Abstract
Protoporphyrinogen oxidase (EC 1–3-3–4), the 60-kDa membrane-bound flavoenzyme that catalyzes the final reaction of the common branch of the heme and chlorophyll biosynthesis pathways in plants, is the molecular target of diphenyl ether-type herbicides. It is highly resistant to proteases (trypsin, endoproteinase Glu-C, or carboxypeptidases A, B, and Y), because the protein is folded into an extremely compact form. Trypsin maps of the native purified and membrane-bound yeast protoporphyrinogen oxidase show that this basic enzyme (pI > 8.5) was cleaved at a single site under nondenaturing conditions, generating two peptides with relative molecular masses of 30,000 and 35,000. The endoproteinase Glu-C also cleaved the protein into two peptides with similar masses, and there was no additional cleavage site under mild denaturing conditions. N-terminal peptide sequence analysis of the proteolytic (trypsin and endoproteinase Glu-C) peptides showed that both cleavage sites were located in putative connecting loop between the N-terminal domain (25 kDa) with the βαβ ADP-binding fold and the C-terminal domain (35 kDa), which possibly is involved in the binding of the isoalloxazine moiety of the FAD cofactor. The peptides remained strongly associated and fully active with the Km for protoporphyrinogen and the Ki for various inhibitors, diphenyl-ethers, or diphenyleneiodonium derivatives, identical to those measured for the native enzyme. However, the enzyme activity of the peptides was much more susceptible to thermal denaturation than that of the native protein. Only the C-terminal domain of protoporphyrinogen oxidase was labeled specifically in active site-directed photoaffinity-labeling experiments. Trypsin may have caused intramolecular transfer of the labeled group to reactive components of the N-terminal domain, resulting in nonspecific labeling. We suggest that the active site of protoporphyrinogen oxidase is in the C-terminal domain of the protein, at the interface between the C- and N-terminal domains.
Protoporphyrinogen oxidase (EC 1–3–3–4) is the penultimate enzyme of the heme biosynthesis pathway and the final enzyme of the common branch of the heme and chlorophyll biosynthesis pathways in plants. It catalyzes the oxidative O2-dependent aromatization of the colorless protoporphyrinogen IX to the highly conjugated protoporphyrin IX. Studies of the structure and function of protoporphyrinogen oxidase have been stimulated by the discovery that diphenyl ether-type herbicides are very potent inhibitors of protoporphyrinogen oxidase activity of yeast, mammal and plant mitochondria, and plant chloroplasts in vitro (1, 2). The phytotoxicity of diphenyl ether-type herbicides is light-dependent and involves intracellular peroxidation caused by protoporphyrin IX, the heme and chlorophyll precursor, leading to cell damage and lysis (3, 4) .
The topological features at the active site of the protein must be studied if we are to understand the interactions of diphenyl ether-type herbicides with protoporphyrinogen oxidase. Radiolabeled acifluorfen (5) has been used to demonstrate that several chemically unrelated inhibitors of protoporphyrinogen oxidase bind to the same site on the plant enzyme (6). Specifically bound tritiated acifluorfen is also competitively displaced by protoporphyrinogen IX, the substrate of protoporphyrinogen oxidase (7). This is consistent with kinetic studies showing that diphenyl ethers are competitive inhibitors (for the tetrapyrrole substrate) of plant, mouse, and yeast protoporphyrinogen oxidase (8). The binding domains for substrates or inhibitors of an enzyme can be characterized by covalent attachment of radioactive derivatives by photoactivation. We have assessed previously the potential of a diazoketone derivative of [3H]acifluorfen (DZ-[3H]AF) (5) for use as a photoaffinity-labeling reagent for protoporphyrinogen oxidase. This compound is a specific ligand for the membrane-bound and purified protoporphyrinogen oxidase of yeast (9) and has been used to characterize a yeast strain previously described as heme-deficient (10) on the basis of a lack of protoporphyrinogen oxidase activity (11). The aim of this work was to map peptides on the yeast protein that were specifically labeled by DZ-[3H]AF. However, protoporphyrinogen oxidase seems to have a very stable structure and is extremely resistant to proteolytic cleavage. This unexpected property of the enzyme was used to characterize the domain structure of the protein and identify the most probable location of the active site.
MATERIALS AND METHODS
Materials.
Protoporphyrin IX, disodium salt, was purchased from Serva, endoproteinase Glu-C (V8-protease, EC 3.4.21.19 from Staphylococcus aureus V8) and carboxypeptidase B (EC 3.4.17.2 from porcine pancreas) were purchased from Boehringer Mannheim. Carboxypeptidase A [EC 3.4.17.1 from bovine pancreas type II-phenylmethylsulfonyl fluoride (PMSF)] and carboxypeptidase Y (EC 3.4.16.1 from Baker’s Yeast), trypsin (EC 3.4.21.4, Type XIII from bovine pancreas), and α-toluenesulfonyl fluoride were obtained from Sigma; acifluorfen sodium salt was from ChemService (Westchester, PA); and the tritiated diazoketone derivative of acifluorfen was synthesized by R. Mornet, Université d’Angers, France (12). Diphenyleneiodonium chloride was obtained from the Alexis Corporation (San Diego). All inhibitors were dissolved in dimethyl sulfoxide to give 10-mM stock solutions.
Purification of Recombinant Yeast Protoporphyrinogen Oxidase.
Recombinant yeast protoporphyrinogen was produced in a heterologous expression system. The HEM14 gene was inserted under control of a T7 RNA polymerase-dependent promoter into the pSBETa plasmid. This pET3 derivative, carrying the ArgU gene (13), was expressed in Escherichia coli strain BL21(DE3). The T7 RNA polymerase gene was induced by adding 1 mM isopropyl β-d-thiogalactoside (IPTG) to cell cultures with an A600 of 1–1.5. The induction medium was a minimal medium (M63) containing 0.4% glucose, 0.1% casamino acids (Difco), and kanamycin (70 μg/ml). Induction media were inoculated with cells from fresh cultures grown in Luria– Bertani medium plus kanamycin at 37°C, and four generations were cultured in the induction medium. The cultures were then chilled on ice for 5 min, IPTG was added, and the cultures were shaken (250 rpm) at 25°C for 15 hr. Protoporphyrinogen oxidase was purified from 5 g (wet weight) induced cells collected by centrifugation and suspended in 0.1 M potassium phosphate, pH 7.2, containing 1 mM EDTA and 70 μg/ml of PMSF, and subjected to sonication on ice for 3 × 15 sec. The membrane fraction was collected by centrifugation for 1 h at 105,000 × g, homogenized (Potter) in 0.1 M potassium phosphate, pH 7.2, containing 1 mM EDTA and 70 μg/ml of PMSF with 0.1 M KCl, and washed by centrifugation. The pelleted membranes were homogenized (Potter) in 0.1 M potassium phosphate, pH 7.2, containing 1 mM EDTA, 0.1 M KCl, 70 μg/ml PMSF, and 2% (wt/vol) n-octyl glucoside. The suspension was subjected to sonication at 4°C, and solubilized membrane proteins were extracted by centrifugation for 1 h at 105,000 × g. Protoporphyrinogen oxidase was purified as described previously (9). The specific activity of the purified protein was 82,000 nmol protoporphyrinogen oxidized⋅h−1⋅mg−1.
Action of Proteases on Protoporphyrinogen Oxidase.
Endoproteinase Glu-C activity was assayed at 25°C in 0.1 M phosphate buffer, pH 7.3. The assay was started by adding 0.5 units of endoproteinase Glu-C (3.2% final concentration, wt/wt). One unit of endoproteinase Glu-C catalyses the hydrolysis of 1 μmol of z-phenylalanine-leucine-glutamate-4 nitroaniline per minute at pH 8.0 at 25°C. Carboxypeptidases A and B were assayed at 25°C in 0.05 M Tris⋅HCl buffer, pH 7.5, containing 0.05 M KCl. The assays were started by adding 0.5 units of carboxypeptidase A or 1.35 units of carboxypeptidase B (3% final concentration, wt/wt). One unit of carboxypeptidase A catalyses the hydrolysis of 1 μmol of hippuryl-l-phenylalanine per minute at pH 7.5 at 25°C. One unit of carboxypeptidase B catalyses the hydrolysis of 1 μmol of hippuryl-l-arginine per minute at pH 7.65 at 25°C. Carboxypeptidase Y was assayed at 25°C in 0.05 M phosphate buffer, pH 6.5, containing 0.05 M KCl. The assay was started by adding 0.9 units of carboxypeptidase Y (3% final concentration, wt/wt). One unit of carboxypeptidase Y catalyses the hydrolysis of 1 μmol of N-CBZ-phenylalanine-alanine per minute at pH 6.75 at 25°C. Trypsin was assayed at 25°C in 0.05 M potassium phosphate buffer, pH 7.3, containing 0.05 M KCl. Proteolysis was started by adding 25 N-α-benzoyl-1-arginine ethyl ester (BAEE) units of trypsin (2% final concentration, wt/wt) to the incubation medium. One BAEE unit of trypsin catalyzes the production of a ΔA253 of 0.001 per min in 3.2 ml at pH 7.6 at 25°C. The control experiments were identical assays without the addition of protease.
Protoporphyrinogen oxidase activity was measured during proteolysis, and the cleavage products were detected by SDS/PAGE. Purified protoporphyrinogen oxidase was diluted 1:2 in 0.1 M potassium phosphate buffer, pH 7.2, containing 8 M urea or 0.4% SDS (final concentration, 4 M urea or 0.2% SDS) for cleavage in denaturing buffer. The reaction was started by adding trypsin (2% final concentration, wt/wt). Control experiments with BSA as the substrate showed that the trypsin was fully active under these assay conditions. Protoporphyrinogen oxidase activity was determined during trypsin proteolysis, and the degree of trypsin cleavage was assessed by SDS/PAGE.
Photolabeling of Binding Sites with Diazo-[3H]Acifluorfen.
Bacterial membranes from protoporphyrinogen oxidase-overproducing cells (10 μg protein⋅ml−1), purified recombinant protoporphyrinogen oxidase (1 ng), or tryptic peptides from protoporphyrinogen oxidase were preincubated with diazo-[3H]acifluorfen (25 nM final concentration) in 0.1 M potassium phosphate buffer, pH 7.2, for 15 min at 30°C in the dark. Nonspecific binding was determined in the presence of 15 μM (final concentration) unlabeled acifluorfen as a competitive ligand. Aliquots (0.5 ml) of the membrane suspensions were transferred to UV-transparent spectrophotometric plastic cuvettes, placed on a Wilmer Lourmat 302 fluorescence table (Prolab, Fontenay sous bois, France), and irradiated for 10 min. The lamps emitted broad-spectrum ultraviolet radiation, with peak emission at 366 nm. The irradiated bacterial membranes were harvested by centrifugation, and the pellet was solubilized in SDS/PAGE sample buffer (14). Labeled, purified yeast protoporphyrinogen oxidase or peptides were precipitated with 10% trichloroacetic acid (wt/vol, final concentration) for 30 min at 4°C and were pelleted by 30-min centrifugation at 15,000 × g. The proteins were denatured in SDS/PAGE sample buffer and separated by SDS/PAGE. The gels were fixed for 30 min in ethanol/acetic acid/water (30/10/60 per volume), treated with Amplify fluorographic reagent (Amersham) for 30 min, dried under vacuum, and used to expose tritium-sensitive films for 1 week at −80°C.
N-Terminal Peptide and Protein Sequence.
Native and protease-treated protoporphyrinogen oxidase molecules were subjected to SDS/PAGE and transferred to Immobilon-P membrane (Millipore) in 10 mM Tris⋅borate, pH 8.8, by electroblotting. Their N-terminal peptide sequences then were determined. The polypeptides were stained on the Immobilon with 0.01% (wt/vol) Coomassie blue R250 in ethanol and were destained with water. The stained bands were cut out and processed for automatic Edman degradation on an Applied Biosystems 390 peptide sequencer by using standard procedures for 6–15 cycles.
Miscellaneous.
Published procedures were used for SDS/PAGE (15) and for the electrophoretic transfer of proteins to nitrocellulose sheets (16). Preparations were incubated with antiprotoporphyrinogen oxidase polyclonal antibodies and detected by enhanced chemiluminescence (Amersham) with peroxidase-conjugated anti-rabbit IgG secondary antibodies, as recommended by the manufacturer.
Protoporphyrinogen oxidase was assayed by measuring the rate of appearance of protoporphyrin fluorescence (17) at 30°C. The incubation mixture was 0.1 M potassium phosphate buffer, pH 7.2, saturated with air, containing 2 μM protoporphyrinogen IX, 3 mM palmitic acid (in dimethyl sulfoxide 0.5% vol/vol final concentration), 5 mM DTT, 1 mM EDTA, and 0.3 mg/ml (final concentration) Tween 80, to ensure maximum fluorescence of the protoporphyrin IX. Protoporphyrinogen was prepared by reducing protoporphyrin IX hydrochloride dissolved in KOH/EtOH (0.04 M/20%) with freshly prepared 3% sodium amalgam (18). Protein concentrations of solutions diluted with 0.1 M NaOH were measured by the micro-Bradford technique on solution. One unit of activity is the amount of enzyme that oxidizes 1 nmol protoporphyrinogen IX to protoporphyrin IX per hour at 30°C.
RESULTS
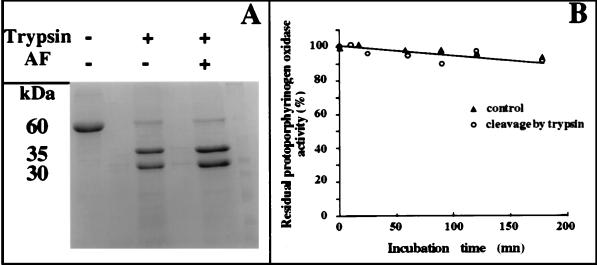
Yeast protoporphyrinogen oxidase (60 kDa) was cleaved rapidly by trypsin into two peptides (30 and 35 kDa) (Fig. 1A). The native protein was almost completely cleaved in less than 5 min; no further peptides were produced in prolonged incubations of up to 2 h. The mixture of peptides was fully active, even after the longest digestion times (Fig. 1B). Protoporphyrinogen oxidase in weak denaturing buffers, containing 4 M urea or 0.2% (final concentrations) SDS, was not cleaved at additional sites by trypsin, and the two main peptides (30 and 35 kDa) were always produced when trypsin was active in these denaturing buffers. Furthermore, in 0.2% SDS-containing buffer, protoporphyrinogen oxidase was fully active even after prolonged trypsin digestion. Only 10% of the initial protoporphyrinogen oxidase activity was retained in 4 M urea, but this activity remained constant during trypsin treatment. Microsequencing analysis of the N-terminal sequences from the native protein and both digestion products was performed. Identical sequences (Met-Leu-Leu-Pro-Leu-Thr-Lys-Leu-) were obtained for the native protein and the 30-kDa peptide and another sequence Ser-Lys-Lys-Thr-Glu-Asn-Leu-His-Gln-Ser-, for the 35-kDa peptide. The sequence of the 35-kDa peptide was sometimes frayed a second, minor sequence, Thr-Glu-Asn-Leu-His-Gln-Ser-Leu-, determined for about 25% of the 35-kDa peptide mixture. The electrophoretic mobility of the 30-kDa peptide was lower than predicted from the calculated mass of the 227-aa peptide, from the initiation methionine to the Arg-227 at the trypsin cleavage site. Therefore, we measured the actual mass of both peptides by mass spectrometry (matrix-associated laser desorption ionization). The measured masses were 25,182 ± 15 and 34,589 ± 15 close to the expected masses (Mr = 25,154 and 34,529). This suggests that some other structure in the 30-kDa peptide changes its electrophoretic mobility.
Figure 1.
Proteolysis of protoporphyrinogen oxidase by trypsin. (A) Coomassie blue staining of proteins on an SDS-polyacrylamide gel. Lanes: 1, purified yeast protoporphyrinogen oxidase after 30-min incubation without trypsin; 2, purified yeast protoporphyrinogen oxidase after 30-min incubation with 25 units trypsin; 3, purified yeast protoporphyrinogen oxidase incubated with 10 μM acifluorfen after 30-min incubation with 25 units trypsin. (B) Protoporphyrinogen oxidase activity measured as a function of incubation time without trypsin (▴) and with 25 units trypsin (○).
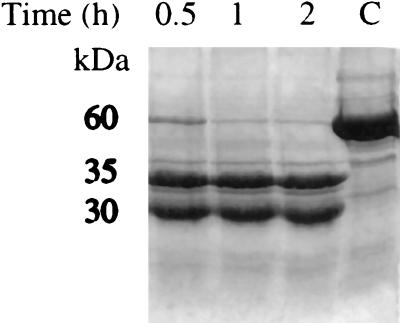
Yeast protoporphyrinogen oxidase (C) was cleaved into two peptides (30 and 35 kDa) by endoproteinase Glu-C (Fig. 2). The native protein was completely cleaved within 30 min, and no further peptides were produced in prolonged incubations of up to 2 h. The mixture of peptides was fully active, even after prolonged digestion. The N-terminal sequence from the 35-kDa peptide was Asn-Leu-His-Gln-Ser-Leu-.
Figure 2.
Kinetics of purified protoporphyrinogen oxidase digestion by endoproteinase Glu-C. Lane C, enzyme incubated for 2 h without protease.
Protoporphyrinogen oxidase was completely resistant to degradation by carboxypeptidases A, B, or Y or mixtures of these proteases. The enzyme’s electrophoretic mobility was unchanged and the enzyme was fully active even after prolonged exposure to carboxypeptidases, whereas control proteins such as BSA were processed efficiently (data not shown).
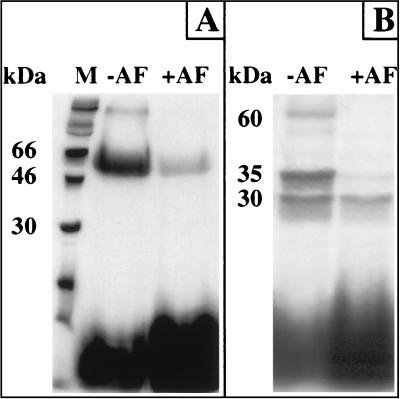
The tritiated diazoketone derivative of acifluorfen (DZ-[3H]AF) specifically labeled the native protoporphyrinogen oxidase in photoaffinity-labeling experiments. The level of nonspecific labeling was consistently less than 10% of that of specific labeling (Fig. 3A). Labeled protoporphyrinogen oxidase also was cleaved by trypsin at a single site, generating the same peptides as the unlabeled protein, as determined by Coomassie blue staining. Fluorography showed that both peptides were labeled but only the 35-kDa peptide appeared to be specifically labeled (Fig. 3B). Quantitative scanning densitometry of fluorograms together with direct counting of the radioactivity in sliced gels showed that all the nonspecific labeling associated to the native protein was recovered on the 30-kDa peptide after trypsin treatment (Fig. 3 A, lane +AF and B, lane +AF). Similarly, in experiments in which the protein was labeled without cold inhibitor, all the radioactivity associated to the native protein was recovered on the tryptic peptides, indicating that trypsin action did not lead to significant loss of the labeled probe. However, the intensity of the nonspecific labeling of the 30-kDa peptide was almost similar to that of the 35-kDa peptide, three to five times more intense than expected based on the nonspecific labeling of native protoporphyrinogen oxidase. Labeled protoporphyrinogen oxidase and free DZ-AF were separated by gel filtration, and the protein was subjected further to trypsin digestion to determine whether some of the remaining free photoaffinity probe interacted with the peptides in the posttrypsin treatment of the samples. The peptides generated under these experimental conditions were labeled the same way as in the previous experiments, suggesting transfer of some of the probe from the 35-kDa peptide to reactive chemical groups on the 30-kDa peptide may be induced by trypsin treatment of protoporphyrinogen oxidase. When the protein was treated with trypsin and then incubated with the photoaffinity probe, the 35-kDa peptide was specifically labeled, but there was less nonspecific labeling of the 30-kDa peptide than observed previously (data not shown).
Figure 3.
Fluorographic detection after SDS/PAGE analysis of purified yeast protoporphyrinogen oxidase in photoaffinity-labeling experiments with DZ-[3H]AF. (A) Intact enzyme. (B) Tryptic fragments of protoporphyrinogen oxidase. Lanes: M, molecular mass markers; −AF, total labeling; +AF, nonspecific labeling (labeling in the presence of 10 μM cold acifluorfen).
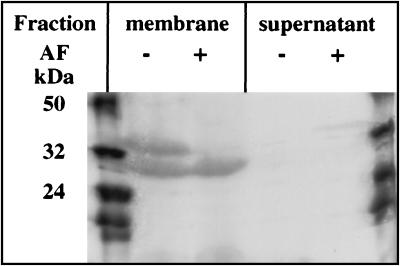
Protoporphyrinogen oxidase associated with the membrane fraction of E. coli cells overexpressing the HEM14 gene was cleaved at a single site by trypsin, as was the purified protein (Fig. 4). Both the 30- and the 35-kDa peptides remained associated with the membrane fraction (Fig. 4B). Photoaffinity labeling of protoporphyrinogen oxidase before trypsin treatment (Fig. 4C) showed that the 35-kDa peptide was specifically labeled whereas the 30-kDa peptide was nonspecifically labeled (Fig. 4D).
Figure 4.
Fluorographic detection after SDS/PAGE of photolabeled tryptic fragments of the membrane-bound protoporphyrinogen oxidase fractionated by ultracentrifugation after the trypsin digestion. Lanes: 1 and 6, molecular mass markers; 2 and 4, total labeling in the membrane fraction (2) and supernatant (4); 3 and 5, nonspecific labeling (labeling in the presence of 10 μM cold acifluorfen) in the membrane fraction (3) and supernatant (5).
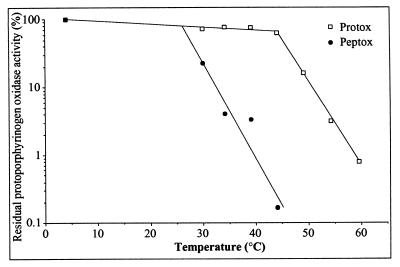
The kinetic properties of the native membrane-bound and purified protoporphyrinogen oxidase were compared with those of the active peptides. The native and the cleaved enzyme had the same overall catalytic properties in terms of affinity for protoporphyrinogen oxidase and IC50 for acifluorfen and diphenyleneiodonium, a novel, mechanism-based inhibitor of protoporphyrinogen oxidase (Table 1). However, the active peptide is significantly more heat-labile than the native protein (Fig. 5).
Table 1.
Kinetic parameters of yeast protoporphyrinogen oxidase inhibition by diphenyl ether-type herbicides (DEPs)
| Inhibitor type | Kinetic constant | Membrane-bound protoporphyrinogen oxidase | Membrane-bound protoporphyrinogen oxidase cleaved by trypsin | Purified protoporphyrinogen oxidase | Purified protoporphyrinogen oxidase cleaved by trypsin |
|---|---|---|---|---|---|
| DPEs | IC50 of AF, nM | 7.50 ± 0.40 | 9.20 ± 0.45 | 8.25 ± 0.40 | 4.60 ± 0.25 |
| IC50 of AFM, nM | 3.10 ± 0.15 | 3.15 ± 0.15 | 3.90 ± 0.20 | 2.05 ± 0.10 | |
| DPI | Ki, nM | 50 ± 2.5 | 81 ± 4.0 | 88 ± 4.5 | 66 ± 3.5 |
| Ki*, nM | 2.9 ± 0.15 | 1.2 ± 0.05 | 2.6 ± 0.15 | 2.9 ± 0.15 | |
| KmO2, nM | 1.7 ± 0.10 | 1.3 ± 0.10 | 2.0 ± 0.10 | 4.8 ± 0.25 |
AF, acifluorfen; AFM, acifluorfen methyl; DPI, 2-2′ diphenyleneiodonium. Ki is the inhibition constant in the E + I 〈=〉 EI reaction; Ki* is the inhibition constant in the EI 〈=〉 EI* isomerization reaction (20).
Figure 5.
Effect of temperature on purified protoporphyrinogen oxidase (□) and the tryptic peptides (•) of protoporphyrinogen oxidase. The proteins were consecutively incubated for 5 min at each temperature, and the initial velocity of the enzyme reaction was measured at 30°C on aliquots of the denatured samples.
DISCUSSION
The goal of this research was to determine the structural and functional properties of yeast protoporphyrinogen oxidase on the basis of its proteolytic digestion patterns and identification of binding sites for photoaffinity probes. Proteolysis studies have shown that the enzyme may fold into two domains, a 30-kDa amino-terminal domain containing the βαβ ADP-binding fold (19) thought to be involved in the interaction between the adenosine diphosphate moiety of the FAD cofactor of the enzyme, and a 35-kDa carboxyl-terminal domain. There is an exposed region linking these two domains (Arg-227; Glu-232) that is susceptible to proteolysis. The limited proteolysis of protoporphyrinogen oxidase by endoproteinase Glu-C and trypsin was unexpected because (i) endoproteinase Glu-C is a very active processing enzyme and protoporphyrinogen oxidase contains 33 aspartic acid and 22 glutamic acid residues as potential endoproteinase Glu-C cleavage sites, (ii) protoporphyrinogen oxidase is a basic protein (pI > 8.5) with 48 lysine and 24 arginine among its 539 aa, all of which are potential trypsin cleavage sites. A theoretical peptide map for this protein predicts fragments no larger than 1,903 Da (residues 14–32) for trypsin digestion and no larger than 3,877 Da (residues 1–36) for endoproteinase Glu-C digestion. The protoporphyrinogen oxidase structure was resistant to trypsin digestion in the presence of denaturing agents such as 4 M urea and 0.2% SDS, showing that the protein is folded into an extremely compact form. The profile of catalytic activity during proteolysis showed there is a correlation between the maintenance of normal activity levels and the strong association of the two peptides, despite very rapid cleavage of the polypeptide chain. We could not separate the two peptides under nondenaturing conditions, either by gel filtration or by ion-exchange chromatography, in which the peptides were eluted in a single peak of protein activity with the same Rf as the native protein. The catalytic properties of the peptides were similar to those of the native protein in terms of affinity constants for both protoporphyrinogen and dioxygen (Km = 0.1 μM and 2.3 μM, respectively) and inhibition constants for acifluorfen (9), a diphenyl ether-type herbicide, and diphenyleneiodonium (Ki = 10 nM and 0.1 μM, respectively) (20). This shows that the structure and function of the active site, including the isoalloxazin moiety of the flavin, were not affected by cleavage of the polypeptide chain. The close association of the two domains of protoporphyrinogen oxidase made it impossible to clearly identify which of the domains carried the active site of the enzyme. Therefore, we used a photoactivated radiolabeled probe for the active site of protoporphyrinogen oxidase.
Photoaffinity ligands are extremely powerful tools for studying the topological features of binding sites. We have shown previously that a diazoketone derivative of acifluorfen (DZ-[3H]AF) is a highly specific ligand for both membrane-bound and purified protoporphyrinogen oxidase (21). DZ-[3H]AF binding to protoporphyrinogen oxidase is competitively displaced by acifluorfen or diphenyl ether-type inhibitors of the enzyme. All these compounds inhibit the enzyme by competing with protoporphyrinogen, its substrate (7). DZ-[3H]AF was synthesized by modification of the carboxylic group of acifluorfen (12). Quantitative Structure Activity Relationship studies on diphenyl ether series have shown that within the diphenyl ether moiety of acifluorfen, the A phenyl ring and its substituents are the components critical for interaction with the active site of the enzyme (22–25). Thus, the diazoketone group of DZ-[3H]AF presumably interacts with the amino acids that are most peripheral to the hydrophobic core of the active site. Trypsin treatment of the photoaffinity-labeled purified protoporphyrinogen oxidase showed that only the 35-kDa carboxyl-terminal domain of the enzyme was specifically labeled. However, there was an unexpected reaction in which the radiolabeled probe was transferred from the 35-kDa carboxyl-terminal domain to the 30-kDa amino-terminal domain, leading to some apparent nonspecific labeling of the 30-kDa amino-terminal domain. This was shown by differences in the ratio of specific to nonspecific labeling in the intact and cleaved proteins (10/1 vs. 2/1). DZ-[3H]AF may bind to the protein via a primary covalent bond involving a labile ether or thioether formed by reaction of the carbene of DZ-[3H]AF with the hydroxyl group of a serine or a threonine or the thiol group of a cysteine residue of the protein. Trypsin, by generating two new flexible ends in the protein, may expose a more reactive group (amino- or guanido-) in the 30-kDa amino-terminal domain that may attack such a labile bond through nucleophilic substitution to generate a more stable derivative of DZ-[3H]AF. The primary targets of the carbene reaction have not been identified, but the connecting loop between the two domains does contain one cysteine and numerous serine and threonine residues that could act as donor groups in this intramolecular transfer reaction.
A key question is whether the extreme resistance of the purified protein to proteases action is an intrinsic property of the protein or results from protection of the protein because of its conformation in the detergent micelles that keep the purified protein in a soluble form. The membrane-bound protoporphyrinogen oxidase from E. coli cells overproducing the protein was treated with trypsin. The same cleavage site was accessible to trypsin as in the purified protein, showing that the putative loop linking the two domains was accessible to trypsin and therefore was not buried in the membrane-lipid bilayer. Both the 30-kDa amino-terminal domain and the 35-kDa carboxyl-terminal domain remained bound to the membrane fraction. However, the close association between these peptides made it impossible to determine whether both domains are anchored directly to the membrane or whether only one of the domains interacts with the membrane and holds the second domain in place through the strong association of the two domains.
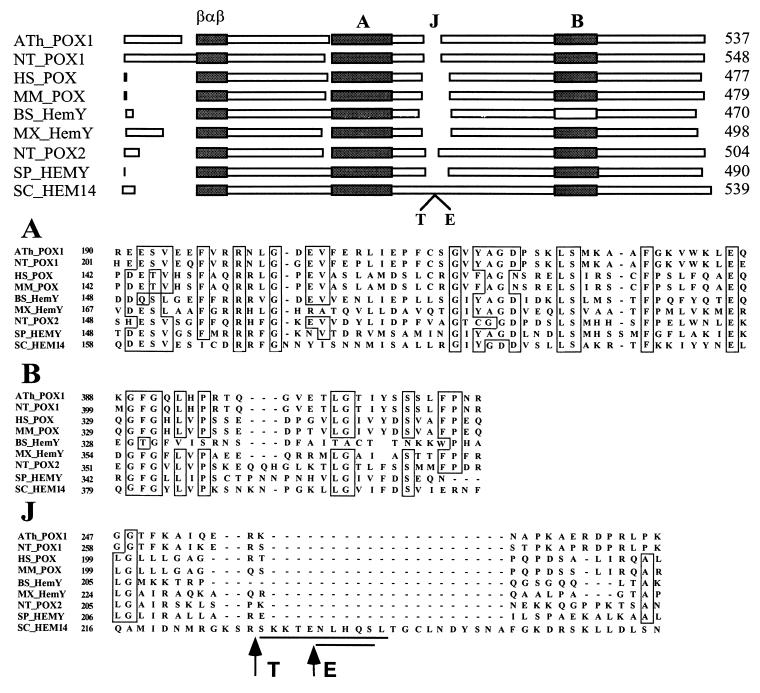
It is thought that the most conserved residues in homologous proteins are likely to be important functional groups or involved in the conservation of specific structural features of the protein. Primary sequence alignment of protoporphyrinogen oxidases identifies two main blocks of sequence similarity (Fig. 6), in addition to the amino-terminal βαβ ADP-binding fold already identified. These two blocks are on either side of the loop bearing the protease cleavage sites. Hydrophobic cluster analysis (26) of block A reveals the occurrence of probable, highly amphipathic α-helices. This block exhibits striking similarities to the amphipathic helices in interaction with the lipid bilayer in prostaglandin-H2 synthase-1 (27, 28), where the hydrophobic surfaces of the helices are facing outward, away from the body of the protein, forming a large, hydrophobic patch on the exterior of the protein whereas the opposite side of the helices contains charged residues that are well positioned to interact with phospholipid head groups. The binding surface is not deep enough to extend beyond one leaflet of the lipid bilayer. Similar membrane anchoring, through a large nonpolar plateau, was found in the structure of squalene cyclase (29) that, together with prostaglandin H2 synthase-1, appears to form a new family of monotopic, membrane-bound proteins (28). Therefore, it is tempting to speculate that protoporphyrinogen oxidase may be a member of this family, owing to the strong hydrophobicity of this integral membrane-bound protein despite the lack of typical trans-membrane segment. The Bacillus subtilis HemYp protoporphyrinogen oxidase is of particular interest. Its protoporphyrinogen-oxidizing activity has been studied (30, 31), and, unlike the other enzymes, it is neither substrate-specific (uroporphyrinogen III and coproporphyrinogen III are both efficiently oxidized to their corresponding porphyrin forms) (32) nor inhibited by diphenyl ether-type herbicides (33). Analysis of the differences in primary structure between HemYp and the other enzymes shows that, whereas the first block of homology is conserved in this protein, it lacks the second block of homology. The role of some of the residues in this block has been demonstrated by the characterization of two missense mutations in the yeast gene (Leu-422 → Pro and Lys-424 → Glu) that abolished enzyme activity (9). This suggests that some of the determinants of specificity for protoporphyrinogen oxidase are located in this structure that therefore should be used for investigating the molecular basis of the interaction of diphenyl ether-type herbicides with protoporphyrinogen oxidase.
Figure 6.
Organization of protoporphyrinogen oxidases from various origins and clustal-w 1.7 alignment of the peptide sequences of characteristic domains A, B, and J. ATh, Arabidopsis thaliana (34); NT, Nicotiana tabacum (35); HS, Homo sapiens (36); MM, Mus musculus (37); BS, Bacillus subtilis (30); MX, Myxococcus xanthus; SP, Schizosaccharomyces pombe; SC, Saccharomyces cerevisiae (9).
The demonstration that the specific binding site for DZ-[3H]AF is associated with the C-terminal domain of the protein, as well as the sequence analysis, suggests that the active site of protoporphyrinogen oxidase is located in the C-terminal domain of the protein, at the interface between the C- and N-terminal domains.
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique, the Université Paris 7-Denis-Diderot, the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche (Actions Coordonnées Concertées-Sciences du Vivant #5), and the Association de Recherche contre le Cancer. We thank Prof. A. Delfour, Institut Jacques Monod, Paris, and J. C. d’Alayer, Institut Pasteur, Paris, for their help in protein microsequencing, and J. P. Le Caer, Ecole Supérieure de Physique et de Chimie Industrielles, Paris, for mass spectrometry analysis. The English text was edited by Dr. J. Knight.
ABBREVIATION
- DZ-[3H]AF
diazoketone derivative of [3H]acifluorfen
References
- 1.Matringe M, Camadro J M, Labbe P, Scalla R. Biochem J. 1989;260:231–235. doi: 10.1042/bj2600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witkowski D A, Halling B P. Plant Physiol. 1989;90:1239–1242. doi: 10.1104/pp.90.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandmann G, Nicolaus B, Böger P. Z Naturforsch. 1990;45c:512–517. [Google Scholar]
- 4.Scalla R, Matringe M. Rev Weed Sci. 1994;6:103–132. [Google Scholar]
- 5.Mornet R, Gouin L, Matringe M, Scalla R, Swithenbank C. J Labelled Compounds Radiopharm. 1992;31:175–182. [Google Scholar]
- 6.Varsano R, Matringe M, Magnin N, Mornet R, Scalla R. FEBS Lett. 1990;272:106–108. doi: 10.1016/0014-5793(90)80459-v. [DOI] [PubMed] [Google Scholar]
- 7.Matringe M, Mornet R, Scalla R. Eur J Biochem. 1992;209:861–868. doi: 10.1111/j.1432-1033.1992.tb17358.x. [DOI] [PubMed] [Google Scholar]
- 8.Camadro J M, Matringe M, Scalla R, Labbe P. Biochem J. 1991;277:17–21. doi: 10.1042/bj2770017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camadro J M, Labbe P. J Biol Chem. 1996;271:9120–9128. doi: 10.1074/jbc.271.15.9120. [DOI] [PubMed] [Google Scholar]
- 10.Urban-Grimal D, Labbe-Bois R. Mol Gen Genet. 1981;183:85–92. doi: 10.1007/BF00270144. [DOI] [PubMed] [Google Scholar]
- 11.Camadro J M, Urban-Grimal D, Labbe P. Biochem Biophys Res Commun. 1982;106:724–730. doi: 10.1016/0006-291x(82)91771-5. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor N, Mornet R, Matringe M, Clair D, Scalla R, Fujimoto T T, Swithenbank C. Bioorg Med Chem. 1994;2:339–342. doi: 10.1016/s0968-0896(00)82190-2. [DOI] [PubMed] [Google Scholar]
- 13.Schenk P M, Baumann S, Mattes R, Steinbiss H H. BioTechniques. 1995;19:196–198. , 200. [PubMed] [Google Scholar]
- 14.Gondal J A, Eiseman J L, Alvares A P. J Pharmacol Exp Ther. 1987;241:540–546. [PubMed] [Google Scholar]
- 15.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Haid A, Suissa M. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 17.Camadro J M, Matringe M, Scalla R, Labbe P. In: Target Assays for Modern Herbicides and Related Phytotoxic Compounds. Böger P, Sandmann G, editors. Boca Raton, FL: CRC/Lewis Publishers; 1993. pp. 29–34. [Google Scholar]
- 18.Poulson R, Polglase W J. J Biol Chem. 1975;250:1269–1274. [PubMed] [Google Scholar]
- 19.Wierenga R K, Terpstra P, Hol W G J. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 20.Arnould S, Berthon J L, Hubert C, Dias M, Cibert C, Mornet R, Camadro J M. Biochemistry. 1997;36:10178–10184. doi: 10.1021/bi970549j. [DOI] [PubMed] [Google Scholar]
- 21.Camadro J M, Matringe M, Thome F, Brouillet N, Mornet R, Labbe P. Eur J Biochem. 1995;229:669–674. doi: 10.1111/j.1432-1033.1995.tb20512.x. [DOI] [PubMed] [Google Scholar]
- 22.Clark R D. J Agric Food Chem. 1996;44:3643–3652. [Google Scholar]
- 23.Lyga J W, Russell, Patera R M, Plummer M J, Halling B P, Yuhas D A. Pestic Sci. 1994;42:29–36. [Google Scholar]
- 24.Margaron P, Gregoire M J, Scasnar V, Ali H, van Lier J. Photochem Photobiol. 1996;63:217–223. doi: 10.1111/j.1751-1097.1996.tb03017.x. [DOI] [PubMed] [Google Scholar]
- 25.Nevalainen T, Kolehmainen E. Environ Toxicol Chem. 1994;13:1699–1706. [Google Scholar]
- 26.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J P. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picot D, Garavito R M. FEBS Lett. 1994;346:21–25. doi: 10.1016/0014-5793(94)00314-9. [DOI] [PubMed] [Google Scholar]
- 28.Picot D, Loll P J, Garavito R M. Nature (London) 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 29.Wendt K U, Poralla K, Schulz G E. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 30.Hansson M, Hederstedt L. J Bacteriol. 1992;174:8081–8093. doi: 10.1128/jb.174.24.8081-8093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansson M, Hederstedt L. J Bacteriol. 1994;176:5962–5970. doi: 10.1128/jb.176.19.5962-5970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansson M, Gustafsson M C, Kannangara C G, Hederstedt L. Biochim Biophys Acta. 1997;1340:97–104. doi: 10.1016/s0167-4838(97)00030-7. [DOI] [PubMed] [Google Scholar]
- 33.Hansson M, Hederstedt L. Eur J Biochem. 1994;220:201–208. doi: 10.1111/j.1432-1033.1994.tb18615.x. [DOI] [PubMed] [Google Scholar]
- 34.Narita S, Tanaka R, Ito T, Okada K, Taketani S, Inokuchi H. Gene. 1996;182:169–175. doi: 10.1016/s0378-1119(96)00545-8. [DOI] [PubMed] [Google Scholar]
- 35.Lermontova I, Kruse E, Mock H P, Grimm B. Proc Natl Acad Sci USA. 1997;94:8895–8900. doi: 10.1073/pnas.94.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura K, Taketani S, Inokuchi H. J Biol Chem. 1995;270:8076–8080. doi: 10.1074/jbc.270.14.8076. [DOI] [PubMed] [Google Scholar]
- 37.Taketani S, Yoshinaga T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, Inokuchi H. Eur J Biochem. 1995;230:760–765. doi: 10.1111/j.1432-1033.1995.0760h.x. [DOI] [PubMed] [Google Scholar]