Abstract
The complete modification of protein amines by reductive glutaraldehydation is achieved in solution or in the gel in less than 15 minutes. Fragmentation of glutaraldehyde-modified proteins and peptides generates a1 fragment ions with enhanced intensity in MS/MS spectra. A method based on reductive glutaraldehydation and LC-MS/MS analysis has been developed to determine the N-terminal residue of proteins with free N-termini.
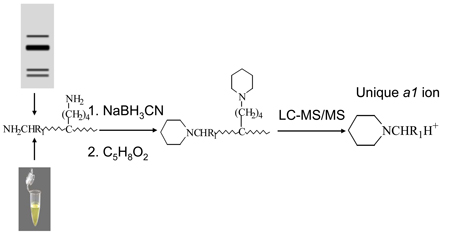
In the present work, reductive alkylation of proteins and peptides with glutaraldehyde (reductive glutaraldehydation) is reported. The reaction is highly efficient and forms piperidine at the N-terminus as well as the side chain of lysine residues. The complete modification of protein amines was achieved by reductive glutaraldehydation in solution or in the gel in less than 15 minutes. The glutaraldehyde-modified peptides display an enhanced intensity in mass spectra and show higher retention time in reversed phase chromatography in comparison to unmodified peptides. Fragmentation of glutaraldehyde-modified proteins and peptides generates a1 fragment ions with enhanced intensity in MS/MS spectra. Thus a method based on reductive glutaraldehydation and LC-MS/MS analysis has been developed to determine the N-terminal residue of proteins with free N-termini.
Reactions of aldehydes and ketones with primary or secondary amines under reductive conditions are among the most useful tools in organic synthesis.1–3 The reaction involves the initial formation of a Schiff base, followed by reduction to form alkylated amine products. Reductive alkylation with formaldehyde has been successfully applied to modifications of proteins and peptides.4–8 Hsu et al showed the reductive methylation of peptides with formaldehyde is a simple and fast process leading to complete conversion of free amines to their mono- and di-methylated forms within five minutes.9 Using regular and stable isotope incorporated formaldehydes, reductive methylation has been applied for quantitative proteomics.9–11 More importantly, it has been found that a1 fragment ions of dimethylated peptides and proteins have enhanced ion intensities in MS/MS spectra.12 Since the a1 ions are not frequently observed in MS/MS spectra of unmodified peptides and proteins, the combination of reductive methylation and mass spectrometry has been proven to be a powerful method for determination of the protein and peptide N-terminal residue.13–14 An elegant approach based on fragmentation of dimethylated proteins was established to study proteolytic processes.15 Reductive dimethylation was also applied to de novo sequencing of neural peptides.16 However, it has also been reported that reductive methylation with sodium cyanoborohydride generates side products and results in incomplete modification of proteins.15–18
Glutaraldehyde is a widely used cross-linking reagent and has broad applications in many areas including histochemistry, microscopy and cytochemistry, and biomedical and pharmaceutical sciences.19–26 Glutaraldehyde is a dialdehyde with a chain length of five carbon atoms. The high crosslinking efficiency of glutaraldehyde is attributed to its multimeric structures in solution.27–28 It has been highly recommended not to use any solutions containing formaldehyde or glutaraldehyde to fix the gel in proteomic analysis.29 Reductive alkylation of tryptic peptides with glutaraldehyde was recently reported.30 In this article, Pinto et al found that the reaction generated the six-membered heterocyclic products in basic solution with 2 hour incubation. The modified peptides showed the enhanced a1 fragment ion in MS/MS that can be used for quantitation of peptides. However, details of reductive alkylation of proteins with glutaraldehyde have not been reported. In the present study, we have characterized the reaction products of proteins with glutaraldehyde in the presence of sodium cyanoborohydride. We found that the reaction is a highly efficient process to completely convert the α-amino groups as well as ε amine groups (lysine) in proteins and peptides, to piperidine derivatives.. A 5-hydroxy-pentanyl derivative was formed when proline was the N-terminal residue. The glutaraldehyde-modified peptides display higher retention time in comparison to unmodified peptides. Collision-induced dissociation of the glutaraldehyde-modified proteins and peptides results in enhanced intensities of a1 ions whose masses are unique for each amino acid residue. We demonstrate that reductive glutaraldehydation is applicable to identification of the N-terminal residue of intact proteins in solutionor in the gel.
Experimental Section
Materials
Horse heart cytochrome C, angiotensin II, bovine serum albumin, lysozyme, bovine hemoglobin, insulin, ribonuclease A, sodium cyanoborohydride, formaldehyde and glutaraldehyde were purchased from Sigma Aldrich and used without further purification. All other peptides were made at the Proteomics Resource Center of the Rockefeller University.
Reductive glutaraldehydation of proteins and peptides in solution or in the gel
The reaction was studied by mixing the proper quantity of solutions of synthetic peptides and proteins with sodium cyanoborohydride followed by addition of glutaraldehyde at different concentrations. In most cases, the concentration of peptides or proteins was 1 or 10 µM and concentrations of sodium cyanoborohydride and glutaraldehyde were 100 mM and 0.5%, respectively. The reaction mixtures were incubated at room temperature for a desired time in water or aqueous solution containing 0.1 % (v/v) acetic acid (pH~4.0) or in phosphate buffer. For in-gel modification, gel slices were reduced with 1, 4-dithiothreitol (DTT) and alkylated with iodoacetamide followed by incubation with 500 mM sodium cyanoborohydride and 2.5% glutaraldehyde at 37 °C for the desired time. Reactions were stopped by addition of 1M Tris-HCl. Sodium cyanoborohydride is a toxic chemical and formaldehyde and glutaraldehyde are carcinogenic and mutagenic. All reactions were carried out in a fume hood.
Mass spectrometric analysis
The reaction products were analyzed by MALDI-TOF and LC-MS/MS mass spectrometry. In MALDI-TOF analysis, α-cyano-4-hydroxysinnipinic acid was used as the matrix and samples were analyzed with a Voyager DE STR MALDI-TOF mass spectrometer. For LC-MS/MS analysis, the reaction products were separated on a 5 cm × 75 micron i.d. reversed-phase C18 column (self packed) using a 40 minute linear gradient of 5 to 60% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min with a Dionex capillary/nano-HPLC system and analyzed by an Applied Biosystems QSTAR XL mass spectrometer using information-dependent, automated data acquisition. For in-gel digestion, samples of selected proteins alone and its reaction products were digested with Sequence Grade Modified Trypsin (Promega) in ammonium bicarbonate buffer at 37 °C overnight. All the digestion products were analyzed by MALDI-TOF and nano-LC-MS/MS.
Results and Discussion
Characterization of Reductive Glutaraldehydation of Peptides and Proteins
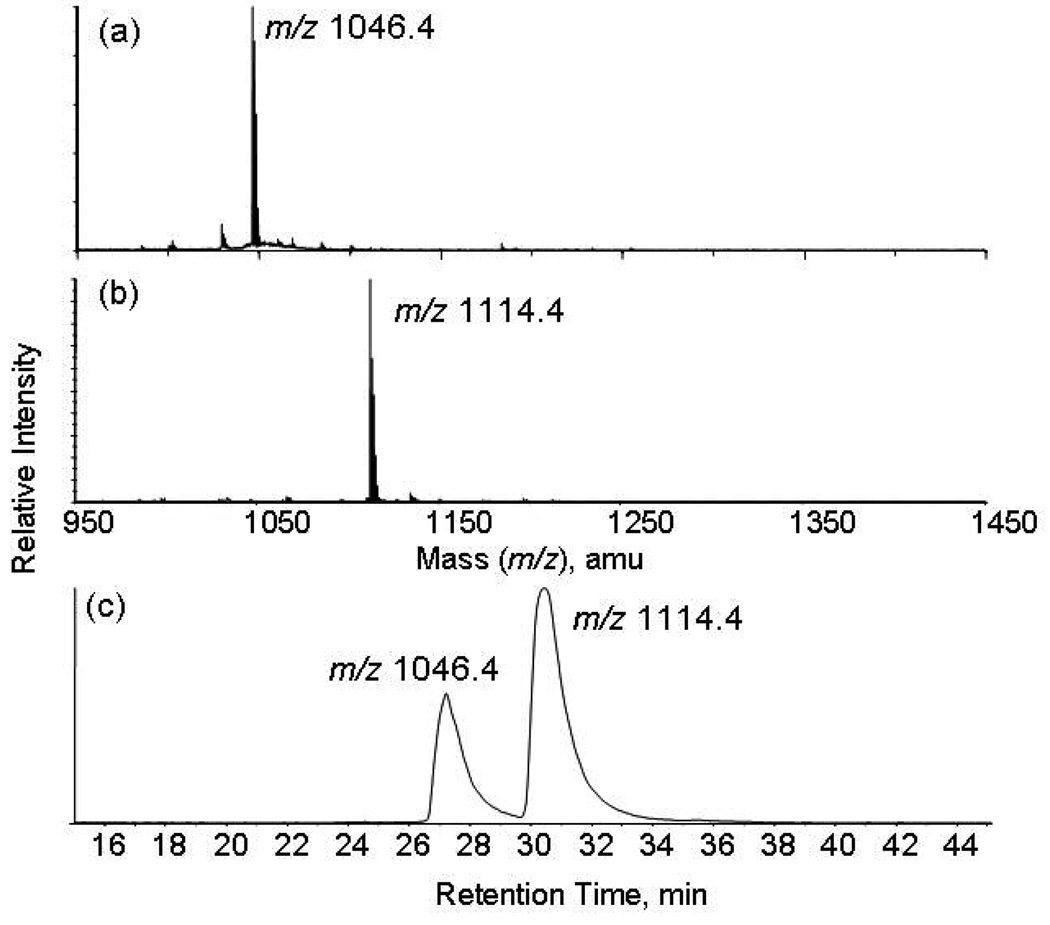
The MALDI-TOF mass spectra of angiotensin II (DRVYIHPFHL) and its reaction products with 0.5% glutaraldehyde and 100 mM sodium cyanoborohydride at room temperature for 10 minutes showed one major reaction product (Figure 1b). The mass of the product at 1114.4 was 68 amu higher than that of angiotensin II at 1046.4 corresponding to formation of a piperidine at the N-terminus. This was confirmed by MS/MS analysis of the modified peptide in which the mass of a1 ion was up shifted by 68 amu consistent with the previous report.30 The modification changes the N-terminal residue from a primary amine to a tertiary amine that has a higher proton affinity. It is expected that the glutaraldehyde-modified peptides have higher ionization efficiency than the unmodified peptides in formation of protonated species. Indeed, the glutaraldehyde-modified peptide showed the enhanced ion intensity in the MS spectrum as shown in Figure 1c that displays the extracted ion chromatogram of un- and glutaraldehyde-modified angiotensin II at the same concentration. The intensity of peak I in Figure 1c corresponding to the extracted ion current of the unmodified peptide is approximately fifty percent lower than that of the modified peptide. The glutaraldehyde-modified angiotensin II displays a higher retention time as seen in Figure 1(c) suggesting that the modified peptide has a higher hydrophobicity in comparison to unmodified angiotensin II. Our results also showed that the retention time of glutaraldehyde-modified angiotensin II is higher than that of dimethylated angiotensin II formed by reductive methylation with formaldehyde.
Figure 1.
MALDI-TOF mass spectra of reaction products of angiotensin II with glutaraldehyde and sodium cyanoborohydride. (a) angiotensin II alone; (b) angiotensin II with 0.5% glutaraldehyde and 100 mM sodium cyanoborohydride in water at room temperature for 10 minutes; (c) The extracted ion chromatogram of angiotensin II and glutaraldehyde-modified angiotensin at the same concentration analyzed by nano-LC-MS/MS.
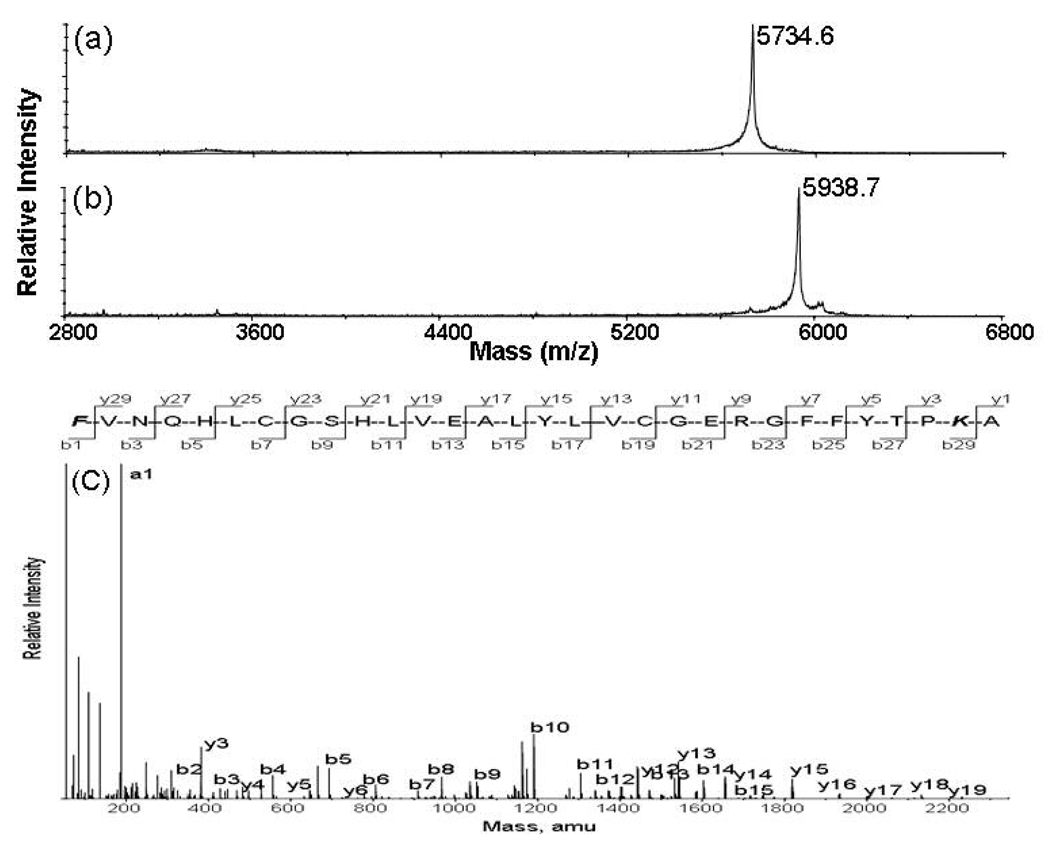
The reactivity of the amino group at N-terminus of proteins and peptides is similar to that on the side chain of lysine residues. Reductive glutaraldehydation also modifies the ε amino group of lysine residues.30 Complete modification of the N-terminal residue and lysine residues at the protein level was demonstrated by the reaction of insulin with glutaraldehyde under similar reductive conditions. The MALDI-TOF mass spectra of insulin and the reaction products with glutaraldehyde are displayed in Figure 2. The mass of the product is 204 amu higher than that of insulin corresponding to the mass of the fully modified insulin with three piperidines since insulin has two N-termini and one lysine residue. No other major reaction products were observed in Figure 2b indicating that the reduction conditions with sodium cyanoborohydride do not compromise the integrity of disulfide bonds. Reduction of the disulfide bonds with DTT generated the mono-glutaraldehyde-modified alpha chain and diglutaraldehyde-modified beta chain in which both amines at the N-terminal and lysine residue were modified. The MS/MS spectrum of the modified insulin beta chain is displayed in Figure 2c. The fragmentation pattern confirms that both the N-terminus and the lysine side chain of insulin beta chain were modified and major fragment ions observed in MS/MS spectrum correspond to b and y ion series. A distinct peak at m/z 188 was observed with enhanced intensity corresponding to the a1 ion of glutaraldehyde-modified phenylalanine. Studies on other modified peptides with different N-terminal residues show that fragmentation of glutaraldehyde-modified peptides generated a1 ions with an enhanced intensity and their masses are 68 amu higher than that of a1 ion of unmodified peptides.
Figure 2.
MALDI-TOF mass spectra of reaction products of bovine insulin with glutaraldehyde and sodium cyanoborohydride. (a) insulin alone; (b) insulin with 0.5% glutaraldehyde and 100 mM sodium cyanoborohydride in water at room temperature for 10 minutes; and (c) MS/MS of the glutaraldehyde-modified insulin β chain observed 884.5 for MH44+ ion. Both N-terminus and the lysine residue were modified and the a1 at 188.06 was observed with enhanced intensity.
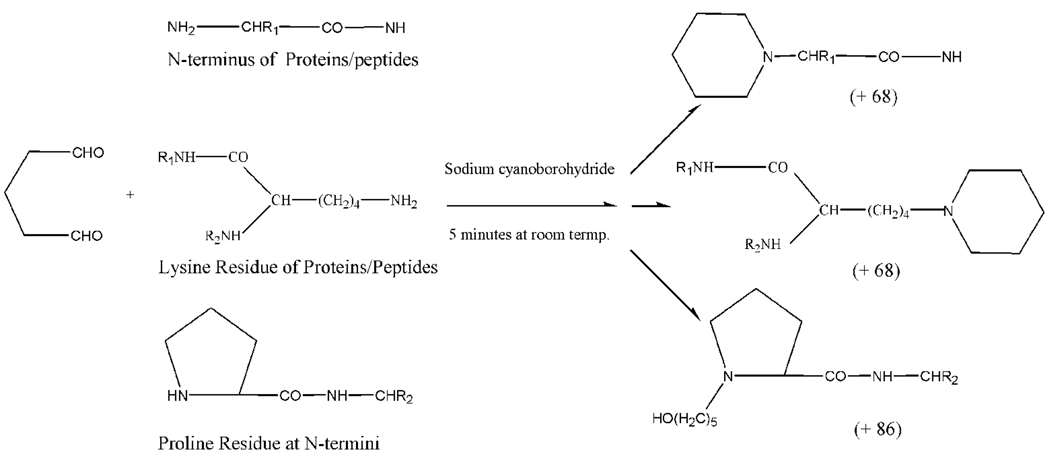
Proline contains a secondary amine. Reductive glutaraldehydation of peptides with proline as the N-terminal residue generated a product whose mass is 86 amu higher than that of unmodified peptides. This mass shift corresponds to formation of 5-hydroxy-pentanyl derivative at the N-terminal proline residue. This product is produced by further reduction of the the unreacted aldehyde group of glutaraldehyde. Peptides with the N-terminal glutaraldehyde-modified proline residue also display the enhanced a1 ions in MS/MS spectra. The formation of piperidine or 5-hydroxy-pentanyl derivative is the typical reaction product of amines with aldehyde under reductive conditions. The intermediate formed in this reaction is a Schiff base that is further reduced to its amine form by sodium cyanoborohydride‥ The reaction is highly efficient and all amines of proteins and peptides can be completely modified with glutaraldehyde in less than 15 minutes at room temperature. The reaction was not pH-dependent and the identical reaction products were observed for reactions in acidic (pH 4), neutral, or basic solution (pH 8). The structures of reaction products are illustrated in Figure 3. Glutaraldehyde is the most widely used cross-linking reagent. However, in our experiments no intra- or inter-crosslinked proteins or peptides were observed when proteins or peptides were treated with sodium cyanoborohydride followed by the addition of glutaraldehyde. In summary, reductive glutaraldehydation of proteins and peptides results in formation of piperidine at the N-termini and lysine side chains or 5-hydroxy-pentanyl derivative for peptides with proline as the N-terminal residue. Fragmentation of glutaraldehyde-modified proteins and peptides display enhanced a1 fragment ions in MS/MS spectra.
Figure 3.
Structures of glutaraldehyde-modified amino groups for free N-terminus, lysine, and N-terminal proline residue.
Determination of the protein N-terminal residue by reductive glutaraldehydation in solution and in the gel
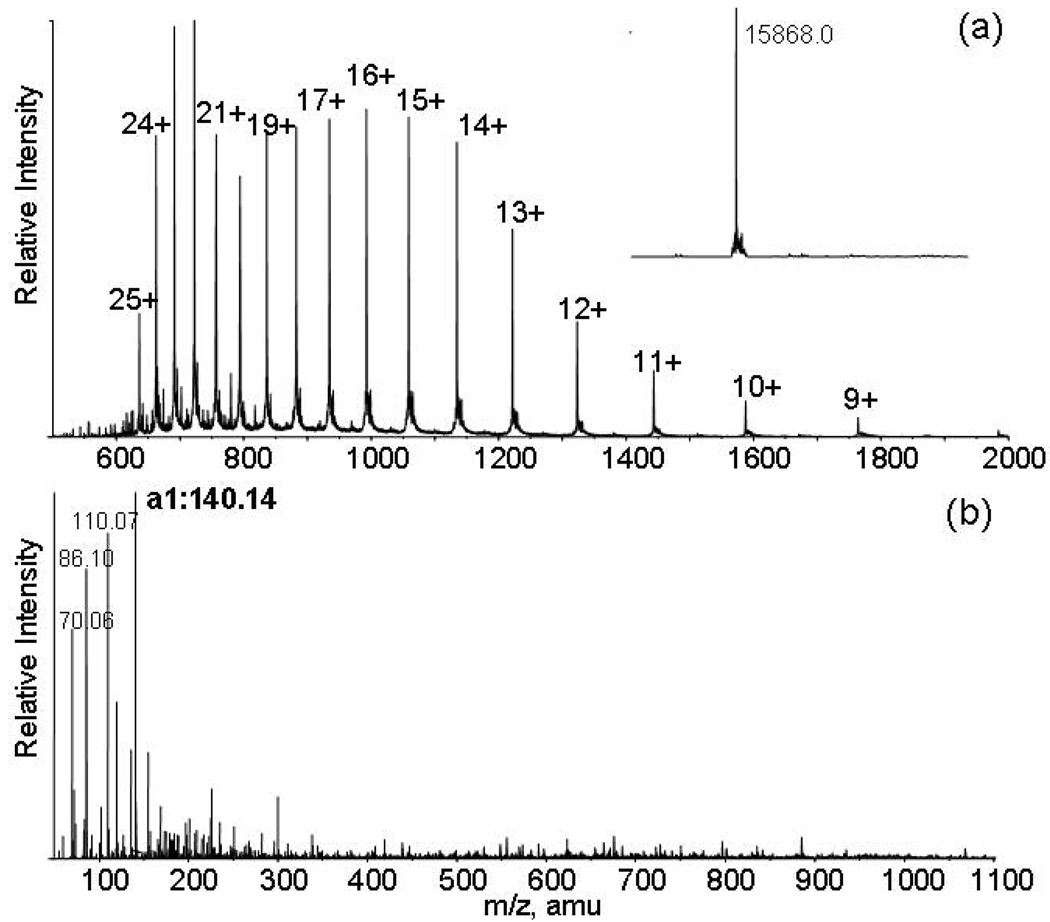
Determination of the N-terminal amino acid residue of proteins is important for characterization of protein isoforms originating from alternative splicing and proteolytic processes. To explore the applicability of reductive glutaraldehydation to determination of protein N-terminal residue, reactions of bovine hemoglobin α chain with 0.5% glutaraldehyde and 100 mM sodium cyanoborohydride was carried out at room temperature for 15 minutes and the reaction products were analyzed by LC-MS/MS. The bovine hemoglobin α chain has 141 residues including 11 lysine residues and a free N-terminus. The masses of bovine hemoglobin α chain and the fully glutaraldehyde-modified protein were calculated to be 15053 and 15869 amu, respectively. The mass spectra of the reaction products of bovine hemoglobin α chain with glutaraldehyde and sodium cyanoborohydride is displayed in Figure 4. The protein charge envelope observed covers upto 25 charge states and deconvolution of the charge envelope determines the mass of the sole product to be 15868. This demonstrates the full conversion of all amines of hemoglobin α chain to piperidines. The time course study shows that full conversion efficiency can be achieved within 5 minute incubation of proteins with 0.5% glutaraldehyde and 100 mM sodium cyanoborohydride at room temperature.
Figure 4.
LC-MS/MS mass spectra of reactions products of bovine hemoglobin with glutaraldehyde and sodium cyanoborohydride in water at room temperature for 15 minutes: (a) the charge distribution of glutaraldehyde-modified hemoglobin α chain and the deconvoluted spectrum as the insert. The mass matches to full modifications of the N-terminus and all lysine residues; (b) MS/MS of the glutaraldehyde-modified hemoglobin α chain observed at 992.73 for MH1616+ ion. The a1 at 140.08 was observed with enhanced intensity.
The MS/MS spectra of glutaraldehyde-modified hemoglobin α chain at different charge states were acquired and showed similar fragmentation patterns. One of the MS/MS spectra for MH1616+ at 992.7 amu is displayed in Figure 4(b). A peak with enhanced intensity was observed at 140.14 corresponding to the mass of the a1 ion of the modified valine residue. Similar results were observed for reactions of glutaraldehyde with other proteins such as ribonuclease A, lysozyme and bovine serum albumin under similar reaction conditions. These results demonstrate that a combination of reductive glutaraldehydation and LC-MS/MS is a useful tool to determine the protein N-terminal residue. The masses of a1 ions for all glutaraldehyde-modified amino acid residues are listed in Table 1. The masses of a1 ions are 138 for glutaraldehyde-modified proline due to formation of 5-hydroxy-pentanyl derivative and 237 for modified lysine because it has two amino groups. It is worth mentioning that masses of a1 ions for glutaraldehyde-modified amino residues do not overlap with any masses of immonium ions. The assignment of the a1 ion are based on that a fragment ion has an enhanced intensity as well as a unique mass matching to the calculated mass of the glutaraldehyde-modified residue.
Table 1.
Masses of glutaraldehyde-modified a1 ions
| Residue | Mass of a1 | Residue | Mass of a1 |
|---|---|---|---|
| Glycine | 98.03438 | Aspartic acid | 156.0398 |
| Alanine | 112.05003 | Glutamine | 169.0715 |
| Serine | 128.04494 | Lysine | 237.1079 |
| Proline | 156.06568 | Glutamic acid | 170.0555 |
| Valine | 140.08133 | Methionine | 172.0534 |
| Threonine | 142.06059 | Histidine | 178.0718 |
| Cysteine | 144.0221 | Phenylalanine | 188.0813 |
| Isoleucine | 154.09698 | Arginine | 197.114 |
| Leucine | 154.09698 | Tyrosine | 204.0762 |
| Asparagine | 155.05584 | Tryptophan | 227.08 |
This method is applicable to studying in vitro proteolytic processing by determination of the N-terminal residue and the mass of glutaraldehyde-modified proteolytic products. It can also be applied to evaluation of the secondary structures and stability of a protein of interest by determining the N-terminal residues and masses of the glutaraldehyde-modified partial digestion products. For proteins with unknown sequences, ambiguities may raise due to the fact that masses of some internal fragments may overlap with those from glutaraldehyde-modified residue. Two methods can be used to overcome this limitation. One is by the accurate mass measurement of the modified N-terminal residue and the other is by comparison of the MS/MS spectra of un- and glutaraldehyde-modified proteins. Detection and fragmentation of intact proteins with high masses is still a challenge. The combination of reductive alkylation and LC-MS/MS is mainly applicable to analysis of small and medium size proteins that can be efficiently separated and electrospray-ionized to generate abundant multiply charged gas phase ions.
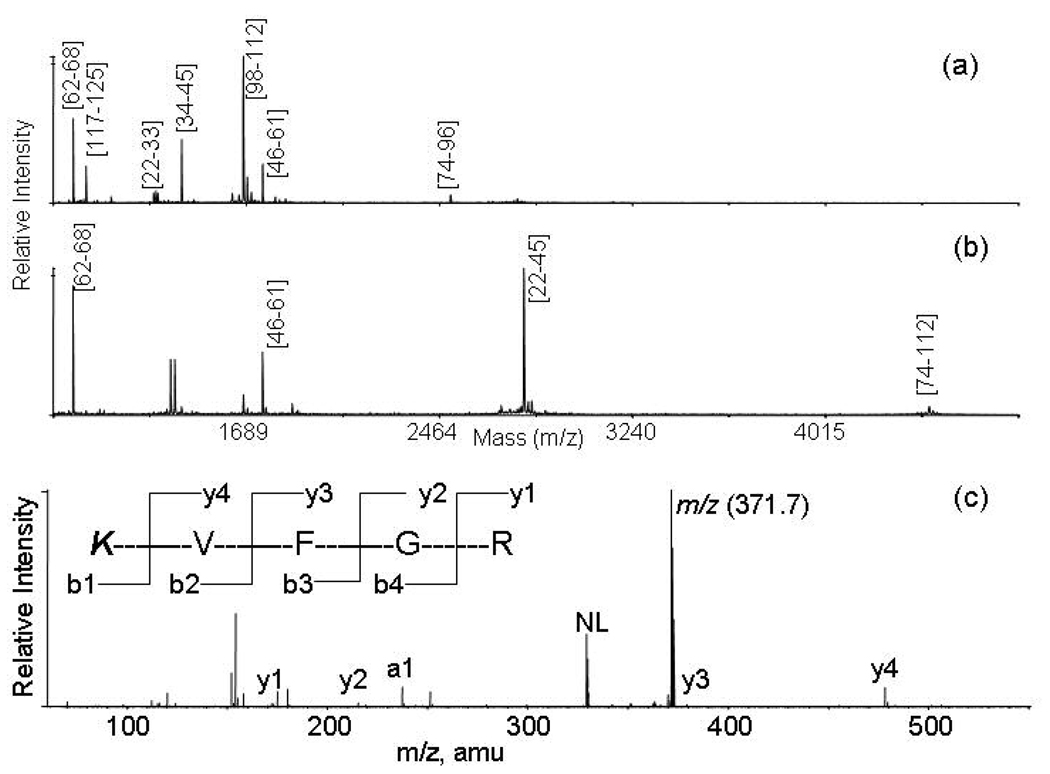
Previous studies mainly focused on modifications of amino groups of proteins and peptides in solution. For proteins with high masses and of low abundance, gel electrophoresis followed by in gel digestion is the method of choice for protein analysis. To test the efficiency of reductive glutaraldehydation of gel separated proteins, two SDS-PAGE gel pieces containing nearly identical amount of lysozyme (80 nanograms) were in-gel reduced and alkylated. One piece was further treated with 50 microliter phosphate solution (pH 8) containing 2.5% glutaraldehyde and 500 mM sodium cyanoborohydride at room temperature for 30 minutes. Then, un- and glutaraldehyde-treated gel pieces were washed and digested with trypsin overnight at 37 °C. Figure 5 shows the MALDI-TOF spectra of tryptic peptides from un- and glutaraldehyde-treated lysozyme in the mass range between 900 and 4800. Seven peptides were observed from un-modified lysozyme in this mass range and four from the glutaraldehyde-modified protein. The peptides [22–33] and [74–96] were only observed from the untreated samples indicating the complete conversion of K33 and K96 to their piperidine counterparts. However, a small peak corresponding to the peptide [98–112] was observed in both samples indicating K97 is not fully modified. Based on the peak intensity, it was estimated that more than 95% of K97 was modified. Similar results were also found for other gel separated proteins such as cytochrome c, and bovine serum albumin. These results show that 95% in-gel labeling efficiency can be achieved under the present experimental conditions. Using LC-MS/MS, we found that a doubly protonated species at 371.7 was only observed from the glutaraldehyde-treated sample. The MS/MS spectrum of this peptide is displayed in Figure 5(c) in which the fragment ion at 237 matches to mass of a1 ion for the doubly modified lysine residue and the sequence was identified as KVFGR the N-terminal tryptic peptide of lysozyme. This demonstrates that the N-terminus of a gel separated protein can be identified by a combination of in-gel reductive glutaraldehydation, digestion and LC-MS/MS analysis.
Figure 5.
MALDI-TOF mass spectra of tryptic peptides from unmodified and in-gel glutaraldehyde-modified lysozyme: (a) unmodified lysozyme; (b) glutaraldehyde-modified lysozyme; and (c) MS/MS of the MH22+ ion at 371.76 that was only observed from the digestion products of glutaraldehyde-modified lysozyme. The mass is corresponding to doubly glutaraldehyde-modified N-terminal peptides. The a1 at 237.1 was observed. The peak labeled with NL corresponds to loss of 85, the mass of piperidine from the precursor ion. Unlike a1 ions, the neutral loss of the piperidine molecule was only observed in a few MS/MS spectra and may be a sequence dependent process.
Conclusion
Reductive glutaraldehydation is a rapid process resulting in formation of piperidine at both N-termini and side chain of lysine residues. The modified peptides have higher proton affinity and show higher retention time in the reversed phase LC chromatography in comparison with unmodified or dimethylated peptides. The glutaraldehyde-modified peptides and proteins generate enhanced a1 fragment ions that have unique masses. We demonstrated that this reaction is highly efficient to modify proteins in solution and in the gel that will be a useful tool for both top-down and bottom up proteomics. A simple method based on a combination of reductive glutaraldehydation and LC-MS/MS analysis allows identification of protein N-terminal residue for proteins in solution or in the gel. Reductive glutaraldehydation can be applied to modification of amine-containing small molecules and metabolites and may have an application in metabolomic analysis. We are in the process of synthesizing isotope-incorporated glutaraldehyde for quantitative proteomics.
Acknowledgement
The authors gratefully acknowledge Dr Brian Chait and Joseph Fernandez for valuable discussions. The support from Amy Wilkerson at the Office of Research Support of the Rockefeller University is greatly appreciated. This work was supported by the NIH (Grant DE 018385) and Chinese Ministry of Science and Technology to LZ (Grant 2006CB504200).
REFERENCES
- 1.Emerson WS. Org. React. (N. Y.) 1948;4:174. [Google Scholar]
- 2.Borch RF, Bernstein MD, Durst HD. J. Am. Chem. Soc. 1971;93:2897. [Google Scholar]
- 3.Gomez S, Peters JA, Maschmeyer T. Advanced Synthesis & Catalysis. 2002;344:1037. [Google Scholar]
- 4.Lundblad RL. The evolution from protein Chemistry to Proteomics. Boca Raton, FL: CRC Press: Taylor & Francis Group; 2006. Chapter 2. [Google Scholar]
- 5.Lundblad RL, Noyes CM. Chemical Reagents for Protein Modification. Vol. 1. Boca Raton, FL: CRC Press; 1984. Chapter 10. [Google Scholar]
- 6.Hermanson GT. Bioconjugate Techniques. San Diego, CA: Academic Press; 1996. [Google Scholar]
- 7.Regnier FE, Julka S. Proteomics. 2006;6:3968–3979. doi: 10.1002/pmic.200500553. [DOI] [PubMed] [Google Scholar]
- 8.Leitner A, Lindner W. Proteomics. 2006;6:5418–5434. doi: 10.1002/pmic.200600255. [DOI] [PubMed] [Google Scholar]
- 9.Hsu J-L, Huang S-Y, Chow N-H, Chen S-H. Anal. Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 10.Melanson JE, Avery SL, Pinto DM. Proteomics. 2006;6:4466–4474. doi: 10.1002/pmic.200600112. [DOI] [PubMed] [Google Scholar]
- 11.Ji C, Li L, Gebre M, Pasdar M, Li L. J. Proteome Res. 2005;4:1419–1426. doi: 10.1021/pr050094h. [DOI] [PubMed] [Google Scholar]
- 12.Hsu JL, Huang SY, Shiea JT, Huang WY, Chen SH. J. Proteome Res. 2005;4:101–108. doi: 10.1021/pr049837+. [DOI] [PubMed] [Google Scholar]
- 13.Shen PT, Hsu JL, Chen SH. Anal Chem. 2007;79:9520–9530. doi: 10.1021/ac701678h. [DOI] [PubMed] [Google Scholar]
- 14.Ji C, Guo N, Li L. J. Proteome Res. 2005;4:2099–2108. doi: 10.1021/pr050215d. [DOI] [PubMed] [Google Scholar]
- 15.Hsu JL, Chen SH, Li DT, Shi FK. J. Proteome Res. 2007;6:2376–2383. doi: 10.1021/pr060639n. [DOI] [PubMed] [Google Scholar]
- 16.Fu Q, Li L. Anal. Chem. 2005;77:7783–7795. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 17.Gidley MJ, Sanders JK. Biochem. J. 1982;224:331–334. doi: 10.1042/bj2030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krusemark CJ, Ferguson JT, Wenger CD, Kelleher NL, Belshaw PJ. Anal. Chem. 2008;80:713–720. doi: 10.1021/ac7019317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson PJ. J. Histochem. Cytochem. 1967;15:652–661. doi: 10.1177/15.11.652. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood D. Histochemie. 1967;11:289–295. doi: 10.1007/BF00305805. [DOI] [PubMed] [Google Scholar]
- 21.Sabatini DD, Bensch K, Barrnett RJ. J. Cell. Biol. 1963;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiernan JA. Microsc. Today. 2000;1:8–12. [Google Scholar]
- 23.Hopwood D. Histochem. J. 1972;4:267–303. doi: 10.1007/BF01005005. [DOI] [PubMed] [Google Scholar]
- 24.Quiocho FA, Richards FM. Biochemistry. 1966;5:4062–4076. [Google Scholar]
- 25.Richards FM, Knowles JR. J. Mol. Biol. 1968;37:231–233. doi: 10.1016/0022-2836(68)90086-7. [DOI] [PubMed] [Google Scholar]
- 26.Schejter A, Bar-Eli A. Arch. Biochem. Biophys. 1970;136:325–330. doi: 10.1016/0003-9861(70)90202-x. [DOI] [PubMed] [Google Scholar]
- 27.Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. Biotechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 28.Wine Y, Cohen-Hadar N, Freeman A, Frolow F. Biotechnol Bioeng. 2007;98:711–718. doi: 10.1002/bit.21459. [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko A, Wilm M, Vorm O, Mann M. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 30.Locke SJ, Leslie AD, Melanson JE, Pinto DM. Rapid Commun Mass Spectrom. 2006;20:1525–1530. doi: 10.1002/rcm.2512. [DOI] [PubMed] [Google Scholar]