Abstract
Purpose
O6-methylguanine-methyltransferase (MGMT) promoter methylation has been shown to predict survival of patients with glioblastomas if temozolomide is added to radiotherapy (RT). It is unknown if MGMT promoter methylation is also predictive to outcome to RT followed by adjuvant procarbazine, lomustine, and vincristine (PCV) chemotherapy in patients with anaplastic oligodendroglial tumors (AOT).
Patients and Methods
In the European Organisation for the Research and Treatment of Cancer study 26951, 368 patients with AOT were randomly assigned to either RT alone or to RT followed by adjuvant PCV. From 165 patients of this study, formalin-fixed, paraffin-embedded tumor tissue was available for MGMT promoter methylation analysis. This was investigated with methylation specific multiplex ligation-dependent probe amplification.
Results
In 152 cases, an MGMT result was obtained, in 121 (80%) cases MGMT promoter methylation was observed. Methylation strongly correlated with combined loss of chromosome 1p and 19q loss (P = .00043). In multivariate analysis, MGMT promoter methylation, 1p/19q codeletion, tumor necrosis, and extent of resection were independent prognostic factors. The prognostic significance of MGMT promoter methylation was equally strong in the RT arm and the RT/PCV arm for both progression-free survival and overall survival. In tumors diagnosed at central pathology review as glioblastoma, no prognostic effect of MGMT promoter methylation was observed.
Conclusion
In this study, on patients with AOT MGMT promoter methylation was of prognostic significance and did not have predictive significance for outcome to adjuvant PCV chemotherapy. The biologic effect of MGMT promoter methylation or pathogenetic features associated with MGMT promoter methylation may be different for AOT compared with glioblastoma.
INTRODUCTION
Expression of the DNA repair protein O6-methylguanine-methyltransferase (MGMT, previously known as alkyltransferase) results in resistance of tumors to alkylating and methylating agents.1 Epigenetical silencing by methylation of the promoter of the MGMT gene located on chromosome 10q26 results in loss of MGMT expression, which potentially renders cells vulnerable for methylating and alkylating chemotherapy. Studies have shown improved survival of patients with glioblastoma without MGMT expression or with epigenetically silenced MGMT treated with radiotherapy (RT) and alkylating chemotherapy.2,3 A recent European Organisation for the Research and Treatment of Cancer (EORTC) study on combined chemoirradiation with temozolomide in glioblastoma showed improved survival with the addition of temozolomide to 60 Gy of RT.4 In that study, it was also shown that the improved outcome after temozolomide treatment was in particular associated with the presence of methylated MGMT promoter in the tumor.5 Because of the modest survival improvement of the patients without a methylated MGMT treated with RT plus temozolomide (best visible when using PFS as end point and no confounding effect of with second-line therapy), this study strongly suggested that methylation of the MGMT promoter could be used to predict which glioblastoma patients benefit from combined chemoirradiation. Since these reports, several studies on newly diagnosed glioblastomas treated with alkylating or methylating chemotherapy have confirmed the major prognostic significance of MGMT promoter methylation status.6 However, in the absence of a RT control arm, these studies do not allow formal assessment of the actual predictive value of MGMT promoter methylation status for outcome to chemotherapy.7 A prospective validation of the predictive value of MGMT promoter methylation is ethically no longer feasible. At present, supportive data for this predictive value are only available from retrospective studies evaluating patients with glioblastomas treated before the addition of alkylating agent therapy became standard of care.8,9 Furthermore, there are no data yet on the impact of MGMT promoter methylation from controlled studies on anaplastic oligodendroglial tumors (AOD).
The randomized controlled EORTC study 26951 on patients with AOD or anaplastic oligoastrocytoma (AOA) investigated the addition of six cycles of adjuvant standard procarbazine, lomustine, and vincristine (PCV) chemotherapy subsequent to radiotherapy with 59.4 Gy in 33 fractions.10 This study showed that the addition of PCV chemotherapy to RT improves progression-free survival (PFS) but not overall survival (OS). In this study, we investigated the correlation between MGMT methylation status and outcome to therapy of patients from this EORTC cohort. To evaluate the methylation status of multiple CpG dinucleotides in the MGMT promoter, we applied semi-quantitative methylation specific-multiplex ligation-dependent probe amplification (MS-MLPA) analysis, which correlates well with methylation-specific polymerase chain reaction (MS-PCR).11
PATIENTS AND METHODS
Patients were eligible for EORTC study 26951 if they had been diagnosed by the local pathologist with AOD or AOA with at least 25% oligodendroglial elements according to the 1994 edition of the WHO classification of brain tumors12; had at least three of five anaplastic characteristics (high cellularity, mitoses, nuclear abnormalities, endothelial proliferation, or necrosis); were between 16 and 70 years old; had an Eastern Cooperative Oncology Group performance status (PS) of 0 to 2 and had not undergone prior chemotherapy or RT to the skull. Clinical and molecular details of this study have been published elsewhere.10,13 All molecular studies were performed using selected areas enriched for a high tumor cell percentage. DNA was extracted from formalin-fixed, paraffin-embedded tissues as previously described.14 Loss of chromosomal arms 1p and 19q was analyzed with fluorescent in situ hybridization with locus specific probes, using probes to 1p36 (D1S32), centromere 1 (pUC1.77), 19p (equivalent amounts of BAC RPCI 11-959O6, −957I1, and −153P24), 19q (BAC 426G3) as described elsewhere.13 For statistical analysis, results on MGMT promoter methylation were correlated to clinical characteristics (age, PS, involved lobe [frontal v other]), molecular features (polysomy chromosome 7, EGFR amplification, loss of chromosome 1p/19q, loss of chromosome 10, loss of chromosome 10q), and histologic features (diagnosis [pure v mixed], and presence or absence of necrosis and endothelial proliferation). PFS and OS were measured from the day of random assignment. Patients provided written informed consent according to national and local regulations for this study.
MS-MLPA for Analysis of MGMT Promoter Methylation
The methylation status of the MGMT promoter was assessed by MS-MLPA analysis using the assay ME011 (MRC Holland, Amsterdam, the Netherlands) as described elsewhere.11 Briefly, the MLPA kit contains eight control probe sequences and 21 methylation-sensitive probes of which three recognize CpG dinucleotides within the MGMT promoter (MGMT 1: 2239-L1261; MGMT 2: 5670-L5146; MGMT 3: 7188-L5144). The methylation-sensitive probes contain a restriction site for HhaI, which only digests unmethylated DNA. Comparison of a digested DNA sample (yielding only signal of methylated DNA) to its undigested counterpart (yielding signal of both methylated and unmethylated DNA; ie, total DNA) provides insight into the degree of methylation. After hybridization of the probe mix to the tumor DNA, the sample is split in two parts. One is subjected to a simple ligation step joining both adjacently hybridized fragments of a probe set, whereas for the other part of the sample ligation is combined with a HhaI digestion leaving only the methylated sequences intact. Subsequent PCR amplification exponentially amplifies all ligated, but undigested, probes. The signal obtained with the part of the sample that has been subjected to both ligation and digestion represents the amount of methylated DNA present in the tumor. For fragment analysis, PCR products were separated by capillary gel electrophoresis (ABI PRISM 3130 × l, Applied Biosystems, Foster City, CA) and quantified using GeneMarker software version 1.7 (SoftGenetics, State College, PA). The MS-MLPA results were normalized by dividing the peak height of each MGMT probe signal by the mean peak height of the eight control fragments within the same sample. To estimate the degree of methylation, normalized values of each MGMT probe within digested DNA samples were divided by normalized values of corresponding undigested samples. This results in methylation ratios for the individual MGMT probes which were averaged (MGMTav). Methylation analyses were performed in duplicate or triplicate and the average ratios of each experiment and for each probe were calculated. For all analyses with outcome, the MGMTav score was used. Analyses were done with MGMTav as a binary variable (using the manufacturer cutoff MGMTav > 0.25 considered as indicative of methylation) and as a continuous variable.
Statistical Analysis
Kaplan-Meier technique was used to estimate PFS and OS. The prognostic significance of the MGMT 1, MGMT2, MGMT3 (MGMT1-3), and MGMTav for PFS and OS were first univariate analyzed. For multivariate analysis, the following major prognostic clinical variables were used: type of surgery (resection or biopsy), WHO performance status (0 to 2); age (< 50, ≥ 50), location (frontal v nonfrontal), the central diagnosis (AOD or AOA), endothelial abnormalities, necrosis, and the molecular factors combined 1p/19q loss, EGFRamp, CHR7poly, CHR10loss, and CHR10qloss. Association between factors except for PS was assessed by the Spearman correlation coefficient; Fisher's exact test was used for inference. For PS (score 0 to 2), the Wilcoxon rank sum test was used. Survival analyses were performed with the log-rank test and the Cox regression analysis with and without forward stepwise selection (5% confidence). Peto's technique was used for interaction tests. Internal validation was performed by bootstrap resampling technique (5% confidence) to assess the generalizability of the models. Factors with a probability of inclusion in regression models of less than 60% based on 1,000 bootstrap samples were considered not confirmed as independent prognostic factor. This analysis was purely exploratory and no adjustment for multiplicity was performed.
RESULTS
A total of 368 patients were included in the clinical trial. Sufficient formalin-fixed, paraffin-embedded tumor tissue was available for MGMT promoter analysis for 165 patients and reliable MS-MLPA results were obtained from 152 cases. In all cases, the MS-MLPA assay was run at least twice, in some three times. However, in 22 cases only one result was obtained (in 118 two assessments were available and in 12 three assessments were available). The MGMTav (but also the results of the individual probes MGMT 1 to 3) showed a continuous distribution without a clear clustering of results. Spearman correlation coefficients between MGMT 1 to 3 varied between 0.32 and 0.61. Test-retest variability for MGMTav was good, with Spearman correlation coefficients between 0.89 and 0.95.
The baseline and survival characteristics in these 152 patients were similar to those of the entire study cohort. Table 1 summarizes the baseline characteristics of these 152 patients. At central pathology review 86 of these 152 patients had been diagnosed as AOD, 37 with as AOA, nine as a low-grade glioma, 17 as high-grade astrocytic glioma, and in three patients other diagnoses were made. Thirty-nine of 152 tumors showed 1p/19q codeletion; in eight no 1p/19q result could be obtained. In 121 of 152 cases (80%), the MS-MLPA average was consistent with methylation (cutoff > 0.25), which included 38 of 39 1p/19q codeleted samples (97%). Of all available molecular (combined 1p/19q loss, EGFRamp, CHR7poly, CHR10loss, and CHR10qloss) and histological features, MGMTav was only correlated to the presence or absence of 1p/19q codeletion (Fisher's test P < .001).
Table 1.
Baseline Characteristics of the Patients With Successful MGMT Analysis (n = 152)
| Characteristic | No. | % |
|---|---|---|
| Treatment | ||
| RT/PCV | 84 | 55.3 |
| RT | 68 | 44.7 |
| Surgery | ||
| Resection | 133 | 87.5 |
| Biopsy | 19 | 12.5 |
| Performance status | ||
| 0 | 58 | 38.2 |
| 1 | 70 | 46.1 |
| 2 | 21 | 13.8 |
| Missing | 3 | 2.0 |
| Age, years | ||
| < 50 | 86 | 56.6 |
| ≥ 50 | 66 | 43.4 |
| Location | ||
| Other | 79 | 52.0 |
| Frontal | 73 | 48.0 |
| 1p/19q | ||
| No loss | 105 | 69.1 |
| Loss | 39 | 25.7 |
| Missing | 8 | 5.3 |
| EGFR amplification | ||
| No | 106 | 69.7 |
| Yes | 32 | 21.1 |
| Missing | 14 | 9.2 |
| Trisomy 7 | ||
| No | 94 | 61.8 |
| Yes | 42 | 27.6 |
| Missing | 16 | 10.5 |
| 10q loss | ||
| No | 123 | 80.9 |
| Yes | 18 | 11.8 |
| Missing | 11 | 7.2 |
| 10 loss | ||
| No | 119 | 78.3 |
| Yes | 21 | 13.8 |
| Missing | 12 | 7.9 |
Abbreviations: MGMT, O6-methylguanine-methyltransferase; RT, radiotherapy; PCV, procarbazine, lomustine, and vincristine; EGFR, epidermal growth factor receptor.
Survival Analysis
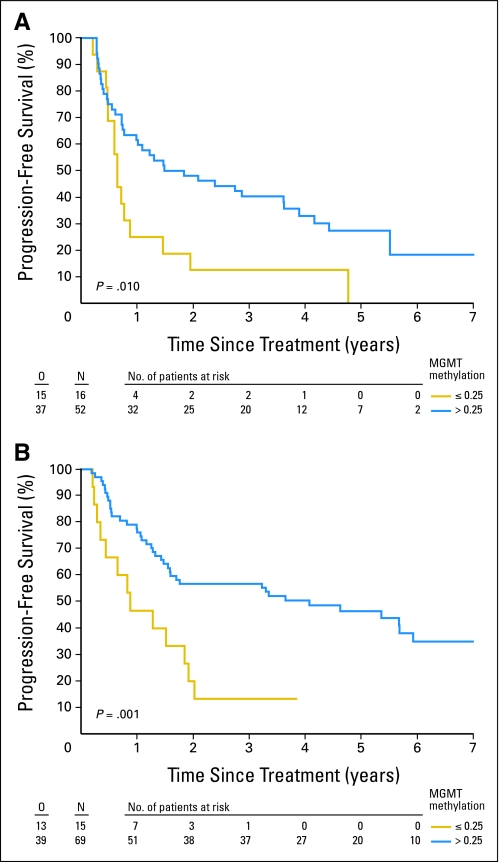
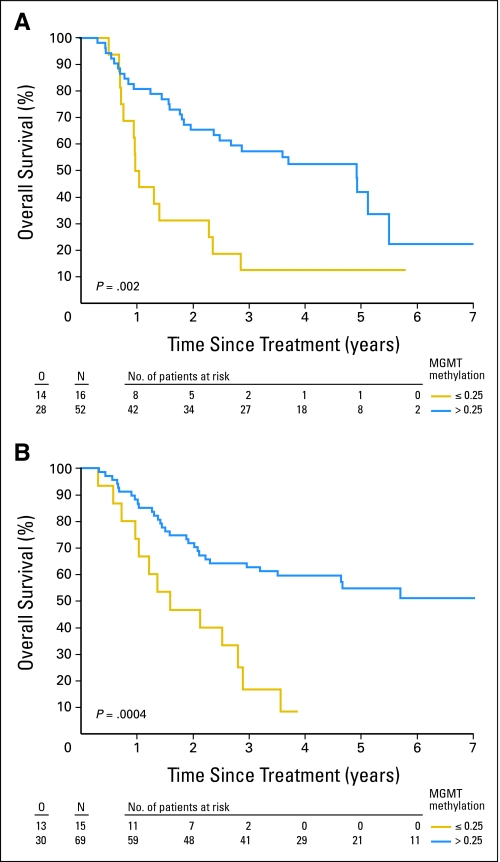
In univariate analysis, all the MGMT probes in the MS-MLPA and the MGMTav correlated with PFS and OS. Median and 2-year PFS in patients with MGMTav ≤ 0.25 was 8.6 months and 16%, respectively, in contrast to 34 months and 53%, respectively, for patients with MGMTav higher than 0.25. Median and 2-year OS with MGMTav 0.25 was 15.9 months and 39%, respectively, in contrast to 61 months and 69%, respectively, for patients with MGMTav higher than 0.25. Neither for PFS nor for OS tests for interaction with treatment (RT v RT/PCV) were significant (P = .49 and .90, respectively).
In multivariate Cox analysis for PFS and OS, MGMTav was entered as a continuous variable together with the previously established clinical and molecular prognostic factors. For both PFS and OS, MGMTav, surgery (biopsy v resection), age, 1p/19q codeletion, and necrosis were statistically significant (P < .05). With stepwise selection, MGMTav, surgery, 1p/19q, and necrosis entered the model for both PFS and OS (Table 2). With bootstrap validation, MGMTav was confirmed in 86% of the times for PFS and for OS in 87% of the times.
Table 2.
Cox Stepwise Selection for PFS and OS With MGMT Entered As a Continuous Variable
| Variable | PFS |
OS |
||
|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |
| MGMTav | 0.28 | 0.13 to 0.60 | 0.24 | 0.10 to 0.56 |
| Surgery | 2.11 | 1.17 to 3.81 | 2.19 | 1.20 to 3.99 |
| 1p/19q loss | 0.39 | 0.22 to 0.70 | 0.28 | 0.14 to 0.58 |
| Necrosis | 2.73 | 1.66 to 4.47 | 3.81 | 2.10 to 6.89 |
Abbreviations: MGMT, O6-methylguanine-methyltransferase; PFS, progression-free survival; OS, overall survival; HR, hazard ratio; av, averaged.
PFS and OS in Relationship to Treatment and MGMT Status
In both treatment arms, patients with tumors with MGMTav higher than 0.25 survived significantly longer. In Table 3 the median and 2-year PFS and OS according to the 0.25 cutoff are summarized. In univariate analysis, the hazard ratio (HR) for PFS in patients with MGMTav higher than 0.25 in the RT/PCV arm was 0.35 (95% CI, 0.18 to 0.68; P = .0011), and in the RT arm 0.46 (95% CI, 0.25 to 0.84; P = .0105). Figures 1A and 1B show the PFS in the RT and the RT/PCV arms based on the MGMTav cutoff of 0.25 (Figures 2A and 2B for OS).
Table 3.
Median and 2-Year PFS and OS in Patients With Methylated Tumors (MGMTav > 0.25) and Unmethylated Tumors (MGMTav ≤ 0.25) in Patients Randomly Assigned to RT and to RT Followed by Adjuvant PCV
| MGMTav | RT |
RT/PCV |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PFS |
OS |
PFS |
OS |
|||||||||||||
| Median Duration (months) | 95% CI | 2 Year (%) | 95% CI | Median Duration (months) | 95% CI | 2 Year (%) | 95% CI | Median (months) | 95% CI | 2 Year | 95% CI | Median (months) | 95% CI | 2 Year | 95% CI | |
| ≤ 0.25 | 7.8 | 7.1 to 17.6 | 12.5 | 2.1 to 32.8 | 12.3 | 11.5 to 28.5 | 31.3 | 11.4 to 53.7 | 10.5 | 5.2 to 23.0 | 20.0 | 4.9 to 42.4 | 19.0 | 12.3 to 34.5 | 46.7 | 21.2 to 68.8 |
| > 0.25 | 17.9 | 11.9 to 43.4 | 48.1 | 34.1 to 60.8 | 59.3 | 30.0 to 66.2 | 65.4 | 50.8 to 76.6 | 49.0 | 19.1 to 71.2 | 56.8 | 44.1 to 67.6 | Not reached | Not reached | 71.7 | 59.2 to 81.0 |
Abbreviations: PFS, progression-free survival; OS, overall survival; MGMT, O6-methylguanine-methyltransferase; RT, radiotherapy; PCV, procarbazine, lomustine, and vincristine; av, averaged.
Fig 1.
Progression-free survival for the patients randomly assigned to (A) radiotherapy and for the patients randomly assigned to radiotherapy followed by (B) adjuvant procarbazine, lomustine, and vincristine depending on methylation status (unmethylated: O6-methylguanine-methyltransferase averaged [MGMTav] ≤ 0.25, methylated: MGMTav > 0.25). O, number of observed events; N, sample size.
Fig 2.
Overall survival for the patients randomly assigned to (A) radiotherapy and for the patients randomly assigned to radiotherapy followed by (B) adjuvant procarbazine, lomustine, and vincristine depending on methylation status (unmethylated: O6-methylguanine-methyltransferase averaged [MGMTav] ≤ 0.25, methylated: MGMTav > 0.25). O, number of observed events; N, sample size.
Prognostic Significance MGMT Status in Relationship to Histology
In further exploratory analysis, we investigated the relationship between HR and histology. For this analysis the central pathology review diagnosis was interpreted according to the WHO 2007 classification.15 In this classification, AOA with necrosis are considered glioblastoma. According to this classification, 97 tumors were diagnosed as an oligodendroglial tumor (AOD and AOA without necrosis), 40 glioblastoma (GBM; and AOA with necrosis), and three anaplastic astrocytoma (Table 4 summarizes the MGMT findings according to this classification). The HR reduction for PFS in the oligodendroglial tumors was highly significant (HR, 0.26; 95% CI, 0.14 to 0.49; P < .0001) but not for the GBM (HR, 0.93; 95% CI, 0.46 to 1.89; P = .84). Similarly, the risk reduction was also highly significant for OS in the oligodendroglial group (HR, 0.17; 95% CI, 0.09 to 0.33; P < .0001), but not for the high-grade astrocytic tumors (HR, 0.87; 95% CI, 0.41 to 1.84; P = .71). For both PFS and OS, statistical tests for heterogeneity were highly significant (interaction tests: P = .0006 for PFS and P < .00001 for OS).
Table 4.
Median MGMT and MGMT According to the Cutoff of 0.25 in Centrally Confirmed AOD/AOA Without Necrosis and GBM/AOA With Necrosis, and the Number of Observed Events (death)
| Condition | No. | Median MGMTav | MGMTav ≤ 0.25 | Observed Events | MGMTav > 0.25 | Observed Events |
|---|---|---|---|---|---|---|
| AOD/AOA without necrosis | 97 | 0.56 | 16 | 15 | 81 | 35 |
| GBM/AOA with necrosis | 40 | 0.48 | 11 | 10 | 29 | 22 |
Abbreviations: MGMT, O6-methylguanine-methyltransferase; AOD, anaplastic oligodendroglial tumors; AOA, anaplastic oligoastrocytoma; GBM, glioblastoma; av, averaged.
DISCUSSION
Malignant glioma are a heterogeneous group of tumors in terms of morphology, tumor genetics, and prognosis. The frequencies of MGMT methylation in the different subtypes of malignant glioma that are part of the studied patient cohort here vary considerably.
In this study on AOD and AOA, MGMT promoter methylation as determined by MS-MLPA was observed in 80% of tumors and was strongly correlated with 1p/19q codeletion. Moreover, MGMT promoter methylation was prognostic for both PFS and OS. Even in the control arm treated with RT only, MGMT promoter methylation was correlated with a statistically and clinically significant increased PFS. The other statistically significant independent prognostic factors were 1p/19q codeletion, extent of resection, and the presence of tumor necrosis. The prognostic significance in the RT arm cannot be explained mechanistically by a function related to the MGMT gene. In the current understanding, MGMT does not play a role in the repair of RT-induced DNA damage. Mechanistically, a modest predictive effect of MGMT methylation may be expected in the RT/PCV arm as a result of the alkylating agent lomustine that is part of PCV chemotherapy.
The high levels of methylation in anaplastic oligodendroglial tumors (88%) has been observed before, in particular in 1p/19q codeleted tumors, which is in accordance with our findings.16,17 Similarly, nearly all oligodendrogliomas with 1p/19q loss in our series were methylated according to the MGMTav 0.25 cutoff. Reduced MGMT expression and MGMT promoter methylation have been proposed as the explanation for the sensitivity of oligodendroglial tumors to chemotherapy.18 At the other extreme are GBMs, in which the MGMT promoter is found methylated in fewer than 50% according to the literature.5 From a molecular analysis of EORTC study 26951, it is clear that a significant percentage of tumors included in this study had molecular characteristics compatible with a GBM (often with morphologic features of an anaplastic oligoastrocytoma with necrosis).13 In our data set, with relatively few glioblastoma patients, it appears that the impact of MGMT promoter methylation on PFS and OS is different for high-grade oligodendroglial tumors as opposed to GBMs. In the former group, a strong risk reduction was found for both PFS and OS, but not for the GBM; despite the relatively small number GBM, tests for interactions were statistically significant (P ≤ .0006).
A randomized controlled study is the most ideal platform to distinguish between factors of prognostic and of predictive significance. By comparing the investigational parameter in the two treatment arms, the effect of the parameter on outcome in the two different treatment modalities can be explored. By looking at the impact of the MGMT promoter methylation on PFS, the influence of subsequent salvage treatments is ruled out (and in this study 82% of patients in the RT control arm had received salvage chemotherapy at the time of progression).
The results of Hegi et al5 strongly suggested MGMT promoter methylation was predictive for outcome to combined chemoirradiation with temozolomide in GBM, with virtually no clinical benefit in MGMT unmethylated tumors. In contrast, in this study a prognostic significance for MGMT methylation in anaplastic oligodendroglioma was found, and although our study did not have sufficient power to efficiently assess marker predictivity, these data do not suggest a strong predictive value of MGMT methylation in anaplastic oligodendroglioma. The findings of this study are corroborated by the recently reported NO4 study on anaplastic glioma which randomized between initial management with radiotherapy or chemotherapy (PCV or temozolomide).19 In that study, the prognostic significance of MGMT methylation status (as determined by MS-PCR) was at least equivalent to 1p/19q codeletion, and the increase in PFS in methylated patients was observed similarly in the RT and chemotherapy treated patients. Thus, there are now two independent studies that suggest that in grade 3 tumors MGMT promoter methylation is of prognostic value even in patients treated with RT. The results of the NO4 study and these results suggest that the clinical significance for MGMT promoter methylation may be different for grade 3 (oligodendroglial) tumors than for glioblastoma, but clearly prospective and properly controlled studies are needed for confirmation.
Several technologies are currently available to assess MGMT promoter methylation.11,20–23 Most methylation assays including the MS-MLPA assess several CpG dinucleotides in the promoter region and the first exon of the MGMT gene. However, the CpG island in this region of the MGMT gene contains almost 100 CpGs. It is currently not clear how many or which CpGs within this region must be methylated for silencing of the MGMT gene.
We used MS-MLPA to assess methylation status, which is a semiquantitave assay to assess the methylation status of multiple CpGs simultaneously. The assay contains three different MGMT probes investigating the methylation status of different CpGs in the MGMT promoter region, one of which is inside the area amplified with MS-PCR as used elsewhere. MS-MLPA was found to correlate well with MS-PCR for MGMT promoter methylation (which was used in the randomized EORTC trial on GBM; κ score, 0.84).11 A disadvantage of the quantitative aspect of this assay is that interspersed unmethylated normal tissue (vessels, microglia, normal brain cells) will affect the calculated ratio. For implementation in a routine diagnostic setting, clear cutoff levels are preferable; however, the continuous distribution of MS-MLPA ratios for MGMT suggests that a different approach for interpretation is required, making matters more complex. Although a technical cut-off level has been established for MS-PCR, this test has also an intermediate zone between clearly unmethylated and methylated tumors.22
In conclusion, MGMT promoter methylation has prognostic significance in anaplastic oligodendroglial tumors. Despite this prognostic information the clinical relevance of MGMT methylation in the management of anaplastic gliomas is unclear; at present the methylation status has no clear implications for treatment decisions. The provocative finding of a prognostic effect in anaplastic glioma, even in patients managed with RT only, may indicate underlying genetic or epigenetic alterations associated with MGMT methylation that molecularly define a more favorable anaplastic glioma subtype. Our results confirm that anaplastic oligodendroglioma is a different disease with distinct biology from glioblastoma, requiring development of specific treatment strategies and separate trials.
Acknowledgment
This study was supported by the European Organisation for the Research and Treatment of Cancer (EORTC) Translational Research Fund Grant No. TRF 01/02; by AstraZeneca EORTC Translational Research Grant No. AZ/01/02; by the Dutch Cancer Society Grant No. DDHK 2005 to 3416 and EMC 2007 to 3932; by Grants No. 2U10CA11488-25 through 2U10CA11488-35 from the National Cancer Institute, Bethesda, MD; and by a donation from the Dutch Cancer Society from the Netherlands through the EORTC Charitable Trust.
Footnotes
The content of this article is solely the responsibility of the authors and does not necessarily reflect the official views of the National Cancer Institute.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Monika Hegi, Oncomethylome Sciences (C) Stock Ownership: None Honoraria: None Research Funding: Martin J. van den Bent, Oncomethylome Sciences; Monika Hegi, Oncomethylome Sciences Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Martin J. van den Bent, Thierry Gorlia, Winand N.M. Dinjens, Johan M. Kros
Administrative support: Martin J. van den Bent, Denis Lacombe
Provision of study materials or patients: Martin J. van den Bent, Marc Sanson, Cathleen R. van der Lee-Haarloo, Ahmed Ibdaih, Alba A. Brandes, Martin J.B. Taphoorn, Marc Frenay, Winand N.M. Dinjens
Collection and assembly of data: Martin J. van den Bent, Hendrikus J. Dubbink, Cathleen R. van der Lee-Haarloo, Thierry Gorlia, Winand N.M. Dinjens, Johan M. Kros
Data analysis and interpretation: Martin J. van den Bent, Hendrikus J. Dubbink, Monika Hegi, Judith W.M. Jeuken, Thierry Gorlia, Johan M. Kros
Manuscript writing: Martin J. van den Bent, Hendrikus J. Dubbink, Marc Sanson, Cathleen R. van der Lee-Haarloo, Monika Hegi, Judith W.M. Jeuken, Ahmed Ibdaih, Alba A. Brandes, Martin J.B. Taphoorn, Marc Frenay, Denis Lacombe, Thierry Gorlia, Winand N.M. Dinjens, Johan M. Kros
Final approval of manuscript: Martin J. van den Bent, Hendrikus J. Dubbink, Marc Sanson, Cathleen R. van der Lee-Haarloo, Monika Hegi, Judith W.M. Jeuken, Ahmed Ibdaih, Alba A. Brandes, Martin J.B. Taphoorn, Marc Frenay, Denis Lacombe, Thierry Gorlia, Winand N.M. Dinjens, Johan M. Kros
REFERENCES
- 1.Gerson SL. MGMT: Its role in cancer etiology and cancer therapeutics. Nature Rev. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 2.Jaeckle KA, Eyre HJ, Townsend JJ, et al. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: A Southwest Oncology Group study. J Clin Oncol. 1999;16:3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Glas M, Happold C, Rieger J, et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. J Clin Oncol. 2009;27:1257–1261. doi: 10.1200/JCO.2008.19.2195. [DOI] [PubMed] [Google Scholar]
- 7.Herrlinger U, Rieger J, Koch D, et al. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. J Clin Oncol. 2006;24:4412–4417. doi: 10.1200/JCO.2006.06.9104. [DOI] [PubMed] [Google Scholar]
- 8.Criniere E, Kaloshi G, Laigle-Donadey F, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–179. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]
- 9.Zawlik I, Vaccarella S, Kita D, et al. Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: A population-based study. Neuroepidemiology. 2009;32:21–29. doi: 10.1159/000170088. [DOI] [PubMed] [Google Scholar]
- 10.van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant PCV improves progression free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: A randomized EORTC phase III trial. J Clin Oncol. 2006;24:2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 11.Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: An attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 12.Kleihues P, Burger PC, Scheithauer BW. New York, NY: Springer-Verlag; 1993. Histological typing of tumours of the central nervous system. [Google Scholar]
- 13.Kouwenhoven MC, Gorlia T, Kros JM, et al. Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: A report from EORTC study 26951. Neuro Oncol. 2009 doi: 10.1215/15228517-2009-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Sijp JR, van Meerbeeck JP, Maat AP, et al. Determination of the molecular relationship between multiple tumors within one patient is of clinical importance. J Clin Oncol. 2002;20:1105–1114. doi: 10.1200/JCO.2002.20.4.1105. [DOI] [PubMed] [Google Scholar]
- 15.Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. Lyon, France: International Agency for Research on Cancer; 2007. [Google Scholar]
- 16.Möllemann M, Wolter M, Felsberg J, et al. Frequent promotor hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int J Cancer. 2004;113:379–385. doi: 10.1002/ijc.20575. [DOI] [PubMed] [Google Scholar]
- 17.Brandes AA, Tosoni A, Cavallo G, et al. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: A prospective GICNO study. J Clin Oncol. 2006;24:4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 18.Nutt CL, Costello JF, Bambrick LL, et al. O6 methylguanine-DNA methyltransferase in tumors and cells of the oligodendrocyte lineage. Can J Neurol Sci. 1995;22:111–115. doi: 10.1017/s0317167100040178. [DOI] [PubMed] [Google Scholar]
- 19.Wick W, Weller M. NOA-04 randomized phase III study of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. J Clin Oncol. 2008;26:1008s. abstract LBA2007. [Google Scholar]
- 20.Nakagawachi T, Soejima H, Urano T, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–8844. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 21.Mikeska T, Bock C, El-Maarri O, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlassenbroeck I, Califice S, Diserens AC, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everhard S, Tost J, El AH, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]