Abstract
More than 150 genes have been identified that affect skin color either directly or indirectly, and we review current understanding of physiological factors that regulate skin pigmentation. We focus on melanosome biogenesis, transport and transfer, melanogenic regulators in melanocytes and factors derived from keratinocytes, fibroblasts, endothelial cells, hormones, inflammatory cells and nerves. Enzymatic components of melanosomes include tyrosinase, tyrosinase-related protein 1 and dopachrome tautomerase, which depend on the functions of OA1, P, MATP, ATP7A and BLOC-1 to synthesize eumelanins and pheomelanins. The main structural component of melanosomes is Pmel17/gp100/Silv, whose sorting involves adaptor protein 1A (AP1A), AP1B, AP2 and spectrin, as well as a chaperone-like component, MART-1. During their maturation, melanosomes move from the perinuclear area toward the plasma membrane. Microtubules, dynein, kinesin, actin filaments, Rab27a, melanophilin, myosin Va and Slp2-a are involved in melanosome transport. Foxn1 and p53 up-regulate skin pigmentation via bFGF and POMC derivatives including α-MSH and ACTH, respectively. Other critical factors that affect skin pigmentation include MC1R, CREB, ASP, MITF, PAX3, SOX9/10, LEF-1/TCF, PAR-2, DKK1, SCF, HGF, GM-CSF, endothelin-1, prostaglandins, leukotrienes, thromboxanes, neurotrophins and neuropeptides. UV radiation up-regulates most factors that increase melanogenesis. Further studies will elucidate the currently unknown functions of many other pigment genes/proteins.
Keywords: melanin, melanosome, pigmentation, regulation, skin
1. Introduction
Key players that regulate human skin pigmentation include melanocytes in the epidermis that synthesize the melanin and neighbouring keratinocytes that receive and distribute it in upper layers of the skin [1]. Other intrinsic factors that help regulate skin pigmentation include fibroblasts in the dermis that affect overlying melanocytes and keratinocytes, endocrine factors from the blood supply, as well as neural factors and inflammation-related factors. Extrinsic factors that directly and/or indirectly affect skin pigmentation include ultraviolet (UV) radiation [2]. Indeed, more than 150 genes have now been identified that affect pigmentation of the skin, hair and/or eyes based on studies of mouse coat color mutants [3] and melanosomal components identified by proteomics analyses [4] - updated lists of those genes can be found at http://www.espcr.org/micemut. Since it would be impossible to detail all those genes/proteins and their functions here, we briefly review recent advances and topics in understanding physiological factors that regulate human skin pigmentation, focusing on melanocytes, keratinocytes and fibroblasts.
2. Factors in melanocytes that regulate skin pigmentation
2.1 Melanosome biogenesis
Melanocytes are unique cells that produce melanosomes, specific melanin-containing intracellular organelles that share several features with lysosomes in that they contain acid-dependent hydrolases and lysosomal-associated membrane proteins (LAMPs) [5]. In fact, melanosomes belong to a family of cell-specific organelles, termed lysosome-related organelles (LROs), which also include lytic granules observed in cytotoxic T lymphocytes and natural killer cells, MHC class II compartments (MIICs) observed in antigen presenting cells, platelet-dense granules, basophil granules, azurophil granules observed in neutrophils and Weibel-Palade bodies observed in endothelial cells [6]. Several pigmentary disorders, including Chediak-Higashi syndrome and Hermansky-Pudlak syndrome (HPS), which have specific symptoms such as infections related to immunological deficiency (caused by the enlargement of lytic granules, MIICs and azurophil granules) and prolonged bleeding times related to the platelet dysfunction (caused by the absence of apparent platelet-dense granules), respectively, underline the importance of studying the biogenesis of LROs [7].
Melanosomes may be the best tool to study the biogenesis of LROs since they can be morphologically classified into four distinct stages (I-IV) according to their degree of maturation. Intraluminal fibrils begin to form in amorphous spherical stage I melanosomes and generate a meshwork characteristic of stage II melanosomes, both stages lacking melanin pigment and being commonly called early melanosomes. Melanin synthesis begins within the fibrillar stage II melanosomes and the melanins are deposited uniformly on the internal fibrils resulting in the production of stage III melanosomes. In heavily pigmented melanocytes, all structural detail is eventually obscured due to the presence of copious amounts of melanin in stage IV melanosomes. Melanosomes are classified as LROs and recent studies characterizing the proteomes of early melanosomes show that they are derived from the endoplasmic reticulum (ER), coated vesicles, lysosomes and endosomes [4,8,9]. Most of the pigment-specific proteins that affect skin pigmentation are localized in melanosomes [3], and consist of enzymatic components required for melanin synthesis, structural fibrillar components required for melanosome structure and binding of melanin, and other protein components with currently unknown functions [10].
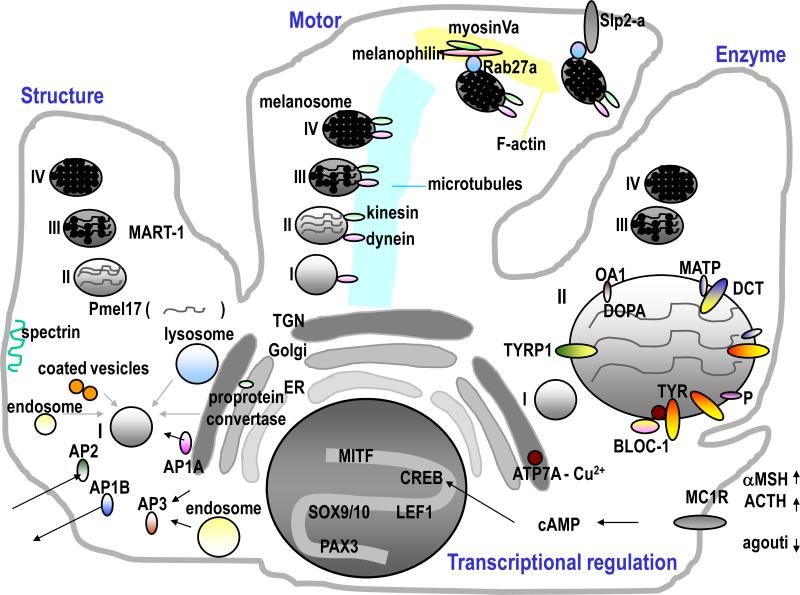
Enzymatic components of melanosomes include tyrosinase (TYR), a critical copper-dependent enzyme required for melanin synthesis, disruption of which is responsible for oculocutaneous albinism (OCA) type 1, tyrosinase-related protein 1 (TYRP1), mutations in which result in OCA3, and dopachrome tautomerase (DCT) (Fig 1, right). Those three enzymes cooperate to synthesize two distinct types of melanins: black-brown eumelanins and yellow-reddish pheomelanins [11]. OA1, a G-protein coupled receptor localized on melanosomal membranes, acts as a selective L-DOPA receptor [12]. P, mutations in which result in OCA2, affects the sorting of TYR, while MATP, mutations in which result in OCA4, affects the sorting of both TYR and TYRP1 [13]. The copper transporter ATP7A, mutations in which result in Menkes disease, transiently colocalizes with TYR in the trans-Golgi network (TGN) and then coexists with TYR within melanosomes. Correct trafficking of those components to melanosomes depends on the function of a sorting component known as Biogenesis of Lysosome-related Organelles Complex (BLOC)-1 as shown in Fig 1 [14]. The disruption of BLOC-1 results in HPS-7 or HPS-8, which suggests in turn that BLOC-1 regulates TYR activity via copper function as a cofactor [15]. Adaptor proteins (APs) also regulate correct protein trafficking in the TGN and in endosomes. β3A-adaptin, a subunit of AP3 [16], is mutated in ‘pearl’ mice, a model for HPS-2 [17].
Fig 1. Factors that affect skin pigmentation within melanocytes.
Schemes show components involved in melanosome structure (left), transport (middle), enzyme (right) and transcriptional (bottom) related factors.
The main structural fibrillar component of melanosomes is Pmel17/gp100/Silv, which is cleaved in a post-Golgi compartment by a furin-like proprotein convertase [18] and is then trafficked to stage I melanosomes to produce the internal fibrils (Fig 1, left) [19]. Indeed the sorting of Pmel17 to melanosomes is very complex and has been intensively investigated by many groups which have revealed its sorting via multiple intracellular transport pathways [20], including retrograde transport from endosomes to the TGN [21]. Thus in addition to its direct sorting from the TGN via AP1A, the indirect sorting of Pmel17 to the plasma membrane occurs via AP1B, which is an epithelial cell-specific complex associated with cellular polarity, after which Pmel17 is internalized and then redirected to melanosomes via AP2 [22]. This trafficking pattern is further supported by a different study that compared the spectrin/ankyrin system, which is involved in the sorting of cargo from the plasma membrane, between non-pigmented melanoma cells (SK-Mel-28) and pigmented melanoma cells (MNT-1). The lack of spectrin in the plasma membrane of SK-Mel-28 cells may be responsible for their hypopigmentation by affecting the trafficking of melanosomal proteins via the early secretory pathways [23]. Other factors, including the content of core 1 O-glycans modified with sialic acid, also influence the sorting of Pmel17 and determine its capacity to form fibrils in melanosomes [24]. MART-1, a melanosomal protein whose gene was initially cloned using melanoma reactive CD8+ T cells, forms a complex with Pmel17 and influences its expression, stability, processing and trafficking, which suggests the role of MART-1 as a chaperone-like structural component rather than an enzymatic component [25].
2.2 Melanosome transport and transfer
Melanosomes move from the perinuclear area of melanocytes where they are produced toward the plasma membrane as they become more melanized due to the functions of microtubules, actin filaments and myosin, which is similar to the movement of other organelles in other types of cells (Fig 1, middle) [26]. Dynein and kinesin mediate that microtubule-dependent intracellular transport [23]. Three different pigment genes are known so far to be involved in the dynamic movement of melanosomes, and mutations in any of those genes elicit a remarkable accumulation of pigments in the perinuclear region of mutant melanocytes due to the disruption of their transport to the cell periphery [27]. Rab27a, melanophilin and myosin Va make a complex to link melanosomes to the F-actin based motors. Rab27a (known as ashen in mice), melanophilin (known as leaden) and myosin Va (known as dilute) and mutations of those genes cause various forms of Griscelli syndrome (types II, III and I, respectively) in humans [28]. Rab27a links synaptoagmin-like protein2-a (Slp2-a) with phosphatidylserine, thereby docking melanosomes at the plasma membrane, which suggests the role of Slp2-a as a regulator of melanosome exocytosis [29].
2.3 Melanogenic regulators in melanocytes
The most well-known receptor on melanocytes that modulates their function is the melanocortin-1 receptor (MC1R), activation of which increases cAMP production that leads to the phosphorylation of cAMP responsive-element-binding protein (CREB) transcription factor family members (Fig 1, bottom) [30]. Agonists of the MC1R include α-melanocyte-stimulating hormone (α-MSH) and adrenocorticotropic hormone (ACTH), while the only known antagonist of the MC1R is agouti signal protein (ASP), which modulates eumelanin versus pheomelanin production. Polymorphisms of MC1R have been extensively investigated with respect to their roles in responses to UV radiation and/or in controlling constitutive skin pigmentation among racial/ethnic groups [31]. Other receptors involved in regulating melanocyte function will be briefly discussed below.
The most critical transcription factor that regulates melanocyte function is microphthalmia transcription factor (MITF), mutations of which result in Waardenburg syndrome type 2 (WS2) [32]. The MITF promoter is itself regulated by various other transcription factors, including PAX3 (which is a neural-crest-associated transcription factor and is associated with WS1 and WS3), SOX9 [33], SOX10 (mutations in which result in WS4), LEF-1/TCF (a downstream regulator of the Wnt/β-catenin signaling pathway) and CREB [34]. Various physiological factors from keratinocytes, fibroblasts and other sources (as discussed below) regulate the expression levels and functions of MITF [35].
3. Factors produced by keratinocytes that regulate skin pigmentation
3.1 Melanosome transfer
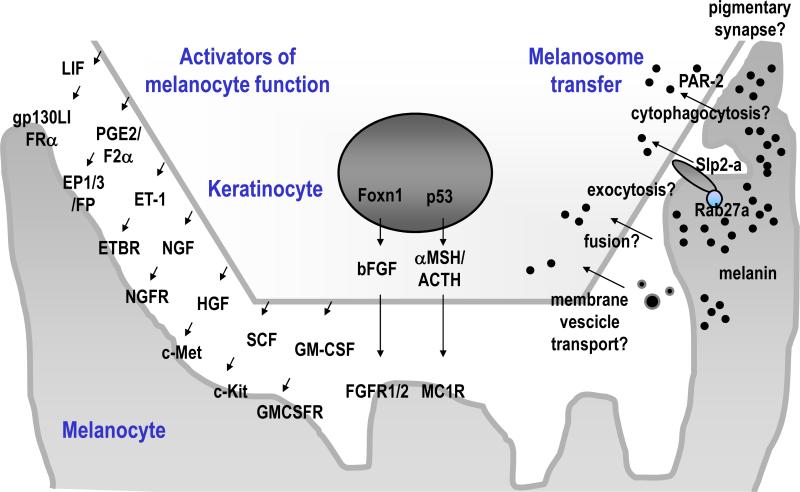
The process by which melanosomes are transferred to keratinocytes is not fully understood at this time although various hypotheses have been proposed, including: exocytosis, cytophagocytosis, fusion and membrane vesicle transport [28]. Similar to synapses in neural systems, a pigmentary synapse must exist between melanocytes and keratinocytes (Fig 2, right). One receptor that is expressed on keratinocytes and seems to be closely involved with melanosome transfer is protease (proteinase)-activated receptor-2 (PAR-2), which is a G-protein coupled receptor. PAR-2 mediates the phagocytosis of melanosomes in a Rho-dependent manner [36]. Various physiological factors, including dickkopf 1 (DKK1), an inhibitor of the Wnt/β-catenin pathway [37], regulate the expression of PAR-2 [38].
Fig 2. Keratinocyte-derived factors that regulate skin pigmentation.
Note that Foxn1 and p53 directly up-regulate skin pigmentation via bFGF and POMC derivatives, respectively. Each of those factors enhances melanocyte growth and function. The right side of the scheme shows several hypotheses for melanosome transfer.
3.2 Regulation of melanocytes by keratinocyte-derived factors
Recently, Foxn1/Whn/Hfh11, a transcription factor expressed by keratinocytes, was identified as a regulator not only of keratinocyte growth and differentiation but also of melanocyte recruitment and induction of pigmentation in the skin via basic fibroblast growth factor (bFGF) production (Fig 2, middle) [39]. In other words, Foxn1 serves as an activator of the pigment recipient phenotype by recruiting melanocytes to their position and by inducing melanosome transfer. Keratinocyte-derived factors that act as activators of melanocytes include not only bFGF but also stem cell factor (SCF/steel factor), hepatocyte growth factor (HGF), granulocyte-macrophage colony-stimulating factor (GM-CSF), nerve growth factor (NGF), α-MSH, ACTH, endorphin, endothelin-1 (ET-1), prostaglandin (PG)E2/PGF2α and leukemia inhibitory factor (LIF) [40]. The corresponding receptors on melanocytes for each of those factors are shown schematically in Fig 2, left. Whether any or all of those factors are regulated by Foxn1 and/or by other factors needs to be clarified in the future.
4. Regulation of melanocyte function by fibroblast-derived factors
4.1 Dickkopf 1 (DKK1) as a regulator of site-specific pigmentation
Skin on the palms of the hands and soles of the feet (palmoplantar areas) is different from skin at other sites of body (termed non-palmoplantar skin) in terms of its hypopigmented and glabrous (non-hairy) nature and its thicker epidermis [41]. Characterization of the differences between palmoplantar and non-palmoplantar skin is useful to elucidate the mechanisms underlying the physiological regulation of pigmentation, hair morphogenesis and thickness of the epidermis and to improve site-specific tissue regeneration [35]. When non-palmoplantar epidermis is transplanted onto a palmoplantar skin defect (e.g. a wound), the palmoplantar phenotype is gradually assumed by the grafted non-palmoplantar epidermis [42]. This clinical significance of epidermal grafting has been recently proven by treating patients with diabetic foot ulcers [43] or with rheumatic ulcers [44] and has disproved the conventional theory that the thicker the graft is the better the results will be [45]. Palmoplantar fibroblasts have the ability to induce the expression of palmoplantar-specific keratin in non-palmoplantar keratinocytes in vitro [46] and microarray studies identified the up-regulated expression of DKK1 in palmoplantar fibroblasts [47]. DKK1 from the mesenchyme seems to be responsible for the site-specificity of palmoplantar skin in terms of its pigmentation [48], glabrous condition [49] and thickness [38]. Studies in mice have shown that the thickened nature of the skin in palmoplantar areas results from the high expression of HOXA13 transcription factor in palmoplantar fibroblasts, followed by the increased secretion of Wnt5a, which suggests the possible involvement of factors in addition to DKK1 in regulating site-specific skin pigmentation [50].
4.2 Regulators of melanocytes by factors derived from fibroblasts
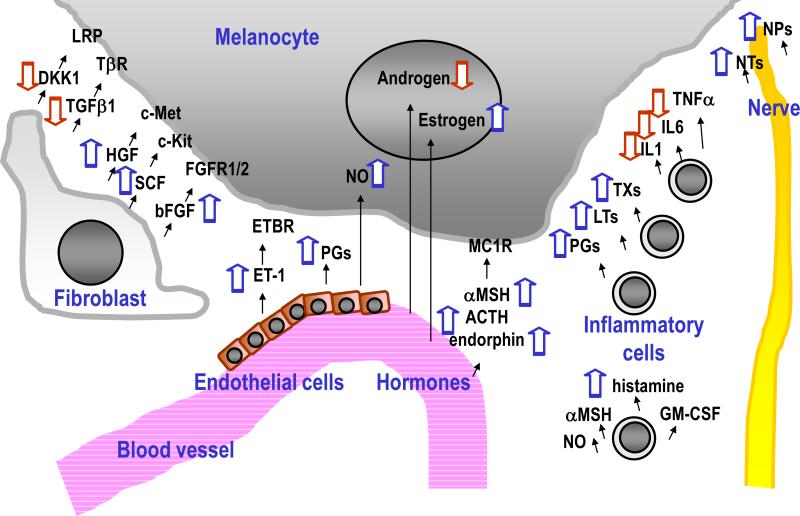
Reagents developed as cosmetic products to reduce skin pigmentation have been reviewed elsewhere [51], but few physiological factors that decrease pigmentation are derived from fibroblasts, except for DKK1 and transforming growth factor-β1 (TGFβ1) (Fig 3, left). Fibroblast-derived factors that act as activators of melanocytes include bFGF, HGF and SCF, all of which are also secreted from keratinocytes [1]. Further studies will probably elucidate other factors derived from fibroblasts that regulate skin pigmentation.
Fig 3. Factors that affect skin pigmentation derived from fibroblasts, blood, inflammatory cells and nerves.
Various factors are shown that up-regulate (blue arrows pointing upward) or down-regulate (red arrows pointing downward) skin pigmentation.
5. Other physiological factors that regulate skin pigmentation
5.1 Regulation of melanocytes by intrinsic factors
Intrinsic factors that regulate pigmentation originate not only from keratinocytes and fibroblasts but also from endothelial cells and hormones via the blood supply, from inflammatory cells and from the nervous system (Fig 3, middle and right). Endothelial cells are sources of ET-1, PGs and nitric oxide (NO). NO not only has a role in smooth muscle relaxation but also initiates melanogenesis, erythema and immunosuppression in response to UV radiation. Hormonal factors that stimulate pigmentation include estrogen (which is related to pregnancy-induced pigmentation), α-MSH, ACTH and endorphin (the latter 3 being produced by cleavage of pro-opiomelanocortin/POMC), while androgens have inhibitory effects on melanocytes [52].
Hyperpigmentation is clinically observed in response to inflammation. Arachidonate derived chemical mediators, especially PGs including PGE2 and PGF2α, leukotrienes (LTs) including LTC4 and LTD4 and thromboxanes (TXs) including TXB2, are responsible for the pigmentation induced by inflammation since those chemicals are enhancers of TYR activity [2]. Other inflammation-related factors that enhance melanogenesis include histamine, NO, GM-CSF and α-MSH. However, not all inflammatory cytokines increase skin pigmentation. Interleukin (IL)-1, IL-6 and tumor necrosis factor-α (TNFα) are known to suppress skin pigmentation, which suggests the importance of further elucidating the relationship between melanogenesis and inflammation.
Since melanocytes and neurons are derived from neural crest cells during embryogenesis, nerve cell-derived factors, including neurotrophins (NTs, such as NGF) and neuropeptides (NPs, such as calcitonin gene-related peptide/CGRP) have stimulatory effects on melanocytes, which supports the neuronal theory to explain the occurrence of segmental vitiligo [53].
5.2 Regulation of melanocytes by extrinsic factors
UV is the most powerful and well-known extrinsic factor that enhances skin pigmentation and most factors discussed above that increase melanogenesis are up-regulated in response to UV radiation [54]. UV facilitates the distribution of melanosomes not only to the supranuclear areas of keratinocytes to protect them against DNA damage but also to the upper epidermis to protect the lower epidermis, which contains melanocytes and keratinocyte stem cells [55]. UV also induces the apoptosis of keratinocytes containing melanin in the upper epidermis to protect against the growth of cells with unrepaired DNA damage [56]. The expression of the oncogene p53 is up-regulated in response to UV and may initiate the tanning cascade [57]. p53 directly regulates the induction of POMC/α-MSH/endorphin, suggesting its central role in the suntan response [58]. Future studies characterizing the short- and long-term effects of UV radiation on the pigmentation of human skin will further elucidate the complicated mechanisms of UV tanning using various biomarkers [59].
6. Concluding remarks
Many research groups are investigating the regulation of skin pigmentation with the goal of developing hypopigmenting and/or tanning cosmetics and also to elucidate the mechanisms of pigmentary disorders in order to cure and/or prevent those diseases. Additionally, anti-cancer studies aiming for the cure of malignant melanoma will never be accomplished without understanding the physiology of normal melanocytes. We have briefly reviewed progress in our knowledge of the regulation of skin color, although to cover all topics in depth was impossible and interested readers should refer to the original references cited for more details. In particular, advances have been remarkable in furthering our understanding of mechanisms underlying the biogenesis of melanosomes and how they are transported and transferred to keratinocytes. Our understanding of how keratinocytes in the epidermis recruit melanocytes and regulate skin pigmentation has also improved dramatically as has our understanding of the important role played by fibroblasts in the underlying dermis. We also reviewed the necessity for further studies in this field focusing on pigment genes/proteins with currently unknown functions and on various factors that affect skin pigmentation from outside of melanocytes.
Acknowledgments
This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, and Technology (Japan; no. 18689028), by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by a grant from the Cosmetology Foundation, and by a grant from the Aichi Cancer Research Foundation. We would like to thank Prof. Dr. Akimichi Morita, MD, PhD, Chairman of Department of Geriatric and Environmental Dermatology, Nagoya City University Graduate School of Medical Sciences, for the critical review of this work.
References
- 1.Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007;282:27557–61. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- 2.Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. Faseb J. 2007;21:976–94. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 3.Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003;16:333–44. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 4.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, Huang H, Canfield VA, Cheng KC, Yang F, Abe R, Yamagishi S, Shabanowitz J, Hearing VJ, Wu C, Appella E, Hunt DF. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–44. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Marks MS. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–97. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell'Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. Faseb J. 2000;14:1265–78. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- 7.Spritz RA, Chiang PW, Oiso N, Alkhateeb A. Human and mouse disorders of pigmentation. Curr Opin Genet Dev. 2003;13:284–9. doi: 10.1016/s0959-437x(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 8.Kushimoto T, Basrur V, Valencia J, Matsunaga J, Vieira WD, Ferrans VJ, Muller J, Appella E, Hearing VJ. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc Natl Acad Sci U S A. 2001;98:10698–703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basrur V, Yang F, Kushimoto T, Higashimoto Y, Yasumoto K, Valencia J, Muller J, Vieira WD, Watabe H, Shabanowitz J, Hearing VJ, Hunt DF, Appella E. Proteomic analysis of early melanosomes: identification of novel melanosomal proteins. J Proteome Res. 2003;2:69–79. doi: 10.1021/pr025562r. [DOI] [PubMed] [Google Scholar]
- 10.Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Costin GE, Valencia JC, Wakamatsu K, Ito S, Solano F, Milac AL, Vieira WD, Yamaguchi Y, Rouzaud F, Petrescu AJ, Lamoreux ML, Hearing VJ. Mutations in dopachrome tautomerase (Dct) affect eumelanin/pheomelanin synthesis, but do not affect intracellular trafficking of the mutant protein. Biochem J. 2005;391:249–59. doi: 10.1042/BJ20042070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez VM, Decatur CL, Stamer WD, Lynch RM, McKay BS. L-DOPA is an endogenous ligand for OA1. PLoS Biol. 2008;6:e236. doi: 10.1371/journal.pbio.0060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costin GE, Valencia JC, Vieira WD, Lamoreux ML, Hearing VJ. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J Cell Sci. 2003;116:3203–12. doi: 10.1242/jcs.00598. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama K, Bonifacino JS. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic. 2002;3:666–77. doi: 10.1034/j.1600-0854.2002.30908.x. [DOI] [PubMed] [Google Scholar]
- 15.Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–6. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dell'Angelica EC, Ooi CE, Bonifacino JS. Beta3A-adaptin, a subunit of the adaptor-like complex AP-3. J Biol Chem. 1997;272:15078–84. doi: 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- 17.Feng L, Seymour AB, Jiang S, To A, Peden AA, Novak EK, Zhen L, Rusiniak ME, Eicher EM, Robinson MS, Gorin MB, Swank RT. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999;8:323–30. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- 18.Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol. 2003;161:521–33. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoashi T, Muller J, Vieira WD, Rouzaud F, Kikuchi K, Tamaki K, Hearing VJ. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J Biol Chem. 2006;281:21198–208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 20.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 21.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–79. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 22.Valencia JC, Watabe H, Chi A, Rouzaud F, Chen KG, Vieira WD, Takahashi K, Yamaguchi Y, Berens W, Nagashima K, Shabanowitz J, Hunt DF, Appella E, Hearing VJ. Sorting of Pmel17 to melanosomes through the plasma membrane by AP1 and AP2: evidence for the polarized nature of melanocytes. J Cell Sci. 2006;119:1080–91. doi: 10.1242/jcs.02804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watabe H, Valencia JC, Le Pape E, Yamaguchi Y, Nakamura M, Rouzaud F, Hoashi T, Kawa Y, Mizoguchi M, Hearing VJ. Involvement of dynein and spectrin with early melanosome transport and melanosomal protein trafficking. J Invest Dermatol. 2008;128:162–74. doi: 10.1038/sj.jid.5701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valencia JC, Rouzaud F, Julien S, Chen KG, Passeron T, Yamaguchi Y, Abu-Asab M, Tsokos M, Costin GE, Yamaguchi H, Jenkins LM, Nagashima K, Appella E, Hearing VJ. Sialylated core 1 O-glycans influence the sorting of Pmel17/gp100 and determine its capacity to form fibrils. J Biol Chem. 2007;282:11266–80. doi: 10.1074/jbc.M608449200. [DOI] [PubMed] [Google Scholar]
- 25.Hoashi T, Watabe H, Muller J, Yamaguchi Y, Vieira WD, Hearing VJ. MART-1 is required for the function of the melanosomal matrix protein PMEL17/GP100 and the maturation of melanosomes. J Biol Chem. 2005;280:14006–16. doi: 10.1074/jbc.M413692200. [DOI] [PubMed] [Google Scholar]
- 26.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 27.Barral DC, Seabra MC. The melanosome as a model to study organelle motility in mammals. Pigment Cell Res. 2004;17:111–8. doi: 10.1111/j.1600-0749.2004.00138.x. [DOI] [PubMed] [Google Scholar]
- 28.Van Den Bossche K, Naeyaert JM, Lambert J. The quest for the mechanism of melanin transfer. Traffic. 2006;7:769–78. doi: 10.1111/j.1600-0854.2006.00425.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda TS, Fukuda M. Rab27A-binding protein Slp2-a is required for peripheral melanosome distribution and elongated cell shape in melanocytes. Nat Cell Biol. 2004;6:1195–203. doi: 10.1038/ncb1197. [DOI] [PubMed] [Google Scholar]
- 30.Rees JL. Genetics of hair and skin color. Annu Rev Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- 31.Rouzaud F, Costin GE, Yamaguchi Y, Valencia JC, Berens WF, Chen KG, Hoashi T, Bohm M, Abdel-Malek ZA, Hearing VJ. Regulation of constitutive and UVR-induced skin pigmentation by melanocortin 1 receptor isoforms. Faseb J. 2006;20:1927–9. doi: 10.1096/fj.06-5922fje. [DOI] [PubMed] [Google Scholar]
- 32.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 33.Passeron T, Valencia JC, Bertolotto C, Hoashi T, Le Pape E, Takahashi K, Ballotti R, Hearing VJ. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc Natl Acad Sci U S A. 2007;104:13984–9. doi: 10.1073/pnas.0705117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–50. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi Y, Hearing VJ, Itami S, Yoshikawa K, Katayama I. Mesenchymal-epithelial interactions in the skin: aiming for site-specific tissue regeneration. J Dermatol Sci. 2005;40:1–9. doi: 10.1016/j.jdermsci.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Scott G, Leopardi S, Parker L, Babiarz L, Seiberg M, Han R. The proteinase-activated receptor-2 mediates phagocytosis in a Rho-dependent manner in human keratinocytes. J Invest Dermatol. 2003;121:529–41. doi: 10.1046/j.1523-1747.2003.12427.x. [DOI] [PubMed] [Google Scholar]
- 37.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–62. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi Y, Passeron T, Hoashi T, Watabe H, Rouzaud F, Yasumoto K, Hara T, Tohyama C, Katayama I, Miki T, Hearing VJ. Dickkopf 1 (DKK1) regulates skin pigmentation and thickness by affecting Wnt/beta-catenin signaling in keratinocytes. Faseb J. 2008;22:1009–20. doi: 10.1096/fj.07-9475com. [DOI] [PubMed] [Google Scholar]
- 39.Weiner L, Han R, Scicchitano BM, Li J, Hasegawa K, Grossi M, Lee D, Brissette JL. Dedicated epithelial recipient cells determine pigmentation patterns. Cell. 2007;130:932–42. doi: 10.1016/j.cell.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 40.Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res. 2005;18:2–12. doi: 10.1111/j.1600-0749.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi Y, Morita A, Maeda A, Hearing VJ. Regulation of skin pigmentation and thickness by dickkopf 1 (DKK1). J Investig Dermatol Symp Proc. 2009;14:73–5. doi: 10.1038/jidsymp.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi Y, Kubo T, Tarutani M, Sano S, Asada H, Kakibuchi M, Hosokawa K, Itami S, Yoshikawa K. Epithelial-mesenchymal interactions in wounds: treatment of palmoplantar wounds by nonpalmoplantar pure epidermal sheet grafts. Arch Dermatol. 2001;137:621–8. [PubMed] [Google Scholar]
- 43.Yamaguchi Y, Yoshida S, Sumikawa Y, Kubo T, Hosokawa K, Ozawa K, Hearing VJ, Yoshikawa K, Itami S. Rapid healing of intractable diabetic foot ulcers with exposed bones following a novel therapy of exposing bone marrow cells and then grafting epidermal sheets. Br J Dermatol. 2004;151:1019–28. doi: 10.1111/j.1365-2133.2004.06170.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi Y, Sumikawa Y, Yoshida S, Kubo T, Yoshikawa K, Itami S. Prevention of amputation caused by rheumatic diseases following a novel therapy of exposing bone marrow, occlusive dressing and subsequent epidermal grafting. Br J Dermatol. 2005;152:664–72. doi: 10.1111/j.1365-2133.2005.06401.x. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi Y, Itami S, Yoshikawa K. Chapter 41: Skin grafting: surgical technique. In: Falabella A, Kirsner R, editors. Wound healing: science and practice. Marcel Dekker; New York: 2005. pp. 535–544. [Google Scholar]
- 46.Yamaguchi Y, Itami S, Tarutani M, Hosokawa K, Miura H, Yoshikawa K. Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial-mesenchymal interactions. J Invest Dermatol. 1999;112:483–8. doi: 10.1046/j.1523-1747.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi Y, Itami S, Watabe H, Yasumoto K, Abdel-Malek ZA, Kubo T, Rouzaud F, Tanemura A, Yoshikawa K, Hearing VJ. Mesenchymal-epithelial interactions in the skin: increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J Cell Biol. 2004;165:275–85. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi Y, Passeron T, Watabe H, Yasumoto K, Rouzaud F, Hoashi T, Hearing VJ. The effects of dickkopf 1 on gene expression and Wnt signaling by melanocytes: mechanisms underlying its suppression of melanocyte function and proliferation. J Invest Dermatol. 2007;127:1217–25. doi: 10.1038/sj.jid.5700629. [DOI] [PubMed] [Google Scholar]
- 49.Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–53. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 50.Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, Ridky TW, Stadler HS, Nusse R, Helms JA, Chang HY. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–7. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solano F, Briganti S, Picardo M, Ghanem G. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects. Pigment Cell Res. 2006;19:550–71. doi: 10.1111/j.1600-0749.2006.00334.x. [DOI] [PubMed] [Google Scholar]
- 52.Tadokoro T, Rouzaud F, Itami S, Hearing VJ, Yoshikawa K. The inhibitory effect of androgen and sex-hormone-binding globulin on the intracellular cAMP level and tyrosinase activity of normal human melanocytes. Pigment Cell Res. 2003;16:190–7. doi: 10.1034/j.1600-0749.2003.00019.x. [DOI] [PubMed] [Google Scholar]
- 53.Grimes PE. New insights and new therapies in vitiligo. Jama. 2005;293:730–5. doi: 10.1001/jama.293.6.730. [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi Y, Hearing VJ. Chapter 6: Melanocyte distribution and function in human skin: effects of UV radiation. In: Hearing VJ, Leong SPL, editors. From melanocytes to malignant melanoma: the progression to malignancy. Humana Press; Totowa: 2006. pp. 101–115. [Google Scholar]
- 55.Tadokoro T, Yamaguchi Y, Batzer J, Coelho SG, Zmudzka BZ, Miller SA, Wolber R, Beer JZ, Hearing VJ. Mechanisms of skin tanning in different racial/ethnic groups in response to ultraviolet radiation. J Invest Dermatol. 2005;124:1326–32. doi: 10.1111/j.0022-202X.2005.23760.x. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi Y, Takahashi K, Zmudzka BZ, Kornhauser A, Miller SA, Tadokoro T, Berens W, Beer JZ, Hearing VJ. Human skin responses to UV radiation: pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. Faseb J. 2006;20:1486–8. doi: 10.1096/fj.06-5725fje. [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi Y, Coelho SG, Zmudzka BZ, Takahashi K, Beer JZ, Hearing VJ, Miller SA. Cyclobutane pyrimidine dimer formation and p53 production in human skin after repeated UV irradiation. Exp Dermatol. 2008;17:916–24. doi: 10.1111/j.1600-0625.2008.00722.x. [DOI] [PubMed] [Google Scholar]
- 58.Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, Fisher DE. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–64. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 59.Coelho SG, Choi W, Brenner M, Miyamura Y, Yamaguchi Y, Wolber R, Smuda C, Batzer J, Kolbe L, Ito S, Wakamatsu K, Zmudzka BZ, Beer J, Miller S, Hearing VJ. Short- and long-term effects of ultraviolet radiation on the pigmentation of human skin. J Investig Dermatol Symp Proc. 2009;14:32–5. doi: 10.1038/jidsymp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]