YUMAC2, YUMOR, YUSAC2, YUSIT1, YUGEN8, and normal melanocytes derived from neonatal foreskin. Sections (5 μm) were cut from the TMA master using a tissue microtome, transferred to glass slides using an ultraviolet cross-linkable tape transfer system (Instrumedics, St Louis, MO), dipped in paraffin, and stored in a nitrogen chamber to prevent antigen degeneration before staining (DiVito KA, Charette LA, Rimm DL, et al: Lab Invest 84:1071-1078, 2004).
Slides were deparaffinized using two xylene exchanges followed by rehydration through an ethanol gradient and washed with tris-buffered saline (triethanolamine-buffered saline). Antigen retrieval was performed by boiling the slides in a sealed pressure cooker containing 6.5 mmol/L sodium citrate, pH 6.0 (except for ATF2 and HDM2, where EDTA, pH 7.5 was used) for 15 minutes. Next, the slides were immersed in absolute methanol containing 0.75% hydrogen peroxide for 30 minutes to neutralize endogenous peroxidase activity, followed by incubation for 30 minutes in 0.3% bovine serum albumin dissolved in 1 mol/L triethanolamine-buffered saline (pH 8.0) to block nonspecific binding.
Fluorescence-based immunohistochemical staining was performed by multiplexing a primary antibody directed against a candidate protein with an S100B antibody of a complementary species (DAKO, Carpinteria, CA) rabbit anti-S100B polyclonal at 1:600 or Biogenex (San Ramon, CA) mouse anti-S100B monoclonal at 1:100, the latter to distinguish melanoma from the surrounding stroma in the absence of counterstain. The selection of protein candidates and their corresponding antibody reagents are listed in Appendix Table A1. External negative controls were obtained by omitting the target protein primary antibody. Primary antibodies were incubated at 4°C overnight. The secondary antibodies, Alexa-546–conjugated goat antibody directed against the anti-S100 antibody (antimouse or antirabbit, 1:200; Molecular Probes, Eugene, OR) diluted into Envision, a horseradish peroxidase–tagged polymer, directed against the protein candidate (neat; DAKO), were applied for 1 hour at room temperature. To visualize the nuclei, 4′,6-diamidino-2-phenylindole (DAPI, 1:100) was included with the secondary antibodies. Finally, a 10-minute Cy5-tyramide (Perkin Elmer Life Sciences, Wellesley, MA) incubation labeled the target. The slides were mounted with 0.6% n-propyl gallate antifade reagent, sealed with a nylon-based lacquer, and stored in the dark until scoring.
Automated quantitative analysis (AQUA) image acquisition and analysis was performed as previously described (Camp RL, Chung GG, Rimm DL. Nat Med 8:1323-1327, 2002). Briefly, stained slides were imaged on a modified computer-controlled epifluorescence microscope (Olympus BX-51 with xy-stage and z controller; Olympus, Tokyo, Japan) illuminated by a high-pressure mercury bulb (Photonic Solutions, Mississauga, ON) with a high-resolution monochromatic camera (Cooke Corporation, Romulus, MI). After user optimization of focus, sets of monochromatic, high-resolution (1,024 × 1,024, 0.5 μm) images were captured for each histospot for each of the DAPI, Alexa-546, and Cy5 fluorescent channels. Two images were captured for each channel—one in the plane of focus and one 8 μm below it. Compartmentalization of each histospot and quantitation of the target protein signal within each compartment are performed as follows. The Alexa-546 signal representing S100B staining is binary gated to indicate whether a pixel is within the tumor mask (on) or not (off). Within the region defined by the tumor, the nuclear compartment is defined as the subset of pixels that demonstrated any DAPI staining within the plane of focus. This was required to compensate for the three-dimensional thickness of the tumor sections, which can blur discrimination of the nuclear boundary. The non-nuclear compartment is then defined as all pixels assigned to the tumor mask but not included within the nuclear compartment. Finally, target antigen expression is automatically determined from Cy5 channel images to obtain relative pixel intensity for the signal emanating from the plane of focus. The final AQUA score for the entire tumor mask or any of its subcellular compartments was calculated as the average AQUA score for each of the individual pixels included in the selected compartment and was reported on a scale of 0 to 255.
Data management and statistical analysis.
Cores whose tumor mask covered less than 5% of the total histospot area were dropped from further analysis. For individuals represented by multiple cores on the TMA, AQUA scores were averaged before analysis. We normalized AQUA scores between parallel runs of the two builds for the validation cohort by first calculating a regression equation between the AQUA scores for cell line controls and then adjusting the scores for the build 1 surgical specimens according to the equation's parameters. The final AQUA score for the validation cohort was then calculated as the mean of build 2 and the adjusted build 1 AQUA scores. Similarly, to normalize the AQUA scores between the discovery and validation cohorts, a regression equation was calculated for the set of 60 samples spotted on both arrays, and the mean values for the validation cohort were adjusted according to the regression equation.
For HDM2, Ki-67, MITF, p21, and PCNA, where previous data support nuclear localization of the target in melanoma (Berger AJ, Kluger HM, Li N, et al: Cancer Res 63:8103-8107, 2003; Denicourt C, Saenz CC, Datnow B, et al: Cancer Res 67:9238-9243, 2007; Camp RL, Dolled-Filhart M, Rimm DL: Clin Cancer Res 10:7252-7259, 2004; Mitchell M: An Introduction to Genetic Algorithms. Cambridge, MA, MIT Press, 1998), the AQUA score for the nuclear compartment was considered. For AP-2α, ATF-2, p16, and p27, where the ratio of non-nuclear to nuclear expression has previously been shown to have prognostic relevance (Berger AJ, Davis DW, Tellez C, et al: Cancer Res 65:11185-11192, 2005; Berger AJ, Kluger HM, Li N, et al: Cancer Res 63:8103-8107, 2003; Denicourt C, Saenz CC, Datnow B, et al: Cancer Res 67:9238-9243, 2007), the natural log of the ratio of non-nuclear to nuclear AQUA scores was evaluated in addition to the nuclear (AP-2α, p16, and p27) or non-nuclear (ATF2) compartment AQUA score. For the remaining markers, the AQUA scores for the total area under the tumor mask were selected.
The genetic algorithm was executed using the X-tile software suite (Camp RL, Dolled-Filhart M, Rimm DL: Clin Cancer Res 10:7252-7259, 2004). Briefly, the algorithm randomly selects a set of markers and, for each marker, chooses a random cut point to binarize the continuous AQUA data, where, by convention, a score of 1 indicates reduced risk and 0 indicates increased risk. Next, for each individual, the binary marker scores are summed and the log-rank statistic for melanoma-specific survival is calculated across all marker sum categories. This initial seed model is then subjected to multiple iterations by either mutation (altering the cut point for an already included marker) or cross over (swapping among the set of eligible markers) until the model converges on a maximum likelihood statistic for melanoma-specific survival.
To develop a multimarker prognostic model from the discovery cohort data, a genetic algorithm using standard methodology (Mitchell M: An Introduction to Genetic Algorithms. Cambridge, MA, MIT Press, 1998; Ooi CH, Tan P: Bioinformatics 19:37-44, 2003) within the X-tile software suite (Camp RL, Dolled-Filhart M, Rimm DL: Clin Cancer Res 10:7252-7259, 2004) with a 33% cross-over and 33% mutation rate constrained to create a multimarker profile that included a minimum of 100 of 192 eligible individuals with complete data across all selected markers was created. Additional algorithm specifications limited individual marker cut points to include ≥ 10% of the available population in each arm and required that each category defined by the marker groupings both contain no fewer than 15% of the available population and, to maintain statistical robustness of the final model, enumerate no fewer than two events of interest. We did not constrain the number of parameters to be included in the selected model. Briefly, the algorithm randomly selects a set of markers and, for each marker, chooses a random cut point to binarize the continuous AQUA data, where, by convention, a score of 1 indicates reduced risk and 0 indicates increased risk. Next, for each individual, the binary marker scores are summed, and the log-rank statistic for melanoma-specific survival is calculated across all marker sum categories. This initial seed model is then subjected to multiple iterations by either mutation (altering the cut point for an already included marker) or cross over (swapping among the set of eligible markers) until the model converges on a set of markers and their respective cut points that yield the highest log-rank χ2 statistic for melanoma-specific survival, typically achieved between 16 and 18 million iterations. Five parallel iterations of the genetic algorithm were executed. Melanoma-specific survival was the end point for all survival analyses; individuals who died from competing causes were censored at the time of death.
Bivariate associations between protein expression in the set of primary tumors with either the metastatic lesions or their associated clinicopathologic criteria were determined using the nonparametric Spearman rank correlation, Mann-Whitney U, or Kruskal-Wallis tests. Bivariate associations between the genetic algorithm output and clinicopathologic criteria were evaluated using χ2 or t tests. Survival curves were calculated using the Kaplan-Meier product-limit method and the log-rank statistic. Univariate and multivariate hazard ratios were calculated using the Cox proportional hazards method, the latter adjusting for known clinicopathologic variables (Breslow thickness, age at diagnosis, sex, stage at diagnosis, microsatellitosis/sentinel lymph node status, tumor site, and receipt of systemic therapy).
Supplemental Results
Bivariate relationships among markers and with clinicopathologic variables.
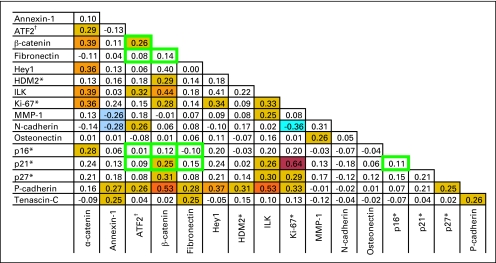
Appendix Figure A1 considers all pairwise Spearman rank correlations (n = 136) between markers for the set of primary tumors. After adjustment for multiple comparisons, 19 positive associations (rs ≥ 0.29; P ≤ .0003) and one negative association (rs ≤ –0.29) achieved significance. The strongest positive correlations were observed between Ki-67 and p21WAFI (rs = 0.64), P-cadherin and β-catenin (rs = 0.53), P-cadherin and integrin-linked kinase (rs = 0.53), with the only significant negative correlation observed between Ki-67 and MMP-1 (rs = −0.36).
Associations between marker AQUA scores and clinicopathologic characteristics among the primary tumors were determined using nonparametric methods (Appendix Table A3). Six markers were significantly associated with Breslow thickness; osteonectin and the ratios for p16INK4A and p27KIP1 increased, whereas ATF2, HDM2, and N-cadherin decreased with increasing tumor thickness. None of the associations with the remaining clinicopathologic parameters achieved significance after adjustment for multiple comparisons.
Individual marker associations with melanoma-specific survival.
Methodologically robust multivariable-adjusted individual-protein hazard ratios (McShane LM, Altman DG, Sauerbrei W, et al: J Natl Cancer Inst 97:1180-1184, 2005) have been published for 12 markers, with our laboratory contributing reports for six. Among the marginal associations for those six markers not previously reported by our group, only our results for osteonectin/SPARC (no association), nuclear p16INK4a (improved survival with increased expression), and p27KIP1 (worsened survival with increased expression) recapitulate previously published results (Alonso SR, Ortiz P, Pollan M, et al: Am J Pathol 164:193-203, 2004; Straume O, Sviland L, Akslen LA: Clin Cancer Res 6:1845-1853, 2000; Alonso SR, Tracey L, Ortiz P, et al: Cancer Res 67:3450-3460, 2007). Unlike previous reports (Alonso SR, Ortiz P, Pollan M, et al: Am J Pathol 164:193-203, 2004; Straume O, Sviland L, Akslen LA: Clin Cancer Res 6:1845-1853, 2000; Ilmonen S, Jahkola T, Turunen JP, et al: Histopathology 45:405-411, 2004; Niezabitowski A, Czajecki K, Rys J, et al: J Surg Oncol 70:150-160, 1999), we did not find a significant association or trend between Ki-67 or tenascin-C with survival, and our significant result for p21WAFI opposed the one published result that indicated a trend toward worse survival with increased p21WAFI levels (Alonso SR, Ortiz P, Pollan M, et al: Am J Pathol 164:193-203, 2004). For the remaining five markers, to our knowledge, these data represent the first report of methodologically robust, multivariable-adjusted survival assessment, with our results for annexin I and Hey-1 being the first instance of immunohistochemistry data in melanoma.
Descriptive statistics for the genetic algorithm-based multimarker prognostic indicator.
One hundred twenty-nine individuals from the discovery cohort (67.2%) possessed complete data for all five selected markers and were included in the training set. Of these, 20 individuals (15.5%) met all five marker conditions, 46 (35.67%) met any four of the five conditions, 42 (32.6%) met three conditions, 19 (14.7%) met two conditions, and the remaining two individuals (1.6%) met one condition only. The latter two classes were combined in the preliminary algorithm-based groupings. Among the 46 individuals who met four of the five conditions, we observed an even distribution of the marker not meeting its cut point, with 11 (23.9%) failing the ATF2 ratio, eight (17.4%) failing β-catenin, nine (19.6%) failing fibronectin, 10 (21.7%) failing the p16INK4A ratio, and eight (17.4%) failing p21WAF1, ruling out any marker-driven selection bias in creating this category.
Bivariate associations between the genetic algorithm-based multimarker prognostic indicator and the clinicopathologic covariates for the Yale Melanoma Discovery Cohort are reported (Appendix Table A5). In those assigned to the high-risk group, compared with the low-risk group, Breslow thickness (2.90 ± 2.10 mm v 2.15 ± 1.88 mm, respectively; P = .04) and percentage receiving nonsurgical therapy (33.3% v 12.1%, respectively; P = .004) were significantly higher. Presence of ulceration (P = .06) and the lack of a brisk lymphocyte infiltrate (P = .08) also trended toward significance.
Fig A1.
Spearman correlation coefficients for all pairwise comparisons among the set of 17 assayed markers (n = 136). Automated quantitative analysis (AQUA) scores used for this analysis represent the total AQUA score under the tumor mask except for the subset of markers indicated by either an asterisk (*), where the nuclear compartment AQUA score, or by a dagger (†), where the non-nuclear compartment AQUA score was selected. Comparisons ringed by a green box represent the subset of comparisons between the markers included in the genetic algorithm-based multi-marker prognostic assay. Colored boxes highlight the subset of comparisons significant at P < .005. Orange boxes indicate positive correlations, whereas blue boxes indicate negative correlations. The richness of color reflects the strength of the association.
Table A1.
Protocols for Immunohistochemical Staining
| Target | Antibody | Provider | Dilution |
|---|---|---|---|
| α-catenin | Mouse monoclonal αCAT-7A4 | Zymed | 1:150 |
| Annexin-1/lipocortin-1 | Mouse monoclonal 29 | Transduction Labs | 1:1,000 |
| AP-2α | Rabbit polyclonal K2403 | Santa Cruz | 1:1,600 |
| ATF-2 | Rabbit polyclonal C19 | Santa Cruz | 1:250 |
| β-catenin | Mouse monoclonal 14 | Transduction Labs | 1:2,500 |
| CD44 | Mouse monoclonal 2C5 | R&D Systems | 1:200 |
| c-Kit | Mouse monoclonal 2E4 | Zymed | 1:50 |
| Connective tissue growth factor | Rabbit polyclonal ab6992 | Abcam | 1:650 |
| E-cadherin | Mouse monoclonal 32 | Transduction Labs | 1:400 |
| Ephrin A1 | Rabbit polyclonal I2203 | Santa Cruz | 1:250 |
| Ephrin receptor Eph A2 | Rabbit polyclonal SC924 | Santa Cruz | 1:200 |
| Fascin | Mouse monoclonal 55K-2 | DAKO | 1:250 |
| Fibronectin (FN1) | Rabbit polyclonal ab299 | Abcam | 1:700 |
| Granulophysin/CD63 | Mouse monoclonal FC-5.01 | Zymed | 1:50 |
| Hairy/enhancer of split-related (HEY1) | Rabbit polyclonal | Santa Cruz | 1:200 |
| Human double-minute-2 (HDM2) | Mouse monoclonal 1B10 | Novocastra | 1:100 |
| Integrin-β3/CD61 | Mouse monoclonal SZ21 | Beckman | 1:50 |
| Integrin-linked kinase | Rabbit polyclonal KAP-ST203 | Stressgen | 1:300 |
| Ki-67 | Mouse monoclonal B56 | Transduction Labs | 1:500 |
| L1-CAM | Mouse monoclonal L1-11A | Lab of P. Altevogt | Neat |
| MAGE-A1 | Mouse monoclonal MA454 | Zymed | 1:90 |
| Metallothionein (MT-1) | Mouse monoclonal M0639 | DAKO | 1:400 |
| Microphthalmia transcription factor (MITF) | Mouse monoclonal C5 + D5 | Zymed | Neat |
| Matrix metalloproteinase-1 (MMP-1) | Mouse monoclonal IM35L | Calbiochem | 1:550 |
| Matrix metalloproteinase-3 (MMP-3) | Rabbit polyclonal AB810 | Chemicon | 1:3,000 |
| Myelin basic protein | Rabbit polyclonal 18-0038 | Zymed | 1:300 |
| N-cadherin | Mouse monoclonal 3B9 | Zymed | 1:150 |
| Osteonectin/SPARC | Mouse monoclonal AON-5031 | Hematologic Technologies | 1:8,000 |
| p120-catenin | Mouse monoclonal 98 | Transduction Labs | 1:400 |
| p16/INK4A | Mouse monoclonal G175-405 | Transduction Labs | 1:500 |
| p21/WAF1/CIP1/CDKN1A | Mouse monoclonal SX118 | Transduction Labs | 1:100 |
| p27/KIP1/CDKN1B | Mouse monoclonal G173-524 | Transduction Labs | 1:300 |
| P-cadherin | Mouse monoclonal 56 | Transduction Labs | 1:250 |
| Proliferating cell nuclear antigen (PCNA) | Mouse monoclonal PC10 | Zymed | 1:10,000 |
| Tenascin-C | Rabbit polyclonal SC20932 | Santa Cruz | 1:600 |
| Tissue inhibitor of metalloproteinase-2 (TIMP-2) | Mouse monoclonal 3A4 | Zymed | 1:75 |
| Tissue inhibitor of metalloproteinase-3 (TIMP-3) | Mouse monoclonal 136-13H4 | Oncogene Research Products | 1:10 |
| Twist | Rabbit polyclonal H-81 | Santa Cruz | 1:250 |
Table A2.
Marker Expression Levels Among Primary and Metastatic Lesions
| Target* | Primary Tumors (n = 192) |
Metastatic Tumors (n = 299) |
P (Mann-Whitney U) | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| α-catenin | 11.53 | 5.86 | 13.10 | 6.72 | .02† |
| Annexin-1/lipocortin-1 | 40.48 | 20.89 | 42.09 | 25.87 | .54 |
| ATF-2, non-nuclear compartment | 65.57 | 39.33 | 46.87 | 35.65‡ | < .0001§ |
| ATF-2, ln(non-nuclear/nuclear compartments) | 1.07 | 0.58 | 1.40 | 0.75‡ | < .0001§ |
| β-catenin | 48.28 | 15.75 | 43.18 | 13.18 | .002† |
| Fibronectin | 47.49 | 13.74 | 64.10 | 24.45‡ | < .0001§ |
| Hairy/enhancer of split-related-1 | 58.93 | 23.89 | 48.23 | 22.93 | < .0001§ |
| Human double-minute-2, nuclear compartment | 65.80 | 21.53 | 52.36 | 20.13‡ | < .0001§ |
| Integrin-linked kinase | 42.95 | 13.60 | 44.17 | 12.91 | .06 |
| Ki-67, nuclear compartment | 18.77 | 6.84 | 23.03 | 9.63 | < .0001§ |
| Matrix metalloproteinase-1 | 36.58 | 18.54 | 33.69 | 22.61 | .008† |
| N-cadherin | 19.48 | 15.33 | 11.82 | 11.42 | < .0001§ |
| Osteonectin/SPARC | 19.89 | 14.42 | 20.14 | 17.12 | .10 |
| p16/INK4A, nuclear compartment | 32.34 | 26.69 | 24.85 | 19.00 | .0006§ |
| p16/INK4A, ln(non-nuclear/nuclear compartments) | −0.15 | 0.32 | −0.07 | 0.25 | < .0001§ |
| p21/WAF1/CIP1, nuclear compartment | 17.76 | 9.06 | 24.14 | 13.60 | < .0001§ |
| p27/KIP1, nuclear compartment | 44.64 | 21.82 | 46.23 | 21.97 | .33 |
| p27/KIP1, ln(non-nuclear/nuclear compartments) | −0.33 | 0.26 | −0.35 | 0.28 | .30 |
| P-cadherin | 35.71 | 7.11 | 33.69 | 6.08 | .007† |
| Tenascin-C | 26.12 | 21.24 | 33.53 | 29.80 | .28 |
Abbreviation: SD, standard deviation.
Each target considers the automated quantitative analysis (AQUA) score under the entire tumor mask, unless otherwise indicated.
Significant at P = .05.
Assay of fibronectin, HDM2, and ATF-2 was restricted to the subset of 198 metastases collected during 1959 to 1994.
Significant at Bonferroni-adjusted P = .0025.
Table A3.
Marker Associations With the Known Clinical Prognostic Characteristics Among the Primary Tumors
| Target |
P |
|||||||
|---|---|---|---|---|---|---|---|---|
| Breslow Thickness (mm) | Sex (male v female) | Stage at Diagnosis | Ulceration (no v yes) | Microsatellitosis (no v yes) | Tumor-Infiltrating Lymphocytes (brisk v nonbrisk) | Chronically Sun Exposed Site (no v yes) | Received Nonsurgical Therapy (no v yes) | |
| α-catenin | .11 | .44 | .10 | .19 | .31 | .07 | .86 | .26 |
| Annexin-1/lipocortin-1 | .95 | .21 | .43 | .13 | .03* | .70 | .08 | .44 |
| ATF-2, non-nuclear compartment | < .0001†‡ | .97 | .25 | .03† | .47 | .01* | .30 | .19 |
| ATF-2, ln(non-nuclear/nuclear compartments) | .03† | .42 | .55 | .65 | .45 | .77 | .03† | .65 |
| β-catenin | .02† | .95 | .14 | .13 | .15 | .38 | .82 | .35 |
| Fibronectin | .49 | .24 | .41 | .40 | .22 | .39 | .09 | .10 |
| Hairy/enhancer of split-related-1 | .40 | .21 | .69 | .40 | .51 | .91 | .15 | .53 |
| Human double-minute-2, nuclear compartment | .006†‡ | .92 | .76 | .23 | .30 | .38 | .81 | .56 |
| Integrin-linked kinase | .18 | .59 | .86 | .91 | .49 | .55 | .59 | .40 |
| Ki-67, nuclear compartment | .19 | .71 | .03* | .35 | .94 | .97 | .79 | .38 |
| Matrix metalloproteinase-1 | .49 | .36 | 1.00 | .75 | .49 | .87 | .29 | .68 |
| N-cadherin | .0006‡ | .66 | .49 | .28 | .71 | .05* | .59 | .13 |
| Osteonectin/SPARC | .004*‡ | .55 | .41 | .39 | .89 | .56 | .87 | .27 |
| p16/INK4A, nuclear compartment | .02† | .96 | .94 | .11 | .30 | .25 | .53 | .33 |
| p16/INK4A, ln(non-nuclear/nuclear compartments) | .007*‡ | .78 | .92 | .01* | .94 | .38 | .51 | .92 |
| p21/WAF1/CIP1, nuclear compartment | .03* | .51 | .63 | .12 | .02* | .64 | .67 | .16 |
| p27/KIP1, nuclear compartment | .89 | .75 | .69 | .97 | .69 | .16 | .82 | .06 |
| p27/KIP1, ln(non-nuclear/nuclear compartments) | .005*‡ | .72 | .52 | .02* | .29 | .55 | .83 | .73 |
| P-cadherin | .04† | .10 | .62 | .15 | .20 | .23 | .62 | .61 |
| Tenascin-C | .04† | .55 | .37 | .27 | .09 | .84 | .31 | .56 |
Significant at P < .05; positive association between increasing automated quantitative analysis (AQUA) score and increased severity of the clinical feature; association with female sex.
Significant at P < .05; negative association between increasing AQUA score and increased severity of the clinical feature.
Significant at Bonferroni-adjusted P < .0025.
Table A4.
Individual Marker Associations With Melanoma-Specific Mortality
| Target | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P* | HR† | 95% CI | P | |
| α-catenin | .43 | .01‡ | ||||
| Quartile 1 (AQUA score 2.56 to 6.82) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 6.83 to 10.95) | 1.21 | 0.66 to 2.24 | 1.28 | 0.68 to 2.42 | ||
| Quartile 3 (AQUA score 10.96 to 14.77) | 1.01 | 0.54 to 1.89 | 0.50 | 0.25 to 0.99 | ||
| Quartile 4 (AQUA score 14.78 to 34.31) | 0.69 | 0.34 to 1.40 | 0.43 | 0.19 to 0.97 | ||
| Annexin-1/lipocortin-1 | .28 | .75 | ||||
| Quartile 1 (AQUA score 4.53 to 21.89) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 21.90 to 41.47) | 1.02 | 0.50 to 2.06 | 1.04 | 0.50 to 2.18 | ||
| Quartile 3 (AQUA score 41.48 to 53.79) | 1.38 | 0.71 to 2.68 | 1.36 | 0.67 to 2.77 | ||
| Quartile 4 (AQUA score 53.80 to 103.66) | 1.75 | 0.90 to 3.40 | 0.97 | 0.47 to 1.99 | ||
| ATF-2, non-nuclear compartment | .001§ | .25 | ||||
| Quartile 1 (AQUA score 13.08 to 36.11) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 36.12 to 59.10) | 0.74 | 0.42 to 1.29 | 0.89 | 0.48 to 1.63 | ||
| Quartile 3 (AQUA score 59.11 to 87.23) | 0.62 | 0.35 to 1.10 | 0.73 | 0.39 to 1.37 | ||
| Quartile 4 (AQUA score 87.24 to 244.30) | 0.25 | 0.12 to 0.54 | 0.47 | 0.21 to 1.04 | ||
| ATF-2, ln(non-nuclear/nuclear compartments) | .20 | .79 | ||||
| Quartile 1 (ratio −1.33 to −0.14) | 1.00 | 1.00 | ||||
| Quartile 2 (ratio −0.13 to +0.04) | 0.57 | 0.30 to 1.08 | 0.95 | 0.48 to 1.90 | ||
| Quartile 3 (ratio +0.05 to +0.34) | 0.61 | 0.33 to 1.12 | 1.23 | 0.62 to 2.46 | ||
| Quartile 4 (ratio +0.35 to +2.38) | 0.58 | 0.32 to 1.05 | 0.87 | 0.45 to 1.66 | ||
| β-catenin | .04‡ | .04‡ | ||||
| Quartile 1 (AQUA score 9.46 to 37.83) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 37.84 to 45.59) | 0.52 | 0.28 to 0.97 | 0.40 | 0.20 to 0.83 | ||
| Quartile 3 (AQUA score 45.60 to 55.94) | 0.64 | 0.34 to 1.18 | 0.86 | 0.44 to 1.67 | ||
| Quartile 4 (AQUA score 55.95 to 104.05) | 0.40 | 0.20 to 0.79 | 0.45 | 0.21 to 0.96 | ||
| Fibronectin | .17 | .33 | ||||
| Quartile 1 (AQUA score 23.00 to 37.06) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 37.07 to 45.67) | 1.24 | 0.64 to 2.41 | 1.05 | 0.50 to 2.22 | ||
| Quartile 3 (AQUA score 45.68 to 57.01) | 0.66 | 0.31 to 1.43 | 0.70 | 0.31 to 1.59 | ||
| Quartile 4 (AQUA score 57.02 to 93.87) | 1.43 | 0.74 to 2.76 | 1.43 | 0.68 to 2.98 | ||
| Hairy/enhancer of split-related-1 | .31 | .22 | ||||
| Quartile 1 (AQUA score 6.09 to 43.14) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 43.15 to 54.77) | 1.37 | 0.72 to 2.59 | 1.13 | 0.56 to 2.26 | ||
| Quartile 3 (AQUA score 54.78 to 74.04) | 0.76 | 0.37 to 1.54 | 0.54 | 0.25 to 1.16 | ||
| Quartile 4 (AQUA score 74.05 to 173.42) | 1.26 | 0.65 to 2.45 | 1.07 | 0.51 to 2.23 | ||
| Human double-minute-2, nuclear compartment | .34 | .24 | ||||
| Quartile 1 (AQUA score 21.84 to 50.09) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 50.10 to 62.04) | 0.65 | 0.33 to 1.28 | 0.79 | 0.37 to 1.68 | ||
| Quartile 3 (AQUA score 62.05 to 76.06) | 0.97 | 0.51 to 1.85 | 1.24 | 0.64 to 2.42 | ||
| Quartile 4 (AQUA score 76.07 to 145.79) | 0.60 | 0.29 to 1.22 | 0.57 | 0.26 to 1.24 | ||
| Integrin-linked kinase | .31 | .29 | ||||
| Quartile 1 (AQUA score 15.82 to 33.17) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 33.18 to 40.70) | 0.78 | 0.42 to 1.46 | 0.54 | 0.28 to 1.05 | ||
| Quartile 3 (AQUA score 40.71 to 50.10) | 0.70 | 0.36 to 1.35 | 0.75 | 0.37 to 1.53 | ||
| Quartile 4 (AQUA score 50.11 to 96.77) | 0.51 | 0.25 to 1.06 | 0.62 | 0.28 to 1.34 | ||
| Ki-67, nuclear compartment | .66 | .21 | ||||
| Quartile 1 (AQUA score 4.25 to 13.83) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 13.84 to 18.08) | 1.12 | 0.60 to 2.09 | 0.91 | 0.47 to 1.77 | ||
| Quartile 3 (AQUA score 18.09 to 22.67) | 0.86 | 0.42 to 1.75 | 0.47 | 0.21 to 1.05 | ||
| Quartile 4 (AQUA score 22.68 to 40.24) | 1.32 | 0.69 to 2.52 | 0.91 | 0.45 to 1.83 | ||
| Matrix metalloproteinase-1 | .46 | .66 | ||||
| Quartile 1 (AQUA score 9.49 to 21.57) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 21.58 to 33.09) | 1.51 | 0.75 to 3.07 | 0.96 | 0.44 to 2.09 | ||
| Quartile 3 (AQUA score 33.10 to 47.86) | 1.16 | 0.56 to 2.39 | 0.97 | 0.44 to 2.16 | ||
| Quartile 4 (AQUA score 47.87 to 91.51) | 1.64 | 0.82 to 3.26 | 1.45 | 0.68 to 3.09 | ||
| N-cadherin | .001‡ | .06 | ||||
| Quartile 1 (AQUA score 4.17 to 8.30) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 8.31 to 14.54) | 0.83 | 0.47 to 1.48 | 0.54 | 0.27 to 1.06 | ||
| Quartile 3 (AQUA score 14.54 to 24.83) | 0.37 | 0.18 to 0.76 | 0.45 | 0.21 to 0.98 | ||
| Quartile 4 (AQUA score 24.84 to 77.09) | 0.32 | 0.16 to 0.66 | 0.38 | 0.18 to 0.82 | ||
| Osteonectin/SPARC | .40 | .23 | ||||
| Quartile 1 (AQUA score 4.92 to 9.69) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 9.70 to 15.45) | 1.78 | 0.89 to 3.55 | 1.83 | 0.89 to 3.76 | ||
| Quartile 3 (AQUA score 15.46 to 23.45) | 1.42 | 0.71 to 2.86 | 0.92 | 0.45 to 1.92 | ||
| Quartile 4 (AQUA score 23.46 to 68.19) | 1.53 | 0.78 to 3.01 | 1.25 | 0.61 to 2.57 | ||
| p16/INK4A, nuclear compartment | .047§ | .04§ | ||||
| Quartile 1 (AQUA score 4.18 to 15.14) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 15.15 to 22.69) | 0.48 | 0.27 to 0.89 | 0.46 | 0.27 to 0.88 | ||
| Quartile 3 (AQUA score 22.70 to 39.78) | 0.56 | 0.31 to 1.01 | 0.42 | 0.22 to 0.81 | ||
| Quartile 4 (AQUA score 39.79 to 158.00) | 0.48 | 0.26 to 0.88 | 0.60 | 0.32 to 1.15 | ||
| p16/INK4A, ln(non-nuclear/nuclear compartments) | .04§ | .15 | ||||
| Quartile 1 (ratio −0.93 to −0.35) | 1.00 | 1.00 | ||||
| Quartile 2 (ratio −0.34 to −0.18) | 0.73 | 0.36 to 1.50 | 0.68 | 0.32 to 1.47 | ||
| Quartile 3 (ratio −0.17 to +0.03) | 1.45 | 0.78 to 2.71 | 1.39 | 0.72 to 2.71 | ||
| Quartile 4 (ratio +0.04 to +1.24) | 1.75 | 0.96 to 3.20 | 1.31 | 0.68 to 2.55 | ||
| p21/WAF1/CIP1, nuclear compartment | .24 | .24 | ||||
| Quartile 1 (AQUA score 7.49 to 12.18) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 12.19 to 15.08) | 0.95 | 0.52 to 1.73 | 0.84 | 0.44 to 1.58 | ||
| Quartile 3 (AQUA score 15.09 to 21.48) | 0.66 | 0.34 to 1.27 | 0.49 | 0.24 to 1.00 | ||
| Quartile 4 (AQUA score 21.49 to 76.02) | 1.28 | 0.71 to 2.31 | 0.74 | 0.38 to 1.46 | ||
| p27/KIP1, nuclear compartment | .67 | .046§ | ||||
| Quartile 1 (AQUA score 10.27 to 28.95) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 28.96 to 39.82) | 1.41 | 0.77 to 2.62 | 2.58 | 1.32 to 4.87 | ||
| Quartile 3 (AQUA score 39.83 to 53.99) | 1.02 | 0.53 to 1.96 | 1.27 | 0.62 to 2.63 | ||
| Quartile 4 (AQUA score 54.00 to 144.49) | 1.10 | 0.59 to 2.07 | 1.53 | 0.75 to 3.11 | ||
| p27/KIP1, ln(non-nuclear/nuclear compartments) | .23 | .36 | ||||
| Quartile 1 (ratio −1.38 to −0.49) | 1.00 | 1.00 | ||||
| Quartile 2 (ratio −0.48 to −0.31) | 0.72 | 0.37 to 1.41 | 0.54 | 0.27 to 1.08 | ||
| Quartile 3 (ratio −0.30 to −0.13) | 0.88 | 0.47 to 1.65 | 0.63 | 0.32 to 1.24 | ||
| Quartile 4 (ratio −0.12 to +0.22) | 1.39 | 0.76 to 2.55 | 0.69 | 0.35 to 1.34 | ||
| P-cadherin | .70 | .49 | ||||
| Quartile 1 (AQUA score 20.02 to 30.95) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 30.96 to 35.24) | 0.75 | 0.40 to 1.42 | 1.14 | 0.56 to 2.33 | ||
| Quartile 3 (AQUA score 35.25 to 39.57) | 0.72 | 0.37 to 1.39 | 0.92 | 0.44 to 1.94 | ||
| Quartile 4 (AQUA score 39.58 to 60.85) | 0.95 | 0.50 to 1.81 | 1.62 | 0.81 to 3.24 | ||
| Tenascin-C | .30 | .046§ | ||||
| Quartile 1 (AQUA score 6.54 to 12.32) | 1.00 | 1.00 | ||||
| Quartile 2 (AQUA score 12.33 to 18.65) | 0.86 | 0.43 to 1.69 | 0.65 | 0.30 to 1.37 | ||
| Quartile 3 (AQUA score 18.66 to 33.95) | 0.71 | 0.35 to 1.43 | 0.92 | 0.40 to 2.09 | ||
| Quartile 4 (AQUA score 33.96 to 142.89) | 1.32 | 0.70 to 2.51 | 1.81 | 0.88 to 3.74 | ||
Abbreviations: HR, hazard ratio; AQUA, automated quantitative analysis.
Univariate and multivariate P values were calculated according to the likelihood ratio test method.
Adjusted for age at diagnosis, sex, Breslow thickness (millimeters), stage at diagnosis, presence of microsatellitosis, sun exposure to anatomic site, and receipt of nonsurgical therapy.
Significant at Bonferroni-adjusted P ≤ .0025.
Significant at P ≤ .05.
Table A5.
Bivariate Associations Between the Genetic Algorithm–Derived Prognostic Indicator and Clinicopathologic Correlates of Melanoma Outcome for the Yale Melanoma Discovery Cohort
| Parameter | Low-Risk Group (n = 66) |
High-Risk Group (n = 63) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Breslow thickness, mm | .04* | ||||
| Mean | 2.15 | 2.90 | |||
| Standard deviation | 1.88 | 2.10 | |||
| Age at diagnosis, years | .33 | ||||
| Mean | 55.36 | 57.94 | |||
| Standard deviation | 15.14 | 14.67 | |||
| Sex | .93 | ||||
| Male | 32 | 48.5 | 31 | 49.2 | |
| Female | 34 | 51.5 | 32 | 48.5 | |
| Stage at diagnosis | .55 | ||||
| Localized | 54 | 83.1 | 52 | 83.9 | |
| Regional spread | 7 | 10.8 | 4 | 6.5 | |
| Distant metastases | 4 | 6.2 | 6 | 9.7 | |
| Ulceration | .06 | ||||
| Absent | 48 | 72.7 | 36 | 57.1 | |
| Present | 18 | 27.3 | 27 | 42.9 | |
| Tumor-infiltrating lymphocytes | .08 | ||||
| Nonbrisk | 49 | 75.4 | 55 | 87.3 | |
| Brisk | 16 | 24.6 | 8 | 12.7 | |
| Histologic subtype | .98 | ||||
| Superficial spreading | 46 | 69.7 | 43 | 68.3 | |
| Nodular | 10 | 15.2 | 12 | 19.1 | |
| Lentigo maligna | 1 | 1.5 | 1 | 1.6 | |
| Acral lentiginous | 4 | 6.1 | 3 | 4.8 | |
| Other | 5 | 7.6 | 4 | 6.4 | |
| Chronically sun-exposed anatomic site | .29 | ||||
| No | 30 | 45.5 | 34 | 54.8 | |
| Yes | 36 | 54.6 | 28 | 45.2 | |
| Received any nonsurgical therapy | .004* | ||||
| No | 58 | 87.9 | 42 | 66.7 | |
| Yes | 8 | 12.1 | 21 | 33.3 | |
| Microsatellitosis | .95 | ||||
| Absent | 50 | 75.8 | 48 | 76.2 | |
| Present | 16 | 24.2 | 15 | 23.8 | |
NOTE. Numbers may not sum to total because of missing values; percentages may not sum to 100% as a result of rounding.
Significant at P < .05.
Footnotes
Supported by Grant No. RO-1 CA114277 (D.L.R.) from the National Institutes of Health.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Annette M. Molinaro, HistoRx (C); Robert L. Camp, HistoRx (C); David L. Rimm, HistoRx (C) Stock Ownership: Robert L. Camp, HistoRx; David L. Rimm, HistoRx Honoraria: None Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Bonnie E. Gould Rothberg, Aaron J. Berger, Robert L. Camp, Harriet M. Kluger, David L. Rimm
Financial support: Harriet M. Kluger, David L. Rimm
Provision of study materials or patients: Aaron J. Berger, Antonio Subtil, Stephan Ariyan, David L. Rimm
Collection and assembly of data: Bonnie E. Gould Rothberg, Aaron J. Berger, Antonio Subtil, Robert L. Camp, William R. Bradley, Stephan Ariyan, Harriet M. Kluger, David L. Rimm
Data analysis and interpretation: Bonnie E. Gould Rothberg, Aaron J. Berger, Annette M. Molinaro, Michael O. Krauthammer, Robert L. Camp, William R. Bradley, Harriet M. Kluger, David L. Rimm
Manuscript writing: Bonnie E. Gould Rothberg, Aaron J. Berger, Harriet M. Kluger, David L. Rimm
Final approval of manuscript: Bonnie E. Gould Rothberg, Harriet M. Kluger, David L. Rimm
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 3.Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: Validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 4.Gimotty PA, Elder DE, Fraker DL, et al. Identification of high-risk patients among those diagnosed with thin cutaneous melanomas. J Clin Oncol. 2007;25:1129–1134. doi: 10.1200/JCO.2006.08.1463. [DOI] [PubMed] [Google Scholar]
- 5.Taylor C. Standardization in immunohistochemistry: The role of antigen retrieval in molecular morphology. Biotech Histochem. 2006;81:3–12. doi: 10.1080/10520290600667866. [DOI] [PubMed] [Google Scholar]
- 6.Gimotty PA, Van Belle P, Elder DE, et al. Biologic and prognostic significance of dermal Ki67 expression, mitoses, and tumorigenicity in thin invasive cutaneous melanoma. J Clin Oncol. 2005;23:8048–8056. doi: 10.1200/JCO.2005.02.0735. [DOI] [PubMed] [Google Scholar]
- 7.Gould Rothberg BE, Bracken MB, Rimm DL. Tissue biomarkers for prognosis in cutaneous melanoma: A systematic review and meta-analysis. J Natl Cancer Inst. 2009;101:452–474. doi: 10.1093/jnci/djp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariyan S, Ariyan C, Farber LR, et al. Reliability of identification of 655 sentinel lymph nodes in 263 consecutive patients with malignant melanoma. J Am Coll Surg. 2004;198:924–932. doi: 10.1016/j.jamcollsurg.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 9.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 10.Kreizenbeck GM, Berger AJ, Subtil A, et al. Prognostic significance of cadherin-based adhesion molecules in cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2008;17:949–958. doi: 10.1158/1055-9965.EPI-07-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell M. An Introduction to Genetic Algorithms. Cambridge, MA: MIT Press; 1998. [Google Scholar]
- 13.Ooi CH, Tan P. Genetic algorithms applied to multi-class prediction for the analysis of gene expression data. Bioinformatics. 2003;19:37–44. doi: 10.1093/bioinformatics/19.1.37. [DOI] [PubMed] [Google Scholar]
- 14.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 15.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 16.Gimotty PA, Botbyl J, Soong SJ, et al. A population-based validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2005;23:8065–8075. doi: 10.1200/JCO.2005.02.4976. [DOI] [PubMed] [Google Scholar]
- 17.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 18.Viros A, Fridlyand J, Bauer J, et al. Improving melanoma classification by integrating genetic and morphologic features. PLoS Med. 2008;5:e120. doi: 10.1371/journal.pmed.0050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 20.Alonso SR, Ortiz P, Pollan M, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: A tissue microarray-based study. Am J Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straume O, Sviland L, Akslen LA. Loss of nuclear p16 protein expression correlates with increased tumor cell proliferation (Ki-67) and poor prognosis in patients with vertical growth phase melanoma. Clin Cancer Res. 2000;6:1845–1853. [PubMed] [Google Scholar]
- 22.Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 23.Evangelou K, Bramis J, Peros I, et al. Electron microscopy evidence that cytoplasmic localization of the p16(INK4A) “nuclear” cyclin-dependent kinase inhibitor (CKI) in tumor cells is specific and not an artifact: A study in non-small cell lung carcinomas. Biotech Histochem. 2004;79:5–10. doi: 10.1080/10520290310001659466. [DOI] [PubMed] [Google Scholar]
- 24.Keller-Melchior R, Schmidt R, Piepkorn M. Expression of the tumor suppressor gene product p16INK4 in benign and malignant melanocytic lesions. J Invest Dermatol. 1998;110:932–938. doi: 10.1046/j.1523-1747.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 25.Sauroja I, Smeds J, Vlaykova T, et al. Analysis of G(1)/S checkpoint regulators in metastatic melanoma. Genes Chromosomes Cancer. 2000;28:404–414. doi: 10.1002/1098-2264(200008)28:4<404::aid-gcc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Maelandsmo GM, Holm R, Fodstad O, et al. Cyclin kinase inhibitor p21WAF1/CIP1 in malignant melanoma: Reduced expression in metastatic lesions. Am J Pathol. 1996;149:1813–1822. [PMC free article] [PubMed] [Google Scholar]
- 27.Karjalainen JM, Eskelinen MJ, Kellokoski JK, et al. p21(WAF1/CIP1) expression in stage I cutaneous malignant melanoma: Its relationship with p53, cell proliferation and survival. Br J Cancer. 1999;79:895–902. doi: 10.1038/sj.bjc.6690143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhoumik A, Ronai Z. ATF2: A transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle. 2008;7:2341–2345. doi: 10.4161/cc.6388. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Deng X, Shyu YJ, et al. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. Embo J. 2006;25:1058–1069. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmann IM, Straume O, Puntervoll HE, et al. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11:8606–8614. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- 31.Maelandsmo GM, Holm R, Nesland JM, et al. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin Cancer Res. 2003;9:3383–3388. [PubMed] [Google Scholar]
- 32.Banerji A, Das S, Chatterjee A. Culture of human A375 melanoma cells in the presence of fibronectin causes expression of MMP-9 and activation of MMP-2 in culture supernatants. J Environ Pathol Toxicol Oncol. 2008;27:135–145. doi: 10.1615/jenvironpatholtoxicoloncol.v27.i2.60. [DOI] [PubMed] [Google Scholar]
- 33.Gaggioli C, Robert G, Bertolotto C, et al. Tumor-derived fibronectin is involved in melanoma cell invasion and regulated by V600E B-Raf signaling pathway. J Invest Dermatol. 2007;127:400–410. doi: 10.1038/sj.jid.5700524. [DOI] [PubMed] [Google Scholar]
- 34.Jaeger J, Koczan D, Thiesen HJ, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 36.Väisänen A, Kallioinen M, Taskinen PJ, et al. Prognostic value of MMP-2 immunoreactive protein (72 kD type IV collagenase) in primary skin melanoma. J Pathol. 1998;186:51–58. doi: 10.1002/(SICI)1096-9896(199809)186:1<51::AID-PATH131>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 37.Väisänen AH, Kallioinen M, Turpeenniemi-Hujanen T. Comparison of the prognostic value of matrix metalloproteinases 2 and 9 in cutaneous melanoma. Hum Pathol. 2008;39:377–385. doi: 10.1016/j.humpath.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Rangel J, Nosrati M, Torabian S, et al. Osteopontin as a molecular prognostic marker for melanoma. Cancer. 2008;112:144–150. doi: 10.1002/cncr.23147. [DOI] [PubMed] [Google Scholar]
- 39.Pacifico MD, Grover R, Richman PI, et al. Development of a tissue array for primary melanoma with long-term follow-up: Discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005;115:367–375. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 40.Pearl RA, Pacifico MD, Richman PI, et al. Stratification of patients by melanoma cell adhesion molecule (MCAM) expression on the basis of risk: Implications for sentinel lymph node biopsy. J Plast Reconstr Aesthet Surg. 2008;61:265–271. doi: 10.1016/j.bjps.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Rangel J, Torabian S, Shaikh L, et al. Prognostic significance of nuclear receptor coactivator-3 overexpression in primary cutaneous melanoma. J Clin Oncol. 2006;24:4565–4569. doi: 10.1200/JCO.2006.07.3833. [DOI] [PubMed] [Google Scholar]
- 42.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 43.Ascierto PA, Kirkwood JM. Adjuvant therapy of melanoma with interferon: Lessons of the past decade. J Transl Med. 2008;6:62. doi: 10.1186/1479-5876-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]