Abstract
The recognition of specific DNA sequences by proteins is thought to depend on two types of mechanisms: one that involves the formation of hydrogen bonds with specific bases, primarily in the major groove, and one involving sequence-dependent deformations of the DNA helix. By comprehensively analyzing the three dimensional structures of protein-DNA complexes, we show that the binding of arginines to narrow minor grooves is a widely used mode for protein-DNA recognition. This readout mechanism exploits the phenomenon that narrow minor grooves strongly enhance the negative electrostatic potential of the DNA. The nucleosome core particle offers a striking example of this effect. Minor groove narrowing is often associated with the presence of A-tracts, AT-rich sequences that exclude the flexible TpA step. These findings suggest that the ability to detect local variations in DNA shape and electrostatic potential is a general mechanism that enables proteins to use information in the minor groove, which otherwise offers few opportunities for the formation of base-specific hydrogen bonds, to achieve DNA binding specificity.
The ability of proteins to recognize specific DNA sequences is a hallmark of biological regulatory processes. The determination of the three-dimensional structures of numerous protein-DNA complexes has provided a detailed picture of binding, revealing a structurally diverse set of protein families that exploit a wide repertoire of interactions to recognize the double-helix1. Nucleotide sequence-specific interactions often involve the formation of hydrogen bonds between amino acid side chains and hydrogen bond donors and acceptors of individual base pairs. It has long been recognized that every base pair has a unique hydrogen bonding signature in the major groove but that this is not the case in the minor groove2. Thus, the expectation has been that the recognition of specific DNA sequences would take place primarily in the major groove through the formation of a series of amino acid- and base-specific hydrogen bonds1. This “direct readout” mechanism is consistent with observations derived from three-dimensional structures of protein-DNA complexes but it is far from the entire story.
In many complexes, the DNA assumes conformations that deviate from the structure of an ideal B-form double helix3–5, sometimes bending in such a way to optimize the protein-DNA interface6 and in some cases undergoing significant conformational changes as in the opening of the minor groove in the complex formed between TBP and the TATA box7,8. The term “indirect readout” was coined9 to describe such recognition mechanisms that depend on the propensity of a given sequence to assume a conformation that facilitates its binding to a particular protein. The bases involved in such mechanisms need not be in contact with the protein and, for example, can be found in linker sequences that connect two half-sites that themselves are bound by individual protein subunits10,11.
We recently described an example of a novel readout mechanism, the recognition of local sequence-dependent minor groove shape12, that is distinct from previously described indirect readout mechanisms. In this case, the sequence-dependence of minor groove width and corresponding variations in electrostatic potential are used by the Hox protein Sex combs reduced (Scr) to distinguish small differences in nucleotide sequence12. Here we report that this mechanism is a widely used mode of protein-DNA recognition that involves the creation of specific binding sites for positively charged amino-acids, primarily arginine, within the minor groove. Minor groove narrowing is found to be correlated with A-tracts13,14, usually defined as stretches of four or more As or Ts that do not contain the flexible TpA step15, but extended here to include as few as three base pairs (see below). Our results offer fundamentally new insights into the structural and energetic origins of protein-DNA binding specificity and thus have important implications for the prediction of transcription factor binding sites in genomes.
Arginine is enriched in narrow minor grooves
Figure 1a reports the percentage of minor groove contacts associated with each amino acid, classified according to the width of the minor groove. Arginine constitutes 28% of all amino acid residues that contact the minor groove and is significantly enriched in narrow minor grooves, defined here by a groove width of <5.0 Å (compared to 5.8 Å in ideal B-DNA). Remarkably, 60% of the residues in narrow minor grooves are arginines as compared to 22% in minor grooves that are defined as not narrow – i.e. width ≥5.0 Å. A smaller enrichment is also observed for lysines but the overall population of lysines within the minor groove is much less than for arginine.
Figure 1. Amino acid frequencies in minor grooves.
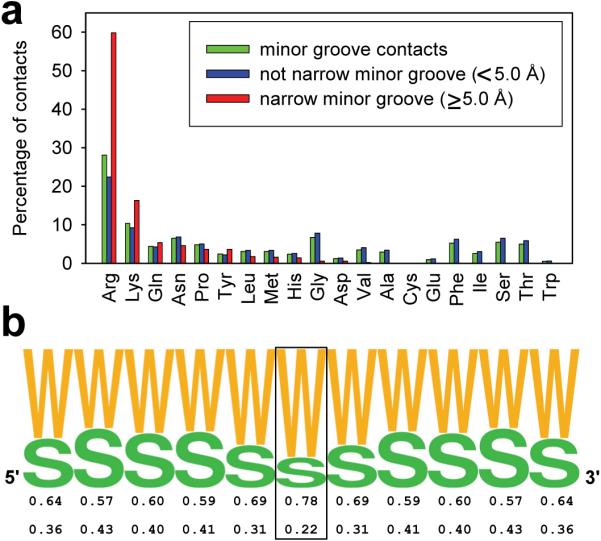
(a) Histograms for each amino acid illustrate the frequency with which they are observed in any minor groove (green), in minor grooves with a width of ≥5.0 Å (blue), and in narrow minor grooves of <5.0 Å width (red). (b) Frequency of AT (W) and GC (S) base pairs in sequences of 229 sites contacted by arginines in narrow minor grooves. The central base pair (boxed) is contacted by arginine. Frequencies are symmetrized by using both complementary strands.
Binding to the minor groove is a characteristic of many, but not all, protein superfamilies and a significant subset of these contact a narrow minor groove (Table 1). Moreover, if the minor groove is contacted, arginines are likely to be involved, and the likelihood that an arginine will be present becomes even greater for narrow minor grooves (Supplementary Table 1).
Table 1.
Protein superfamilies with minor groove contacts.
| SRF-like |
| IHF-like DNA-binding proteins |
| Histone-fold |
| DNA breaking-rejoining enzymes |
| Zn2/Cys6 DNA-binding domain |
| Homeodomain-like |
| p53-like transcription factors |
| lambda repressor-like DNA-binding domains |
| Winged helix DNA-binding domain |
| Leucine zipper domain |
| C-terminal effector domain of the bipartite response regulators |
| Restriction endonuclease-like |
| Glucocorticoid receptor-like (DNA-binding domain) |
| DNA repair protein MutS, domain I |
| Origin of replication-binding domain, RBD-like |
| DNA/RNA polymerases |
| Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment |
| Ribonuclease H-like |
| TATA-box binding protein-like |
Listed are SCOP superfamilies47 that have an arginine-minor groove contact within a distance of <6.0 Å from the base. Superfamilies that use arginine to contact a narrow minor groove (<5.0 Å) have grey shading; those that use arginine to contact a non-narrow minor groove (≥5.0 Å) are unshaded. Only superfamilies with a minimum of ten protein chains in PDB structures bound to DNA at least one helical turn long are included. The percentages of chains with minor groove contacts vary considerably among SCOP superfamilies and are provided in Supplementary Table 1.
Figure 1b compiles the DNA sequence preferences for protein-DNA complexes in which an arginine contacts a narrow minor groove. The figure shows that the base pair that has the shortest contact distance with the arginine guanidinium group has a probability of 78% of being an AT and 22% of being a GC. Neighboring base pairs in both the 5' and 3' directions surrounding the closest contacting base pair also have a strong tendency to be AT. Taken together, these data demonstrate that arginines tend to bind narrow minor grooves in AT-rich DNA.
AT-rich sequences tend to narrow minor grooves
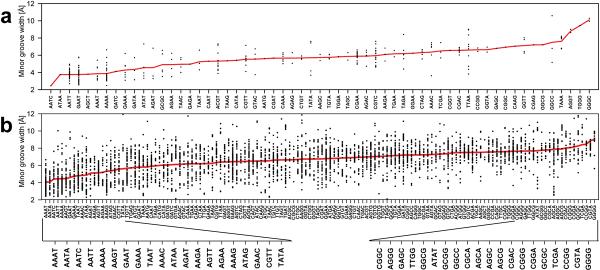
We calculated minor groove widths for all tetranucleotides contained in PDB structures for both free DNA (Figure 2a) and DNA in complexes with proteins (Figure 2b). There is a large spread of values due in part to end effects and to the effects of crystal packing but some trends are nevertheless evident. For example, for free DNA structures most of the tetranucleotides with narrow minor grooves (width <5.0 Å) are AT-rich (Figure 2a and Supplementary Table 2a). Similar behavior is observed in protein- DNA complexes (Figure 2b and Supplementary Table 2b). In contrast, tetranucleotides with wide minor grooves have a strong tendency to be GC-rich.
Figure 2. Distribution of tetranucleotide sequences according to average minor groove width.
Tetranucleotides from structures with a minimum length of one helical turn for which minor groove width can be defined are ordered by average minor groove width (red). The widths of all tetranucleotides are shown (black) and the sequence, average width, and occurrence in our dataset are given in Supplementary Table 2. (a) The 59 unique tetranucleotides from free DNA structures. (b) The set of all 136 unique tetranucleotides derived from protein-DNA complexes.
The correlation between AT content and groove width is not unexpected given the fact that A-tracts are known to produce narrow minor grooves. However, TpA steps have a tendency to widen the minor groove15, so it was of interest to determine whether the distinct properties of A-tracts and TpA steps are reflected in our tetranucleotide data set. We find that 67% of tetranucleotides composed only of AT base pairs have a narrow minor groove but that this number increases to 82% if we exclude TpA steps so as to consider only A-tracts. Even A-tracts of length three have a strong tendency to narrow the minor groove. Forty three percent of the tetranucleotides with a minor groove width of <5.0 Å have an A-tract of length three, a percentage that decreases to 11% of tetranucleotides with canonical minor groove widths (between 5.0 and 7.0 Å) and to 4% of tetranucleotides with minor grooves wider than 7.0 Å (Supplementary Figure 1). Additionally, compared to other AT-rich sequences, A-tracts are specifically enriched in DNAs with narrow minor grooves (Supplementary Figure 1). Thus, although A-tracts are usually thought of as requiring four or more base pairs, in part because a minimum of four is required to rigidify the DNA14, this analysis shows that A-tracts as short as length three are positively correlated with narrow minor grooves.
Arginines recognize enhanced electrostatic potentials
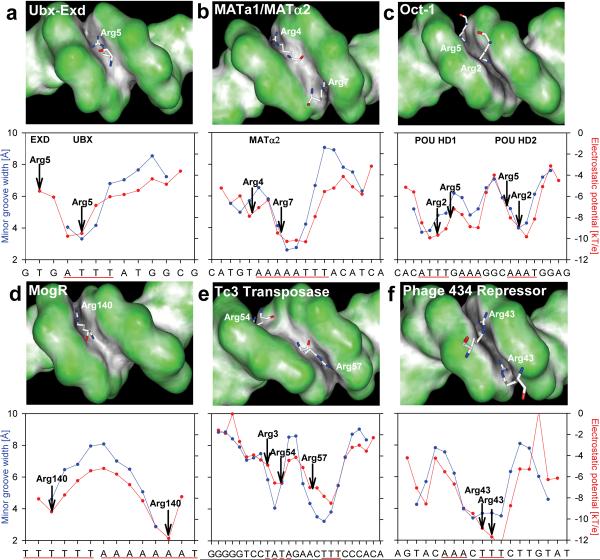
Figure 3 and Supplementary Figure 2 plot minor groove width and electrostatic potential vs. binding site sequence for several complexes whose binding interface includes an arginine inserted into the minor groove. The correlation of width and potential as well as the tendency of arginines to be located close to minima in width and potential is evident. Below we highlight a few specific examples of how arginine-minor groove interactions are used in DNA recognition.
Figure 3. Specific examples of minor groove shape recognition by arginines.
DNA shapes of the binding sites of (a) Ubx-Exd 1b8i16, (b) MATa1/MATα2 1akh17, and (c) Oct-1/PORE 1hf018, (d) the MogR repressor 3fdq19, (e) the Tc3 transposase 1u7822, and (f) the phage 434 repressor 2or123 are shown in GRASP surface representations31,50 with convex surfaces color-coded in green and concave surfaces in grey/black. Plots of minor groove width (blue) and electrostatic potential in the center of the minor groove (red) are shown below. Arginine contacts (defined by the closest distance between the guanidinium groups and the bases) are indicated. A-tract sequences are highlighted by a solid red line, the TATA box in (e) by a dashed line.
Figure 3a represents the ternary complex of the Hox protein Ultrabithorax (Ubx) and its cofactor Extradenticle (Exd) bound to DNA16. In this complex, Arg5 of Ubx, which is a conserved residue across all homeodomains, inserts into a narrow region formed by a four base pair A-tract. Figures 3b provides an example of a long and very narrow A-tract that binds α2-Arg7 from the MATa1/MATα2 complex with DNA17. In contrast, α2-Arg4 inserts into a shallower region at one end of the A-tract where there are local minima in width and potential that are smaller than at the Arg7 site in the center of the A-tract. The two POU-domains of the Oct-1/PORE complex bind to two A-tracts (Figure 3c) where the minima are positioned in such a way to provide binding sites for four arginines, two from each POU domain18.
The location of these A-tracts with respect to other nucleotide sequence features can be used to generate specificity, as previously discussed for the Hox protein Scr12. In the case of Scr binding, the position of a TpA step within an AT-rich region plays a critical role in binding specificity. A similar strategy is used by the MogR transcription factor where two long A-tracts separated by a TpA step produce two arginine binding sites19 (Figure 3d). The unique shape recognized by these two arginines is likely to contribute to the position of the MogR binding site along the DNA sequence. The overall tendency of TpA steps to widen the minor groove is most apparent when they are positioned between two A-tracts (as in Scr12 and MogR19) where the TpA step acts as a `hinge' between more rigid elements15,20. In other contexts, due to their flexibility, TpA steps can also be accommodated in narrow minor grooves21. An example is provided by the bipartite DNA-binding domain of Tc3 transposase where the arginines bind to a narrow region containing a TATA box22 that displays enhanced negative electrostatic potential (Figure 3e).
Although less frequent, arginines also bind narrow grooves associated with non-A- tract sequences. Figure 3f summarizes features of the binding of the 434 repressor to its operator23 which contains seven base pairs that are all AT with the exception of a central CG. (The guanine amino group tends to widen narrow grooves but a single GC base pair can be accommodated with only little disruption.)
Arginine-minor groove interactions in the nucleosome
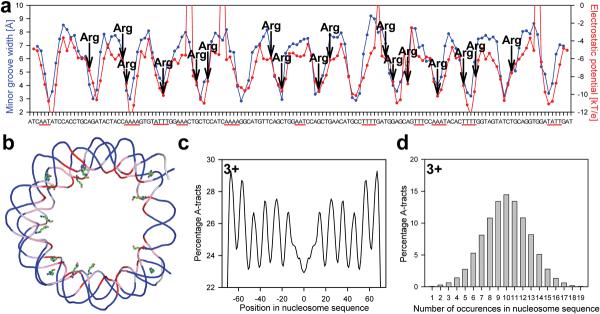
Figure 4a plots minor groove width and electrostatic potential along the DNA sequence of the nucleosome core particle containing recombinant histones and a 147 base pair DNA fragment (PDB code 1kx5)24. There are 14 minima in minor groove width corresponding to regions where the DNA bends so as to wrap around the histone core. As above, there is a striking correlation between width and potential. The variation in width between the narrowest and widest regions is about 5 Å and the difference between the maxima and minima in electrostatic potential is about 6 kT/e (Figure 4a). As a consequence, there should be a strong driving force for basic amino acids to bind to narrow regions and indeed arginines are found in 9 of the 14 minima. These arginines are shown in Figure 4b where the nucleosomal DNA has been color coded by minor groove width. (Although all 14 narrow minor groove regions are contacted by arginines24 only 9 of these satisfy our criteria of <6.0 Å between arginine atoms and base atoms in the groove). A similar repeating pattern of narrow minor grooves that are contacted by arginines is seen in all 35 available nucleosome crystal structures (Supplementary Figure 3a,b).
Figure 4. Minor groove shape recognition in the nucleosome.
(a) Correlation of minor groove width of the nucleosome core particle (PDB code 1kx5)24 (blue) and electrostatic potential (red). Arginine contacts (defined by the closest distance between the guanidinium groups and the bases) are indicated. A-tract sequences are highlighted by solid red lines. (b) Schematic representation of the DNA backbone in the nucleosome color-coded by minor groove width (red ≤4.0 Å, pink >4.0 Å and ≤5.0 Å, light blue >5.0 Å and ≤6.0 Å, dark blue >6.0 Å), including all arginines that contact the minor groove. (c) The distribution of A-tracts of length three base pairs or longer in 23,076 yeast nucleosome-bound DNA sequences29. (d) Histogram of the occurrence of A-tracts of length three or longer in the same dataset29.
Because short A-tracts narrow the minor groove and facilitate the bending of DNA, we would expect to see a periodicity of A-tracts in DNA sequences bound by nucleosomes in vivo. Previous analyses have focused on dinucleotide statistics25,26 although it has been known for some time that there is a periodic pattern of AAA and AAT trinucleotides in nucleosome core DNA27,28. An analysis of DNA sequences bound in vivo by yeast nucleosomes29 reveals a clear periodicity for A-tracts of at least length three (denoted 3+, Figure 4c). Moreover, nucleosomal DNAs contain, on average, 10.0 A-tracts of length 3+ (Figure 4d). Periodicity is also detected for A-tracts of length 4+ and even 5+, although the number per nucleosome decreases to 4.1 and 1.6, respectively (Supplementary Figure 3). Thus, even though long A-tracts tend to be excluded from the nucleosome30, A-tracts of ≤ five base pairs, when present, are used to facilitate bending of the DNA around the histone core.
To evaluate the effect of TpA steps, we compared the periodicities of A-tracts of length three to those of other trinucleotides composed only of AT base pairs. Trinucleotides that contain TpA steps exhibit a much weaker periodic signal than A-tracts of length three, which exclude the TpA step (Supplementary Figure 4). Together, this analysis suggests that many of the sequence periodicities in nucleosomal DNA reflect the presence of short A-tracts that lead to narrow regions in the minor groove that in turn are recognized by a complementary set of arginines present on the surface of the nucleosome core particle.
Effects of groove width on electrostatic potential
The remarkable correlation between minor groove width and electrostatic potential (Figures 3 and 4) is due primarily to the properties of the Poisson-Boltzmann (PB) equation that have been extensively discussed in the literature31. Biological macromolecules are less polarizable than the aqueous solvent and, in the language of classical physics, can be thought of as a low dielectric region embedded in a high dielectric solvent. Solutions of the PB equation for DNA showed that lines of electrostatic potential due to backbone phosphates follow the shape of the DNA and are the most negative within the grooves32. This effect is due to electrostatic focusing, first observed for the protein superoxide dismutase31, where the narrow active site focuses electric field lines away from the protein and into the high dielectric solvent. The same physical phenomenon produces enhanced potentials in grooves, accounting for the strong correlation described above.
In order to establish the source of the effect in quantitative terms, we calculated the potentials for the MogR binding site19 when the dielectric constant is set to 80 both inside the macromolecule and in the solvent (Figure 5, dashed line) and for the case where the two dielectric constants are different (Figure 5, solid line). Strikingly, a significant enhancement of electrostatic potentials is only observed when the dielectric constant of the macromolecule and solvent are different, reflecting the focusing of electric field lines described qualitatively above. The small effect seen when the dielectric constant is the same results from the phosphates being closer to the center of the groove when it is narrow (see Supplementary Figure 5 for a breakdown of the contributions to the net electrostatic potential). Both sets of calculations were carried out at physiological salt concentrations. Although ionic strength has as strong effect on the absolute values of the potentials, the effect remains qualitatively the same (Supplementary Figure 6).
Figure 5. The biophysical origins of the negative potential of narrow minor grooves.
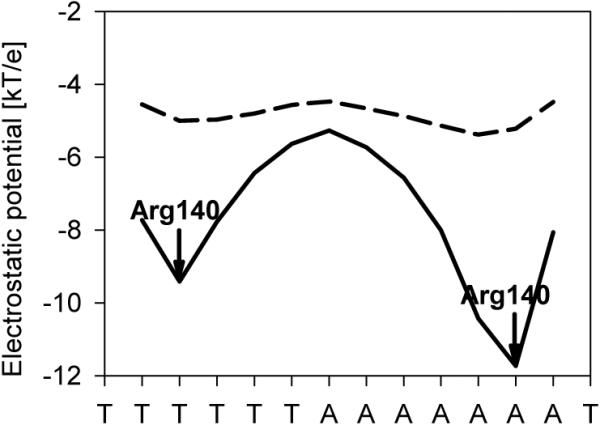
Electrostatic potential in the minor groove of the MogR binding site (PDB code 3fdq)19, calculated in the presence of a dielectric boundary (ε=2 in solute and ε=80 in solvent – solid line) and in the absence of a boundary (ε= 80 in both solute and solvent – dashed line).
Why are arginines preferred over lysines?
It is somewhat surprising that there is such a significant population of arginines in the minor groove, and a large enrichment when the groove is narrow, whereas the effects for lysines are more modest (Figure 1a). Arginines have been known for some time to be enriched relative to lysines in protein-protein33 and protein-DNA interfaces34 and the difference has generally been attributed to the ability of the guanidinium group to engage in more hydrogen bonds than the amino group of lysine35. To evaluate this idea we determined the number of hydrogen bonds formed by all the arginines and lysines in our data set that penetrate the minor groove. Surprisingly, on average, less than one hydrogen bond is formed by either amino acid side chain to DNA (0.9 for arginine and 0.6 for lysine), and the standard deviations are such that this difference is insignificant (Supplementary Table 3).
An alternate explanation derives from the difference in the size of the cationic moieties of the two residues. According to the classical Born model the solvation free energies of ions are proportional to the inverse of their radii31, suggesting that it is energetically less costly to remove a charged guanidinium group from water than it is to remove the smaller amino group of a lysine. To test this quantitatively, we calculated the change in free energy in transferring arginine and lysine from water to a medium of dielectric constant 2 (see Methods for details). The difference in the transfer free energies between the two residues ranges from 2.3 to 6.7 kcal/mole, depending on the force field that was used, with lysine consistently having the higher value (Supplementary Table 4). These results suggest that the higher prevalence of arginines compared to lysines in minor grooves is due, at least in part, to the greater energetic cost of removing a charged lysine from water than to remove a charged arginine.
Concluding remarks
We have shown that there is a dramatic enrichment of arginines in narrow regions of the DNA minor groove that provides the basis for a novel DNA recognition mechanism that is used by many families of DNA-binding proteins. A readout mechanism based on groove width requires a connection between sequence and shape. This connection appears to be provided in part by A-tracts, which have a strong tendency to narrow the groove, producing binding sites for arginines that, when spaced appropriately on the protein surface, offer a complementary set of positive charges that can recognize local variations in shape. Arginines often insert into the minor groove as part of short sequence motifs (e.g. RQR in the Hox protein Scr12, RKKR in POU homeodomains18, RPR in Engrailed36, RGHR in MATa1/MATα217, RRGR in the nuclear orphan receptor37 and RGGR in the human orphan receptor38), thus offering a variety of presentation modes that can contribute to the specificity of DNA shape recognition.
The tendency of A-tracts to narrow the minor groove is due primarily to their ability to assume conformations, through propeller twisting, that lead to the formation of inter- base pair hydrogen bonds in the major groove15. This network is disrupted by TpA steps as strikingly seen in the MogR binding site19. GC base pairs also have a tendency to widen the minor groove14. The combination of these and other factors, such as effects induced by flanking bases that are not directly located within the binding site39, can produce a complex minor groove landscape that offers numerous possibilities for specific interactions with proteins. Indeed, minor groove geometry is no doubt the result of the interplay of intrinsic and protein-induced structural effects.
The physical mechanisms described here are dramatically evident in the nucleosome. The energetic cost of narrowing and bending the DNA in regions where the backbone faces inward will be reduced by the presence of short A-tracts that have an intrinsic propensity to assume such conformations and hence to bend the DNA28. In addition, the penetration of arginines into the minor groove at sites where the DNA bends and the groove is narrow21,40 provides a significant stabilizing interaction
The variations in DNA shape observed in protein-DNA complexes often reflect conformational preferences of free DNA4,10,41. Sequence-dependent conformational preferences have also been observed in computational studies11,21,42 and, most recently, analysis of hydroxyl radical cleavage patterns shows that DNA shape is under evolutionary selection43. Such observations suggest that the role of DNA shape must be taken into consideration when annotating entire genomes and predicting transcription factor binding sites. The biophysical insights described here, together with the increased availability of high-throughput binding data, offer the hope of major progress in understanding how proteins recognize specific DNA sequences and in the development of improved predictive algorithms.
Methods Summary
Minor groove geometry was analyzed with Curves44 for all 1,031 crystal structures of protein-DNA complexes in the PDB that have any amino acid contacting base atoms. Protein side chains contact the minor groove in 69% of those structures that have at least one helical turn of DNA. The probabilities for each amino acid to contact the minor groove were calculated for three groups of DNAs: total, narrow, and not narrow. Proteins were grouped based on 40% sequence identity. The properties of free DNAs and DNAs bound to proteins were analyzed based on the minor groove widths of tetranucleotides, defined at the central base pair step.
All 35 crystal structures of the nucleosome available in the PDB were analyzed. The analysis of nucleosomal DNA is based on 23,076 sequences in an in vivo yeast dataset29. The signal for a sequence motif in nucleosomal DNA is positive for a base pair when the base pair comprises any part of the sequence motif. Frequencies were symmetrized by analyzing both complementary DNA strands.
Electrostatic potentials were obtained from solutions to the non-linear Poisson-Boltzman equation at physiologic ionic strength using the DelPhi program31,45. Regions inside the molecular surface of the DNA were assigned a dielectric constant of 2 while the solvent was assigned a value of 80. The potential is reported at a reference point at the center of the minor groove. The reference point is located close to the bottom of the groove in approximately the plane of a base pair. This definition provides a measure of electrostatic potential as a function of base sequence. Solvation free energies of amino acids were calculated for extended conformations of arginine and lysine side chains and compared for four different force fields.
Methods
Calculation of minor groove width
There were in total 1,031 crystal structures of protein-DNA complexes in the PDB as of June 1, 2008 in which the DNA was contacted by any amino acid side chain at a distance <6.0 Å from base atoms. Of these structures, 567 contained at least one helical turn, and no chemical modifications or deformations that prevent the calculation of minor groove width. Groove geometry was analyzed using Curves44 and minor groove width was calculated as a function of base sequence by averaging all the Curves levels given for each nucleotide.
Statistical analysis of protein-DNA contacts
Of the 567 protein-DNA structures in our dataset, 392 have at least one minor groove contact defined by a distance of <6.0 Å between any base and side chain atoms. To avoid an oversampling bias, proteins in this dataset that shared ≥40% sequence identity were grouped to create 109 groups. The average number of contacts within each group was subsequently averaged over all 109 groups. These averages were divided by the sum of the average number of contacts for all amino acids to calculate the total minor groove contacts, contacts in not narrow minor grooves (≥5.0 Å), and contacts in narrow minor grooves (<5.0 Å), for each amino acid.
Hydrogen bond contacts between amino acid side chains and the DNA bases and phosphates, water molecules, and other protein atoms were identified with the HBplus program46.
Structural annotation of DNA-binding proteins
The proteins in our dataset of protein-DNA complexes were classified in SCOP47 superfamilies. Proteins for which SCOP annotations were not available were annotated manually or using the ASTRAL database48.
Correlation of tetranucleotide structure and sequence
Tetranucleotides in free DNA and protein-DNA complexes were used to analyze the base sequence propensity of minor groove regions as a function of minor groove width. The minor groove width of a tetranucleotide was defined by the average of all Curves44 levels for groove width of the second nucleotide and the first level of the third nucleotide, which describes groove width at the central base pair step. End regions and irregular tetranucleotides were excluded by requiring groove width definitions for at least one Curves level of each of the four nucleotides. Tetranucleotides from nucleosomal DNA were excluded from this analysis because the DNA is strongly deformed and the spacing between narrow regions is fixed at about one helical turn, thus adding a bias to the results. When applied to the 521 protein-DNA complexes in our dataset, these criteria allowed the analysis of all 136 possible unique tetranucleotides. When applied to the 88 free DNA structures in our dataset, the same criteria resulted in the analysis of 59 unique tetranucleotides. In order to increase coverage for the free DNA dataset, NMR structures were included if dipolar coupling data were used in the refinement.
Propensity of sequence motifs in nucleosomes
The structural analysis of nucleosomes includes all 35 crystal structures in the PDB as of May 1, 2009. The sequence analysis was based on 23,076 nucleosome sequences of length 146–148 base pairs in a yeast in vivo dataset29. These nucleosome sites were scanned for sequence motifs such as A-tracts of different length, TpA steps, or other AT- rich regions. A given motif contributed to a positive signal for any base pair that overlapped that motif, thus longer motifs contributed signals to more nucleotide positions. The frequencies of all motifs were symmetrized by analyzing both complementary strands.
Calculations of electrostatic potentials
Electrostatic potentials were obtained from solutions to the non-linear Poisson- Boltzman equation at 0.145 M salt using the DelPhi program31,45. Partial charges and atomic radii were taken from the Amber force field49. The interior of the molecular surface of the solute molecule (calculated with a 1.4 Å probe sphere) was assigned a dielectric constant of ε=2 while the exterior aqueous phase was assigned a value of ε=80. Debye-Hückel boundary conditions and five focusing steps were used with a cubic grid size of 165 (a grid size of 185 was used for the nucleosome).
The electrostatic potential is reported at a reference point close to the bottom of the minor groove approximately in the plane of base pair i. The reference point i is defined as the geometric midpoint between the O4' atoms of nucleotide i+1 in the 5'-3' strand and nucleotide i−1 in the 3'−5' strand12. Where the DNA strongly bends into the major groove the reference point can clash with the guanine amino group and cause large positive potentials (as seen in Figure 4a for three regions of the nucleosome).
Desolvation free energies were calculated with the DelPhi program31,45 for the transfer of arginine and lysine side chains in extended conformations from water to a medium of dielectric constant ε=2. Transfer free energies were calculated for each of the two side chains based on charge distributions and atomic radii taken from Amber49 and three other force fields (see Supplementary Table 3).
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM54510 (R.S.M.) and U54 CA121852 (B.H. and R.S.M.). The authors thank Andrea Califano for many helpful conversations.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8(5):937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travers AA. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- 4.Shakked Z, et al. Determinants of repressor/operator recognition from the structure of the trp operator binding site. Nature. 1994;368(6470):469–473. doi: 10.1038/368469a0. [DOI] [PubMed] [Google Scholar]
- 5.Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J Mol Biol. 2000;300(4):819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 6.Hegde RS, Grossman SR, Laimins LA, Sigler PB. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359(6395):505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 8.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z, et al. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 10.Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine--thymine tract and its role in gene regulation. Proc Natl Acad Sci U S A. 2001;98(15):8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13(10):1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Joshi R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131(3):530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhoff AM, Tullius TD. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- 14.Haran TE, Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys. 2009;42(1):41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 15.Crothers DM, Shakked Z. DNA bending by adenine-thymine tracts. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structures. Oxford University Press; London: 1999. pp. 455–470. [Google Scholar]
- 16.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397(6721):714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Jin Y, Vershon AK, Wolberger C. Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract. Nucleic Acids Res. 1998;26(24):5707–5718. doi: 10.1093/nar/26.24.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remenyi A, et al. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol Cell. 2001;8(3):569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 19.Shen A, Higgins DE, Panne D. Recognition of AT-Rich DNA Binding Sites by the MogR Repressor. Structure. 2009;17:769–777. doi: 10.1016/j.str.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci U S A. 2004;101(5):1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J Mol Biol. 2007;371(3):725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watkins S, van Pouderoyen G, Sixma TK. Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA. Nucleic Acids Res. 2004;32(14):4306–4312. doi: 10.1093/nar/gkh770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 24.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 25.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 28.Travers AA, Klug A. Bending of DNA in nucleoprotein complexes. In: Cozzarelli NR, Wang JC, editors. DNA Topology and its Biological Effects. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1990. pp. 57–106. [Google Scholar]
- 29.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4(11):e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 32.Jayaram B, Sharp KA, Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989;28(5):975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- 33.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 1997;6(1):53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38(7):1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- 35.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29(13):2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 37.Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat Struct Biol. 1999;6(5):471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- 38.Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. 2003;327(4):819–832. doi: 10.1016/s0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- 39.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr Opin Struct Biol. 2009;19(2):171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 41.Locasale JW, Napoli AA, Chen S, Berman HM, Lawson CL. Signatures of protein-DNA recognition in free DNA binding sites. J Mol Biol. 2009;386(4):1054–1065. doi: 10.1016/j.jmb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolstorukov MY, Virnik KM, Adhya S, Zhurkin VB. A-tract clusters may facilitate DNA packaging in bacterial nucleoid. Nucleic Acids Res. 2005;33(12):3907–3918. doi: 10.1093/nar/gki699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324(5925):389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6(4):655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 45.Rocchia W, et al. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 46.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 47.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 48.Brenner SE, Koehl P, Levitt M. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res. 2000;28(1):254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornell WD, et al. A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. J Am Chem Soc. 1995;117(19):5179–5197. [Google Scholar]
- 50.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.