Abstract
Purpose
We evaluated the efficacy of gemcitabine versus gemcitabine and carboplatin in patients with advanced non–small-cell lung cancer (NSCLC) and a performance status (PS) of 2 and assessed if tumoral RRM1 and ERCC1 protein levels are predictive of response to therapy.
Patients and Methods
A randomized phase III trial was conducted in community-based oncology practices. Tumor specimens were collected a priori and shipped to a single laboratory for blinded determination of in situ RRM1 and ERCC1 protein expression levels by an automated quantitative immunofluorescent-based technology.
Results
One hundred seventy patients were randomly assigned. Overall median survival was 5.1 months for gemcitabine and 6.7 months for gemcitabine and carboplatin (P = .24). RRM1 (range, 5.3 to 105.6; median, 34.1) and ERCC1 (range, 5.2 to 131.3; median, 34.7) values were significantly and inversely correlated with disease response (r = −0.41; P = .001 for RRM1; r = −0.39; P = .003 for ERCC1; ie, response was better for patients with low levels of expression). A model for response prediction that included RRM1, ERCC1, and treatment arm, was highly predictive of the treatment response observed (P = .0005). We did not find statistically significant associations between survival and RRM1 or ERCC1 levels.
Conclusion
Single-agent chemotherapy remains the standard of care for patients with advanced NSCLC and poor PS. Quantitative analysis of RRM1 and ERCC1 protein expression in routinely collected tumor specimens in community oncology practices is predictive of response to gemcitabine and gemcitabine and carboplatin therapy. Oncologists should consider including in situ expression analysis for these proteins into their therapeutic decisions.
INTRODUCTION
The combination of two cytotoxic drugs, a platinum and a nonplatinum agent, is the standard of care for first-line treatment of patients with advanced non–small-cell lung cancer (NSCLC) and good performance status (PS, 0 to 1).1 This treatment yields response rates of approximately 25% to 30% and a median overall survival (OS) of 9 to 11 months in otherwise unselected patients.2–4 Because a high rate of toxicity has been observed with dual-agent therapy in patients with a poor PS (PS, 2), single-agent therapy has become an accepted standard. However, subgroup analyses of cooperative group trials have suggested improved survival with dual- versus single-agent therapy for this group of patients.5,6
Promising results on the utility of molecular parameters in predicting efficacy of systemic cytotoxic therapy in NSCLC have been reported. Two molecules involved in DNA synthesis and damage repair, ERCC1 and RRM1, have been associated with efficacy of platinum agents and gemcitabine.7–11 ERCC1 is a component of the nucleotide excision repair complex, and it is responsible for the 5′ incision required for the removal of DNA adducts that are the basis for platinum cytotoxicity.12 RRM1 is the molecular target of gemcitabine and a component of ribonucleotide reductase, which is required for deoxynucleotide production.13 However, with one exception,11 these studies have evaluated gene expression at the RNA level; and the relationship between cellular levels of RNA and protein, the molecule responsible for function, is tenuous.14,15 In addition, most of these studies have demonstrated differential survival by gene expression and not differences in response rates.7,8,10,11
We studied if significant survival differences exist between NSCLC patients with PS 2 treated with gemcitabine alone or gemcitabine and carboplatin, and the predictive utility of RRM1 and ERCC1 protein expression on therapeutic efficacy in tumor specimens collected under standard conditions in community oncology practices.
PATIENTS AND METHODS
Clinical Trial
A randomized phase III clinical trial (USO-03012) was designed and conducted in community oncology practices in patients with previously untreated stage IIIB/IV NSCLC,16 a PS of 2,17 and measurable disease by Response Evaluation Criteria in Solid Tumors (RECIST),18 (registration #: NCT00190710). Up to 6 cycles of therapy was given. gemcitabine and carboplatin was given every 3 weeks at doses of 1,000 mg/m2 of gemcitabine on days 1 and 8 and of carboplatin at an area under the curve of 5 on day 1. Gemcitabine was given every 3 weeks at a dose of 1,250 mg/m2 on days 1 and 8 (Appendix, online only).
Therapeutic Efficacy Assessment
Survival analyses were performed on an intent-to-treat basis. OS was the time interval between the dates of first treatment and death; progression-free survival (PFS) was the time interval between the dates of first treatment and disease progression or death, whichever occurred first. For patients who did not start treatment, the date of registration was used for survival calculations. The data lock occurred on February 28, 2008.
The sum of maximal diameters of all measurable tumor lesions was recorded at baseline and after treatment cycles 2, 4, and 6. These measurements were used to calculate the largest percentage of shrinkage or smallest percentage of growth using the baseline assessment as reference. The best response to therapy was categorized according to RECIST. Adverse events were assessed using National Cancer Institute's Common Terminology Criteria version 3.0.
Specimen Collection and Protein Expression Analysis
Tumor specimens from patients whose diagnosis was performed on histologic or equivalent cytological samples were shipped to a single central laboratory for biomarker analysis. Full specimen sections of 4-μm thickness were cut from paraffin blocks and mounted on adhesive coated or charged glass slides. A fluorescent-based immunohistochemical (IHC) method combined with automated quantitative analysis (AQUA) was used to determine in situ expression levels for RRM1 and ERCC1 as previously described.19 For ERCC1 detection, mouse clone 8F1 (1:300 dilution, lot #9475) produced by Exalpha Biologicals (Maynard, MA) and distributed by Sigma-Aldrich (St Louis, MO) was used. For RRM1 detection, a custom-made rabbit antiserum (R1AS-6b, 1:300 dilution) was used.15
For each patient, two full specimen sections were analyzed, one slide for RRM1 and one slide for ERCC1. For each slide, random spots (spot diameter 0.6 mm) ranging in number from 5 to 66 (median, 21) for ERCC1 and from 3 to 69 (median, 19) for RRM1 were scanned with SpotGrabber (HistoRx, New Haven, CT), and image data were analyzed with AQUA (PM-2000, version 1.2, score range 0 to 255, HistoRx).
Determination of Cross-Analysis-Adjusted RRM1 and ERCC1 Protein Expression Values
In order to adjust for variances in AQUA scores among tumor specimens analyzed in different runs, we incorporated a uniform calibration tissue microarray in each run. The array included three specimen replicates of 10 permanent human cell lines that displayed a range of expression for the target molecules. Representative RRM1 and ERCC1 scores from each experimental tumor specimen and the calibration specimens were obtained using quarter-root power transformations. Adjusted means were obtained for each experimental specimen by averaging their standardized quarter-rooted transformed raw scores. These adjusted means were used for all statistical analyses (Appendix).
Statistical Analyses
OS was the primary end point of the clinical trial. It was estimated that 220 eligible patients were required to detect an improvement from 2.4 months for gemcitabine to 3.8 months for gemcitabine and carboplatin (ie, a 36.8% reduction in the hazard rate of death, with a power of 0.90 and a type 1 error of 0.05). The OS targets were derived from the subset analysis of PS 2 patients in trial Cancer and Leukemia Group B 9730.5 The anticipated patient accrual time was 22 months. OS and PFS were estimated using the Kaplan-Meier method. Tumor response rates were calculated as the sum of the number of observed CRs and PRs divided by the number of patients qualifying for the efficacy analysis (ie, patients who met enrollment criteria and received at least one dose of study medication). Two-sided 95% CI were calculated based on the exact binomial probability.
The primary goal of the biomarker analysis was to determine if RRM1 and ERCC1 protein levels were predictive of best tumor response. First, we determined that no transformation was needed for values of best tumor response. Separate weighted linear regression models for ERCC1 and RRM1 were performed using best measured tumor response as the dependent variable and the adjusted biomarker means as the predictor variables. The number of ERCC1 scores (range, 5 to 66) and RRM1 scores (range, 3 to 69) per patient was used as the weight. Follow-up analyses included a weighted regression for ERCC1 and RRM1, including the study arm and a nonweighted regression including ERCC1, RRM1, and study arm.
In order to compare demographic and clinical features of the treatment arms within the clinical and biomarker groups, and to compare patients with and without biomarker data, the Wilcoxon rank sum test was used for continuous variables while the Fisher's exact test was used for categoric variables, both based on exact distribution theory. We used the maximal χ2 method and log-rank testing for optimal cut point determination with appropriate adjustments of the P values for multiple looks to evaluate OS and PFS differences.20 Only cutpoints within the central 80% of ordered RRM1 and ERCC1 levels were considered.
RESULTS
Clinical Trial
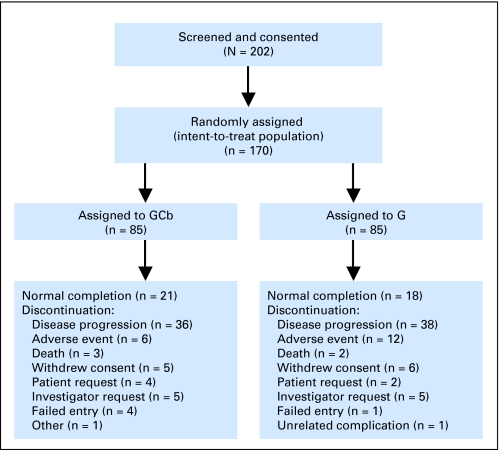
A total of 202 patients from 84 community-based oncology practices were consented between March 2004 and December 2006 (Fig 1). Patient accrual was 50% of the expected rate, and the trial was terminated. Thirty-two patients withdrew consent before randomization. Both arms, with 85 patients each, were demographically well balanced (Table 1). At least one dose of chemotherapy was given to 160 patients, 81 in the gemcitabine and 79 in the gemcitabine and carboplatin arm.
Fig 1.
CONSORT diagram. G, gemcitabine; GCb, gemcitabine and carboplatin.
Table 1.
Patients' Characteristics by Treatment Arm and Biomarker Analysis
| Characteristic | Gemcitabine and Carboplatin (n = 85) |
Gemcitabine (n = 85) |
P | Nonbiomarker Group (n = 101) |
Biomarker Group (n = 69) |
P | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | |||
| Age, years | .820* | .718* | ||||||||
| Median | 72.9 | 75.0 | 73.8 | 75.0 | ||||||
| Range | 46.3-88.4 | 45.2-89.7 | 45.2-89.7 | 49.4-85.5 | ||||||
| Sex | 1.000† | .275† | ||||||||
| Male | 47 | 48 | 60 | 35 | ||||||
| Female | 38 | 37 | 41 | 34 | ||||||
| Histology | .311† | .609† | ||||||||
| Adenocarcinoma | 53 | 45 | 55 | 43 | ||||||
| Squamous carcinoma | 14 | 22 | 23 | 13 | ||||||
| Other | 18 | 18 | 23 | 13 | ||||||
| Stage | .035† | .180† | ||||||||
| IV | 70 | 80 | 86 | 64 | ||||||
| IIIB | 13 | 5 | 14 | 4 | ||||||
| Unknown | 2 | 0 | 1 | 1 | ||||||
| Best measurable response by CT§ | .003† | .977† | ||||||||
| CR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PR | 29 | 43.9 | 10 | 16.4 | 21 | 31.3 | 18 | 30.0 | ||
| SD | 23 | 34.8 | 35 | 57.4 | 30 | 44.8 | 28 | 46.7 | ||
| PD | 14 | 21.2 | 16 | 26.2 | 16 | 23.9 | 14 | 23.3 | ||
| Best measurable response by CT | < .001† | 1.000† | ||||||||
| CR and PR | 29 | 43.9 | 10 | 16.4 | 21 | 31.3 | 18 | 30.0 | ||
| 95% CI | 24.2 to 54.2 | 8.2 to 28.1 | 20.6 to 43.8 | 18.9 to 43.2 | ||||||
| Best confirmed response category‖ | .050† | .668† | ||||||||
| CR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PR | 16 | 21.1 | 5 | 6.3 | 10 | 11.5 | 11 | 15.9 | ||
| SD | 34 | 44.7 | 38 | 47.5 | 39 | 44.8 | 33 | 47.8 | ||
| PD | 20 | 26.3 | 25 | 31.3 | 25 | 28.7 | 20 | 29.1 | ||
| Unknown | 6 | 7.9 | 12 | 15.0 | 13 | 14.9 | 5 | 7.2 | ||
| Best confirmed response | .010† | .240† | ||||||||
| CR and PR | 16 | 21.1 | 5 | 6.3 | 10 | 12.7 | 11 | 17.2 | ||
| 95% CI | 12.5 to 31.9 | 2.1 to 14.0 | 4.8 to 17.5 | 8.2 to 26.7 | ||||||
| OS, months | .242‡ | .312‡ | ||||||||
| Median | 6.7 | 5.1 | 4.9 | 6.0 | ||||||
| 95% CI | 4.9 to 10.0 | 3.9 to 6.3 | 3.9 to 6.7 | 5.1 to 8.4 | ||||||
| PFS, months | .143‡ | .698‡ | ||||||||
| Median | 3.8 | 2.7 | 2.6 | 3.1 | ||||||
| 95% CI | 2.6 to 4.6 | 1.9 to 3.6 | 2.0 to 4.1 | 2.8 to 4.3 | ||||||
Abbreviations: CT, computed tomography; CR, complete response; PR, progressive disease; SD, stable disease; PD, progressive disease; OS, overall survival; PFS, progression-free survival.
Wilcoxon rank-sum test.
Fisher's exact test.
Log-rank test.
Best measurable response by CT was available in 66 patients treated with gemcitabine and carboplatin and 61 patients treated with gemcitabine.
Best confirmed response was available in 70 patients treated with gemcitabine and carboplatin and 68 patients treated with gemcitabine.
The median number of cycles was 4 in the gemcitabine and carboplatin arm, 38% received 6 cycles, and it was 3 in the gemcitabine arm, 13% received 6 cycles. More patients received dose reductions in the gemcitabine and carboplatin arm (55.7% had ≥ 1 dose reduction of gemcitabine and 34.2% had ≥ 1 reduction of carboplatin in the gemcitabine and carboplatin arm versus 13.6% with ≥ 1 reduction in the gemcitabine arm). The median relative dose intensity of gemcitabine was 93.8% in the gemcitabine and carboplatin arm and 97.1% in the gemcitabine arm; and it was 96.5% for carboplatin in the gemcitabine and carboplatin arm.
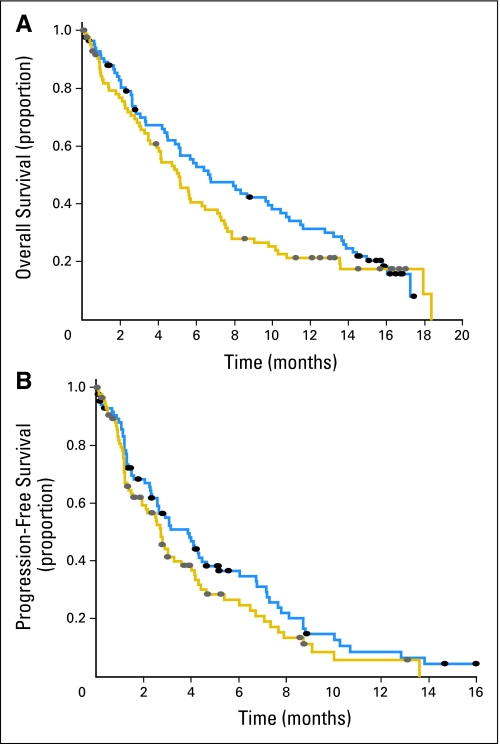
There was a statistically significantly higher confirmed response rate in the group of patients treated with gemcitabine and carboplatin (21.1%) compared with the group treated with gemcitabine (6.3%; P = .01). The best measurable response rates by CT were 43.9% and 16.4%, respectively (P < .001). However, the survival differences between the treatment arms were not statistically significant (median OS, 6.7; 95% CI, 4.9 to 10.0 v 5.1; 95% CI, 3.9 to 6.3 months; P = .24; median PFS, 3.8; 95% CI, 2.6 to 4.6; v 2.7; 95% CI, 1.9 to 3.6 months; P = .14). At 1 year, the OS rate was 31.3% for gemcitabine and carboplatin and 21.2% for gemcitabine (Fig 2).
Fig 2.
(A) Overall survival (OS) and (B) progression-free survival (PFS) estimates by treatment arm. (gold line) treatment with gemcitabine; (blue line) treatment with gemcitabine and carboplatin; (solid oval) censored patients. The differences in median OS and PFS were not statistically significant (log-rank test P = .14).
Common toxicities reported in the patients who received at least one dose of study treatment are summarized in Table 2. The rates of grade 3 or 4 cytopenias were notably higher in the gemcitabine and carboplatin arm compared with the gemcitabine arm. Grade 3 or 4 febrile neutropenia, however, did not occur in either treatment arm. Nonhematologic toxicities were not substantially different between treatment arms. The most common grade 2 or higher nonhematologic toxicities included fatigue, dyspnea, anorexia, and nausea, all of which occurred in more than 10% of patients.
Table 2.
Toxicity Summary
| Toxicity | Occurrences by Grade |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gemcitabine and Carboplatin (n = 79) |
Gemcitabine (n = 81) |
|||||||||||
| 2 |
3 |
4 |
2 |
3 |
4 |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Hematologic | ||||||||||||
| Neutropenia | 9 | 11.4 | 29 | 36.7 | 17 | 21.5 | 6 | 7.4 | 7 | 8.6 | 2 | 2.5 |
| Anemia | 19 | 24.1 | 12 | 15.2 | 0 | 0.0 | 19 | 23.5 | 5 | 6.2 | 1 | 1.2 |
| Thrombocytopenia | 5 | 6.3 | 14 | 17.7 | 21 | 26.6 | 3 | 3.7 | 3 | 3.7 | 0 | 0.0 |
| Febrile neutropenia | — | — | 0 | 0.0 | 0 | 0.0 | — | — | 0 | 0.0 | 0 | 0.0 |
| Nonhematologic | ||||||||||||
| Fatigue | 19 | 24.1 | 3 | 3.8 | 0 | 0.0 | 15 | 18.5 | 13 | 16.0 | 2 | 2.5 |
| Dyspnea | 9 | 11.4 | 12 | 15.2 | 1 | 1.3 | 9 | 11.1 | 15 | 18.5 | 1 | 1.2 |
| Anorexia | 11 | 13.9 | 1 | 1.3 | 0 | 0.0 | 14 | 17.3 | 2 | 2.5 | 0 | 0.0 |
| Nausea | 9 | 11.4 | 0 | 0.0 | 0 | 0.0 | 8 | 9.9 | 2 | 2.5 | 0 | 0.0 |
| Pneumonia | 0 | 0.0 | 5 | 6.3 | 0 | 0.0 | 2 | 2.5 | 6 | 7.4 | 1 | 1.2 |
| Dehydration | 4 | 5.1 | 1 | 1.3 | 0 | 0.0 | 4 | 4.9 | 4 | 4.9 | 0 | 0.0 |
| Diarrhea | 3 | 3.8 | 0 | 0.0 | 0 | 0.0 | 4 | 4.9 | 3 | 3.7 | 0 | 0.0 |
| Vomiting | 4 | 5.1 | 0 | 0.0 | 0 | 0.0 | 3 | 3.7 | 2 | 2.5 | 0 | 0.0 |
| Pleural effusion | 1 | 1.3 | 4 | 5.1 | 0 | 0.0 | 1 | 1.2 | 2 | 2.5 | 0 | 0.0 |
| Dizziness | 4 | 5.1 | 1 | 1.3 | 0 | 0.0 | 2 | 2.5 | 0 | 0.0 | 0 | 0.0 |
| Rash | 3 | 3.8 | 0 | 0.0 | 0 | 0.0 | 1 | 1.2 | 0 | 0.0 | 0 | 0.0 |
| Chest pain | 0 | 0.0 | 2 | 2.5 | 0 | 0.0 | 2 | 2.5 | 1 | 1.2 | 1 | 1.2 |
| Alopecia | 2 | 2.5 | — | — | — | — | 0 | 0.0 | — | — | — | — |
Tumor Specimens and Patient Characteristics
Specimens from 87 patients and 42 different study sites were received by the biomarker laboratory. Eight specimens had been stained, which resulted in 79 specimens useful for analysis. Specimens from 10 patients were inadequate to produce results because the specimen had washed off during the procedure or because of absence of tumor cells. RRM1 data were obtained on 69 and ERCC1 data on 65 patients (four specimens were exhausted after RRM1 analysis).
Of the 69 patients with protein expression data, 34 had been treated with gemcitabine and carboplatin and 35 with gemcitabine. There were no significant differences between the groups of patients with and without biomarker data (Table 1).
Predictive Impact of RRM1 and ERCC1 Levels on Measurable Tumor Response
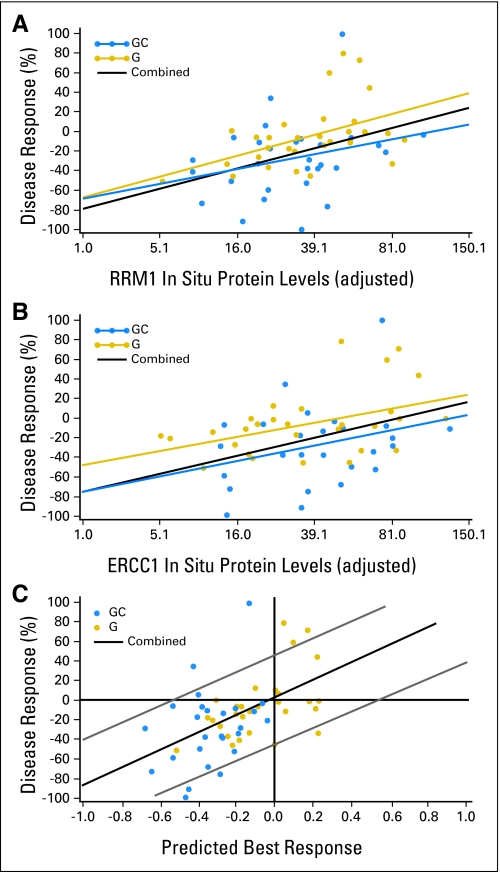
Since full section specimens from different patients were processed at separate time points, we standardized the biomarker values by incorporating calibration specimens in each analysis to adjust for inter-analysis variability. The adjusted values for RRM1 ranged from 5.3 to 105.6 (median, 34.1), and those for ERCC1 ranged from 5.2 to 131.3 (median, 34.7). They were not statistically significantly associated with patients' age, sex, tumor histology, and stage; and they were not significantly different between both treatment arms (Table 3). For comparison, the raw values ranged from 8.2 to 157.3 (median, 46.7) for RRM1 and from 8.2 to 115.2 (median, 40.2) for ERCC1. Best measurable responses ranged from a 100% reduction (−100%) to a 133% increase in tumor sizes. In situ RRM1 levels were significantly and inversely correlated with disease response (r = −0.41; P = .001; Fig 3A) in the 58 patients with both parameters available. Likewise, ERCC1 levels were significantly and inversely correlated with disease response (r = −0.39; P = .003) (Fig 3B) in the 55 patients with both parameters available. RRM1 and ERCC1 expression levels were significantly and positively correlated (r = 0.36; P = .003; N = 65).
Table 3.
RRM1 and ERCC1 Protein Levels by Clinical Characteristics
| Characteristic | Protein Level |
|||||
|---|---|---|---|---|---|---|
| RRM1 (n = 69) |
ERCC1 (n = 65) |
|||||
| Median | Range | P | Median | Range | P | |
| Age, years | .552 | .650 | ||||
| ≥ 75.0 | 33.4 | 8.5-105.6 | 34.7 | 5.2-131.3 | ||
| < 75.0 | 34.2 | 5.3-90.1 | 35.4 | 10.1-102.3 | ||
| Sex | .722 | .401 | ||||
| Male | 36.1 | 5.3-105.6 | 34.4 | 5.2-127.6 | ||
| Female | 33.2 | 8.6-93.5 | 47.3 | 11.5-131.3 | ||
| Histology | .570 | .893 | ||||
| Adeno | 36.9 | 5.3-105.6 | 39.9 | 5.2-131.3 | ||
| Squamous | 23.7 | 9.9-93.5 | 34.1 | 6.0-104.9 | ||
| Large cell and NOS | 34.1 | 14.7-90.1 | 27.9 | 11.5-127.6 | ||
| Stage | .312 | .029 | ||||
| IV | 35.1 | 5.3-105.6 | 34.4 | 5.2-127.6 | ||
| IIIB | 27.2 | 14.8-36.9 | 69.7 | 51.3-131.3 | ||
| Treatment | .196 | .610 | ||||
| Gemcitabine and carboplatin | 32.9 | 6.4-105.6 | 36.7 | 12.8-131.3 | ||
| Gemcitabine | 39.1 | 5.3-90.1 | 33.4 | 5.2-127.6 | ||
| Best measurable response category by CT | .023 | .402 | ||||
| PR | 22.9 | 5.3-81.4 | 34.6 | 10.1-84.7 | ||
| SD | 39.0 | 8.5-105.6 | 34.1 | 5.2-131.3 | ||
| PD | 42.2 | 15.2-89.4 | 51.8 | 18.7-102.3 | ||
NOTE. Wilcoxon rank sum test (with Van der Waerden quantile normal scores) for two groups and Kruskal-Wallis test for three groups. The χ2 rank test P value for associations of best response with RRM1 was < .01, and it was > .10 for best response with ERCC1.27
Abbreviations: NOS, not otherwise specified; CT, computed tomography; PR, partial response; SD, stable disease; PD, progressive disease.
Fig 3.
(A) RRM1 in situ protein levels, (B) ERCC1 in situ protein levels, and (C) predicted tumor response by combining both markers and treatment arm into a model displayed on the x-axis in relation to tumor response displayed on the y-axis (negative values denote tumor shrinkage and positive values tumor growth; [gold circle] treatment with gemcitabine [G]; [blue circle] G and carboplatin [GC]; [black line] both combined: [A] rho = −0.41, P = .001; [B] ρ = −0.39, P = .003; [C] ρ = 0.54, P = .0005).
We evaluated if an interaction between RRM1 or ERCC1 levels and treatment arm existed (ie, if RRM1 or ERCC1 had a differential predictive impact in the gemcitabine or gemcitabine and carboplatin treatment arms). We did not find a statistically significant interaction (P = .64 for RRM1; P = .79 for ERCC1). However, the slope of the best-fit line between RRM1 values and disease response was steeper (0.47) than the ERCC1 response slope (0.34) in the gemcitabine arm (Fig 3A). This difference was less notable in the gemcitabine and carboplatin arm (0.35 for RRM1; 0.29 for ERCC1; Fig 3B).
We generated a model for response prediction that included RRM1, ERCC1, and treatment arm. The predicted response based on the model was positively correlated (r = 0.54; P = .0005) with the observed response (Fig 3C). Of the 42 patients predicted to have a reduction in tumor size as best response, 38 did have an observed reduction (positive predictive value = 0.90); whereas 6 of 13 tumors predicted to increase in size actually grew (negative predictive value = 0.46).
Impact of RRM1 and ERCC1 Levels on Survival
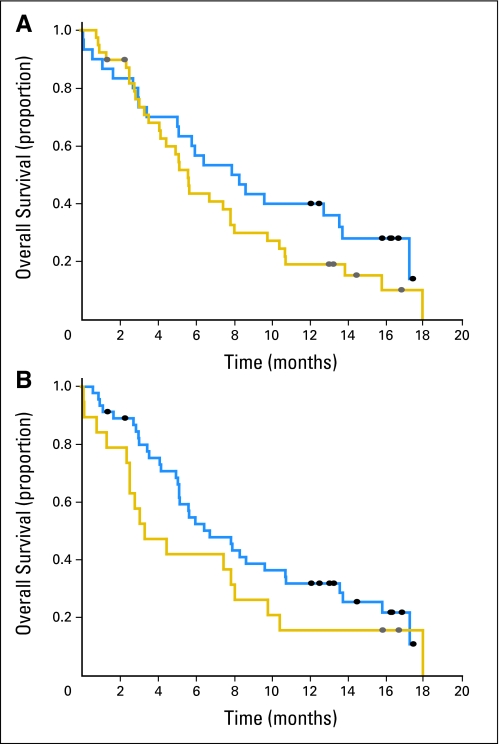
We analyzed if RRM1 or ERCC1 protein levels would be associated with patients' survival using an optimal cut point method. The median OS for the 30 patients with RRM1 levels below 32.1 was 8.2 months (95% CI, 5.1 to 13.6), and it was 5.6 months (95% CI, 4.1 to 7.9) for the 39 patients with RRM1 levels above 32.1 (Fig 4A). The median OS for the 46 patients with ERCC1 levels below 65.0 was 6.8 months (95% CI, 5.2 to 10.8), and it was 3.4 months (95% CI, 2.6 to 8.1) for the 19 patients with ERCC1 levels above this value (Fig 4B). These differences were not statistically significant (P = .16 for RRM1; P = .19 for ERCC1; P values adjusted for multiple looks). We did also not find a statistically significant difference in PFS for RRM1 or ERCC1 between patients with high and low expression levels (median PFS, 4.0 and 3.0 months for low and high RRM1, P = .31; median PFS, 3.6 and 2.9 months for low and high ERCC1, P = .53). A Cox regression analysis also produced statistically nonsignificant associations between survival and RRM1 or ERCC1.
Fig 4.
(A) Differential overall survival for patients dichotomized into high and low groups using the optimal RRM1 level of 32.1 as cut point (P = .16; log-rank test with Miller-Siegmund adjustment).20 (B) Differential survival for patients dichotomized into high and low groups using the optimal ERCC1 level of 65.0 as cut point (P = .19; log-rank test with Miller-Siegmund adjustment).20 (gold line) high protein levels; (blue line) low protein levels.
DISCUSSION
The primary objective of the clinical trial was to demonstrate superior OS for gemcitabine and carboplatin over gemcitabine in patients with advanced NSCLC and PS 2. Although there was a trend toward longer survival in the group of patients treated with gemcitabine and carboplatin compared with gemcitabine, this difference was not statistically significant. We thus conclude that single-agent chemotherapy remains the standard of care for this group of patients. However, the possibility exists that this result may be spurious. Reasons may be early closure of the trial and a higher than anticipated survival in the single-agent treatment arm.
We based our a priori survival estimates on a published subgroup analysis of a randomized phase III trial that reported a median OS of 2.4 months (Cancer and Leukemia Group B 9730) in 50 patients with PS 2 treated with single-agent paclitaxel.5 This estimate was clearly too low as evidenced by our result (5.1 months), a prior single-arm trial of gemcitabine in a similar group of patients (4.8 months),21 a PS 2 subgroup analysis of a randomized phase III Italian trial (3.5 to 4.2 months in 86 patients treated with single-agent vinorelbine or gemcitabine),22 and data from a randomized phase II trial in PS 2 patients showing a median OS of 4.8 months for single-agent gemcitabine.23 It is conceivable that the survival of patients enrolled in trials specifically designed for those with PS 2 may be better than the survival observed in PS 2 subgroup patients from a trial open to patients with PS 0 to 2 as a result of a biased interpretation of the PS classification by enrolling physicians.
It is noteworthy that the survival of patients with PS 2 receiving dual-agent therapy was approximately 2 months longer than the survival of patients receiving single-agent therapy in the three referenced studies5,22,23 and our study, which suggests that a lack of statistical significance in the OS differences may have been caused by the small sample sizes.
Another objective of the trial was to evaluate the predictive utility of RRM1 and ERCC1 protein levels on tumor response to gemcitabine and gemcitabine and carboplatin. Both molecules are promising predictive markers because of their demonstrated impact on gemcitabine and platinum efficacy in experimental models.9,24 Two earlier clinical investigations reported a significant association between RRM1 mRNA levels and disease response.9,24 Both studies had small patient numbers, 35 and 18 respectively, and both utilized specimen procurement conditions that required a sophisticated infrastructure. Two additional studies had reported a significantly better survival for patients with low RRM1 mRNA levels compared with patients with high levels.8,10 Similarly, low ERCC1 mRNA levels were associated with disease response to platinum-based therapy in lung, ovarian, and colorectal cancer.7,9,25,26 Finally, high tumoral ERCC1 levels were associated with lack of benefit from adjuvant cisplatin-based therapy in patients with resected NSCLC.11
Our data demonstrate that in situ RRM1 and ERCC1 protein levels in tumor specimens collected under routine conditions in community oncology practices are significantly predictive of disease response in patients with advanced NSCLC. The result that ERCC1 protein levels are predictive of response to gemcitabine alone can be explained by the highly significant association between ERCC1 and RRM1 levels.8–10,15 In addition, the mechanism of gemcitabine cytotoxicity is not only mediated through targeting RRM1, but also involves direct interference with DNA synthesis through incorporation of gemcitabine, a cytosine analog.27 It is thus possible that ERCC1, through its role in nucleotide excision repair, may impact on gemcitabine efficacy. In addition, we found that the integration of both markers into a model for response prediction was significantly correlated with the observed disease response, having a positive predictive value of 0.90.
The lack of association of RRM1 levels with patients' age, sex, and histology is consistent with prior reports.9,10,15 Likewise, most studies found no significant associations between ERCC1 levels and these parameters.7,9,10,15 However, Olaussen et al11 reported that ERCC1 levels were significantly higher in older patients, men, and squamous cell carcinomas. This discrepancy is possibly explained by the relatively low number of patients included in our study (65 patients with ERCC1 levels) and the other reported studies (between 35 and 184 patients) compared with the study by Olaussen et al (761 patients).
We conclude that single-agent chemotherapy remains the standard of care in patients with poor PS NSCLC. We further conclude that RRM1 and ERCC1 are predictive markers of gemcitabine and gemcitabine and carboplatin efficacy, and that a determination of protein expression levels of both markers in routinely collected specimens from patients with advanced NSCLC in community oncology practices is feasible in approximately half of all patients without requiring a change practice patterns. Future trials that use gemcitabine or a platinum-agent should incorporate RRM1 and ERCC1 expression levels into the analysis plan. In addition, large prospective trials that further validate and refine the predictive utility of both markers are desirable.
Appendix
Detailed Methodology
Clinical trial.
A randomized phase III clinical trial (USO-03012) was designed and conducted in community oncology practices across the US Oncology network in patients with advanced stage non–small-cell lung cancer (NSCLC) and a performance score (PS) of 2. The trial was performed in accordance with principles of good clinical practice, applicable laws and regulations, and the Declaration of Helsinki. Investigators were responsible for obtaining written informed consent from participating patients before treatment initiation and approval from study sites' ethical review boards. The trial had two primary objectives. One was to assess if OS would be superior in patients treated with gemcitabine and carboplatin compared with gemcitabine. The other was to assess if tumoral expression levels of RRM1 and ERCC1 are predictive of response to therapy. A PS of 2 was defined as disease-related symptoms that impact normal daily living activities but with the patient being out of bed or a recliner for more than 50% of the waking hours.17 Eligibility included histologically or cytologically newly diagnosed NSCLC, stage IIIB or stage IV,16 measurable disease by Response Evaluation Criteria in Solid Tumors Group,18 no prior chemotherapy, age ≥ 18 years, and adequate organ function. gemcitabine and carboplatin was given every 3 weeks at doses of 1,000 mg/m2 of gemcitabine on days 1 and 8 and of carboplatin at an area under the curve of 5 on day 1. Gemcitabine was given every 3 weeks at a dose of 1,250 mg/m2 on days 1 and 8. Up to six cycles of therapy were planned unless there was evidence for disease progression or intolerable toxicity.
Specimen collection and protein expression analysis.
Tumor specimens from patients whose diagnosis was performed on histologic specimens or equivalent cytologic specimens, such as cell blocks prepared from a pleural or pericardial effusion, were processed by routine fixation and paraffin embedding. The specimens were firstly used to establish the diagnosis. The protocol did allow for any method of tissue fixation and processing according to standard operating procedures at the local institution. Remaining material was shipped as paraffin blocks or sections on slides to a single central laboratory for biomarker analysis. Full specimen sections of 4-μm thickness were cut from paraffin blocks and mounted on adhesive coated or charged glass slides. Scant cytologic specimens were only used for diagnostic confirmation at the local institution; they were not shipped to the central laboratory.
Determination of cross-analysis–adjusted RRM1 and ERCC1 protein expression values.
In order to adjust for variances in AQUA scores among tumor specimens analyzed in different runs, we incorporated a uniform calibration tissue microarray in each run. The array included triplicate 0.6 mm in diameter measuring specimens of four permanent human cell lines derived from NSCLC, three from colon cancer, and three from breast cancer. The cell lines had been chosen because they displayed a range of expression for the target molecules, and they had been harvested in logarithmic growth phase, embedded in low-melt agarose to simulate tissue, and 10% v/v neutral-buffered formalin fixed and paraffin embedded.
To adjust for potential experimental run effects, we studied the raw, quantitative RRM1 and ERCC1 expression scores in these 10 cell lines using 34 and 39 separate experimental runs for ERCC1 and RRM1, respectively. Spearman correlation analysis produced a correlation coefficient of 0.82 (range, 0.63 to 0.91) for ERCC1 and 0.74 (range, 0.43 to 0.94) for RRM1.
We then determined the most appropriate equation to calculate a representative biomarker score from the multiple expression scores obtained from each experimental tumor specimen and the calibration specimens. A power transformation approach was used with one eighth multiples between 0 (logarithmic transformation) and 1 considered as possible powers (except that one third was used in lieu of three eighths). The optimal transformation was deemed to be the one with the smallest non-negative skewness. Using this criterion, a quarter-root transformation was the best choice.
To adjust the individual tumors' biomarker scores across the multiple runs, the means of the transformed scores from the calibration specimens from each experimental run were obtained. These means were then back-transformed to a raw score. Mathematically this step can be summarized as: Mk = {Σ (xijk)0.25/(3 × 10)}4 where xijk is the ith measurement (each calibration sample in triplicate) from the jth calibration sample in the kth experimental run, and the summation is over i and j. This was done separately for ERCC1 and RRM1, and the individual multiple raw scores from the experimental specimens were then standardized by multiplication with 25/Mk and 50/Mk respectively; these numerators brought the range of adjusted scores close to the range of raw scores observed in the experimental runs. Then, adjusted means were obtained for each experimental specimen by averaging their standardized quarter-rooted transformed raw scores. These adjusted means were used for all statistical analyses.
Footnotes
Supported in part by Grant No. R01 CA102726 from the National Cancer Institute and by Eli Lilly and Company; the Moffitt Cancer Center and G.B. have a licensing agreement with Genzyme Corporation for the use of RRM1 and ERCC1 for response prediction.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00190710.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Coleman Obasaju, Eli Lilly (U); John R. Caton, US Oncology (U); Linda C. DeMarco, US Oncology (C); Mark A. O'Rourke, US Oncology (C); Gail Shaw Wright, US Oncology (C); Kristi A. Boehm, US Oncology (C); Lina Asmar, US Oncology (C); Jane Bromund, Eli Lilly (C); Guangbin Peng, Eli Lilly (C); Matthew J. Monberg, Eli Lilly (C) Consultant or Advisory Role: Gerold Bepler, Eli Lilly (C), Genmab (C) Stock Ownership: Coleman Obasaju, Eli Lilly Honoraria: None Research Funding: Craig Reynolds, Eli Lilly; Gerold Bepler, Eli Lilly, sanofi-aventis Expert Testimony: None Other Remuneration: Gerold Bepler, Genzyme Corporation
AUTHOR CONTRIBUTIONS
Conception and design: Craig Reynolds, Coleman Obasaju, Gerold Bepler
Financial support: Craig Reynolds, Coleman Obasaju, Gerold Bepler
Administrative support: Craig Reynolds, Coleman Obasaju, Gerold Bepler
Provision of study materials or patients: Craig Reynolds, Coleman Obasaju, Xueli Li, Zhong Zheng, John R. Caton, Linda C. DeMarco, Mark A. O'Rourke, Gail Shaw Wright, Kristi A. Boehm, Gerold Bepler
Collection and assembly of data: Craig Reynolds, Coleman Obasaju, Xueli Li, Zhong Zheng, Lina Asmar, Jane Bromund, Guangbin Peng, Gerold Bepler
Data analysis and interpretation: Craig Reynolds, Coleman Obasaju, Michael J. Schell, David Boulware, Gerold Bepler
Manuscript writing: Craig Reynolds, Coleman Obasaju, Michael J. Schell, Matthew J. Monberg, Gerold Bepler
Final approval of manuscript: Craig Reynolds, Coleman Obasaju, Michael J. Schell, Xueli Li, Zhong Zheng, David Boulware, John R. Caton, Linda C. DeMarco, Mark A. O'Rourke, Gail Shaw Wright, Kristi A. Boehm, Lina Asmar, Jane Bromund, Guangbin Peng, Matthew J. Monberg, Gerold Bepler
REFERENCES
- 1.NCCN. Non-Small Cell Lung Cancer: NCCN Clinical Practice Guidelines in Oncology V. 2.2008, 2008. www.nccn.org.
- 2.Kelly K, Crowley J, Bunn PA, Jr, et al. Randomized phase III trial of paclitaxel plus carboplatin versus vinorelbine plus cisplatin in the treatment of patients with advanced non–small-cell lung cancer: A Southwest Oncology Group trial. J Clin Oncol. 2001;19:3210–3218. doi: 10.1200/JCO.2001.19.13.3210. [DOI] [PubMed] [Google Scholar]
- 3.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 4.Fossella F, Pereira JR, von Pawel J, et al. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: The TAX 326 study group. J Clin Oncol. 2003;21:1–9. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 5.Lilenbaum R, Herndon JE, List MA, et al. Single-agent versus combination chemotherapy in advanced non-small-cell lung cancer: The Cancer and Leukemia Group B (study 9730) J Clin Oncol. 2005;23:190–196. doi: 10.1200/JCO.2005.07.172. [DOI] [PubMed] [Google Scholar]
- 6.Langer C, Li S, Schiller JH, et al. Randomized phase II trial of paclitaxel plus carboplatin or gemcitabine plus cisplatin in Eastern Cooperative Oncology Group performance status 2 non-small-cell lung cancer patients: ECOG 1599. J Clin Oncol. 2007;25:418–423. doi: 10.1200/JCO.2005.04.9452. [DOI] [PubMed] [Google Scholar]
- 7.Lord RVN, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 8.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 9.Bepler G, Kusmartseva I, Sharma S, et al. RRM1-modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 10.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 11.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 12.Volker M, Mone MJ, Karmakar P, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Molec Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 13.Stubbe J. Ribonucleotide reductases in the twenty-first century. Proc Natl Acad Sci U S A. 1998;95:2723–2724. doi: 10.1073/pnas.95.6.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Z, Chen T, Li X, et al. The DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 16.American Joint Committee on Cancer. Staging Handbook. ed 6. New York, NY: Springer-Verlag; 2002. [Google Scholar]
- 17.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. [PubMed] [Google Scholar]
- 18.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organisation for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 19.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 20.Miller R, Siegmund D. Maximally selected chi-square statistics. Biometrics. 1982;38:1011–1016. [Google Scholar]
- 21.Neubauer MA, Reynolds CH, Joppert MG, et al. Results of a phase II trial of gemcitabine in patients with non-small-cell lung cancer and a performance status of 2. Clin Lung Cancer. 2005;6:245–249. doi: 10.3816/clc.2005.n.004. [DOI] [PubMed] [Google Scholar]
- 22.Perrone F, Di Maio M, Gallo C, et al. Outcome of patients with a performance status of 2 in the Multicenter Italian Lung Cancer in the Elderly Study (MILES) J Clin Oncol. 2004;22:5018–5020. doi: 10.1200/JCO.2004.04.370. [DOI] [PubMed] [Google Scholar]
- 23.Kosmidis PA, Dimopoulos MA, Syrigos K, et al. Gemcitabine versus gemcitabine-carboplatin for patients with advanced non-small cell lung cancer and a performance status of 2: A prospective randomized phase II study of the Hellenic Cooperative Oncology Group. J Thorac Oncol. 2007;2:135–140. [PubMed] [Google Scholar]
- 24.Nakahira S, Nakamori S, Tsujie M, et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120:1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 25.Dabholkar M, Vionnet J, Bostick-Bruton F, et al. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest. 1994;94:703–708. doi: 10.1172/JCI117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–4304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 27.Huang P, Chubb S, Hertel LW, et al. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]