Abstract
The cation-independent mannose 6-phosphate receptor is a multifunctional protein which binds at the cell surface to two distinct classes of ligands, the mannose 6-phosphate (M6P) bearing proteins and IGF-II. Its major function is to bind and transport M6P-enzymes to lysosomes, but it can also modulate the activity of a variety of extracellular M6P-glycoproteins (i.e., latent TGFβ precursor, urokinase-type plasminogen activator receptor, Granzyme B, growth factors, Herpes virus). The purpose of this review is to highlight the synthesis and potential use of high affinity M6P analogues able to target this receptor. Several M6P analogues with phosphonate, carboxylate or malonate groups display a higher affinity and a stronger stability in human serum than M6P itself. These derivatives could be used to favour the delivery of specific therapeutic compounds to lysosomes, notably in enzyme replacement therapies of lysosomal diseases or in neoplastic drug targeting. In addition, their potential applications in preventing clinical disorders, which are associated with the activities of other M6P-proteins involved in wound healing, cell growth or viral infection, will be discussed.
Keywords: Binding Sites; Glycoproteins; metabolism; Humans; Lysosomes; metabolism; pathology; Mannosephosphates; chemistry; metabolism; therapeutic use; Neoplasms; drug therapy; Receptor, IGF Type 2; agonists; chemistry; metabolism
Keywords: CI-M6PR, lysosome, mannose 6-phosphate analogues, drug targeting, therapy
I. INTRODUCTION
1. LYSOSOMES AND MANNOSE 6-PHOSPHATE ADDRESSING
Lysosomes and associated organelles contain numerous acid hydrolases and constitute a system of acid compartments that establish the communication between intracellular and extracellular medium by means of endocytosis, phagocytosis and exocytosis. The main function of lysosomes in the cell is the degradation of internalized material by lysosomal enzymes, the acid hydrolases [1]. These enzymes are synthesized in the endoplasmic reticulum and during their maturation in the Golgi apparatus they acquire the mannose 6-phosphate (M6P) recognition marker [2–6]. Most of these soluble hydrolases are transported to lysosomes through specific M6P receptors [7]. The receptor-enzyme complexes segregated in the trans-Golgi network are transferred to the lysosomal compartments via the secretory pathway through clathrin-coated vesicles.
In mammalian cells, the targeting of newly synthesized hydrolases, as well as others ligands with M6P residues on their N-linked oligosaccharides, to the lysosomes depends on their recognition by two specific M6P receptors at the trans-Golgi network: the cation-dependent M6P receptor (CD-M6PR) and the cation-independent M6P receptor (CI-M6PR), also called mannose 6-phosphate/insulin-like growth factor II receptor in eutherian mammals, in which it binds IGF-II [8]. These two receptors, the two only members of the P-type lectin family, are type I transmembrane glycoproteins. They are considered sorting receptors because of their routing function. In the pre-lysosomal compartment, acidity induces release of enzymes from both M6PR which are then recycled into the Golgi apparatus. However, the CI-M6PR, which participates to this cellular routing, is also anchored to the cell surface membrane and can internalize extracellular ligands [8–10]. In summary, the pool of lysosomal acid hydrolases comes both from in situ synthesis, which involves the two M6P receptors, and from endocytosis through the CI-M6PR [11, 12].
2. STRUCTURE OF MANNOSE 6-PHOSPHATE RECEPTORS AND LIGAND MULTIPLICITY
Sequence comparison between CD-M6PR and CI-M6PR shows that they are related. In fact, the extracytoplasmic domain of CD-M6PR is homologous to the approximately 145 amino acid long repeating domains present in the CI-M6PR with sequence identity ranging from 14 to 28%. These studies allowed to conclude that these two receptors located on different chromosomes (12p13 and 6q26, respectively) have diverged from a common ancestral gene [13, 14].
The CD-M6PR is known as the cation-dependent M6P receptor because of its ligand binding ability in the presence of divalent cations. This receptor is a 46 kDa single polypeptide chain that contains a putative signal sequence and a transmembrane domain. This is a single membrane-spanning domain, which separates a N-terminal extracytoplasmic region with 5 potential Asn-linked glycosylation sites, from a C-terminal cytoplasmic region without Asn-glycosylation sites. Sequence analysis of the bovine CD-M6PR revealed that it consists of a 28 amino-acid residue N-terminal signal sequence, a 159 amino acid residue luminal domain, a 25 amino acid residue transmembrane domain and a 67 amino acid residue C-terminal cytoplasmic domain. It is highly conserved from mouse to human (93% homology). This receptor appears to be a homodimer at the membrane [15], and either a dimer or a tetramer in solution [13, 16, 17].
The CI-M6PR is a large 300 kDa glycoprotein which binds M6P in a cation-independent manner. The human CI-M6PR is 2491 amino acid-long and includes a N-terminal signal sequence of 40 amino acids, an extracytoplasmic domain consisting of 15 homologous repeat sequences of 134–167 amino acids (2264 amino acids for the full extracytoplasmic region), a transmembrane region of 23 amino acids, and a cytoplasmic domain of 164 amino acids which constitute the C-terminal domain [18]. One of the repeats contains an additional 43-residue segment that is similar to the type II repeat of fibronectin [19]. The extracytoplasmic domain has 19 potential glycosylation sites [20, 21]. Although the quaternary structure of this receptor has not been totally resolved, some results have allowed a description of its membrane dimerization [22]. In addition, a soluble form of the CI-M6PR receptor is naturally released by proteolytic cleavage from the intact cellular receptor and has been detected at low concentrations (0.1 nM) in serum of a variety of mammalian species. On the other hand, the nucleotide sequence of the full-length cDNA and the deduced amino acid sequence for the CI-M6PR, were reported to be strikingly similar (99.4% identical in the amino acid sequence) to those reported for the human insulin-like growth factor II (IGF-II) receptor from HepG2 hepatoma cells [23]. These findings supported the suggestion that CI-M6PR is a multifunctional binding protein, identical to the IGF-II receptor [18], that binds both to M6P and IGF-II [24]. Indeed, within the extracytoplasmic domain, the CI-M6PR binds 2 molecules of M6P and 1 molecule of IGF-II per molecule of receptor [25, 26].
IGF-II is the best characterized non M6P-bearing ligand of CI-M6PR. This hormone plays key roles in metabolic regulation and foetal growth [27] through three types of receptors, two tyrosine kinase receptors (IGF-I receptor and insulin receptor isoform A) and CI-M6PR. The stimulation of the type 1 receptor tyrosine kinase induces a protein phophorylation cascade leading to biological effects, particularly the insulin-mediated growth and increase in CI-M6PR expression [28], which mediates endocytosis and clearance of IGF-II [29]. The lysosomal degradation of IGF-II is critical because elevated levels of IGF-II induce overgrowth. Indeed, mice without CI-M6PR accumulate high amounts of IGF-II and die peri-natally [30–32]. As this phenotype can be reversed by a deficiency in IGF-II, it is directly caused by the over-expression of IGF-II. In humans, the soluble levels of CI-M6PR in serum are higher in infants and fall by 40% in adult life to reach a circulating level of about 700 μg/l [33, 34]. The receptor’s circulating level increases during pregnancy and in case of diabetes, and appears correlated with the elevation of circulating lysosomal enzymes in both cases [33]. Though CI-M6PR circulates at a concentration lower than that of IGF-II, it seems to play a role in IGF-II clearance. It has been demonstrated that this soluble receptor exerts biological effect, notably by binding IGF-II and blocking IGF-II-stimulated DNA synthesis in hepatocytes and fibroblasts, and at physiological concentrations this receptor can block tumour growth mediated by IGF-II [35].
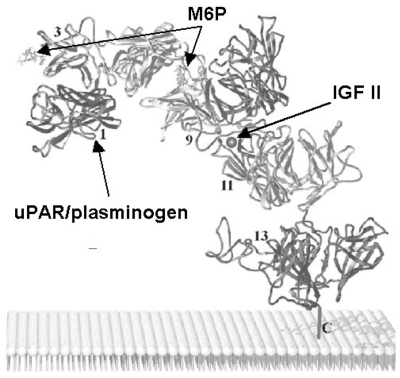
IGF-II and proteins bearing M6P are linked by the same receptor but their binding sites are distinct. In fact, some of the 15 homologous repeating domains of the extracytoplasmic region are responsible for the binding to M6P or IGF-II. The IGF-II binding site is contained within domain 11 (residues 1508–1566) [36] and sequences in domain 13 contribute a 10 fold enhancement in IGF-II affinity [37]. Domains 1–3 and 7–9 contain the two high affinity M6P-binding sites with essential residues localized within domains 3 and 9 [38] (Fig. (1)). In addition, domain 5 of CI-M6PR exhibits significant sequence homology to domains 3 and 9 as well as to the extracytoplasmic region of CD-M6PR, suggesting that CI-M6PR contains a third M6P recognition site that is located in domain 5 and exhibits lower affinity than the carbohydrate-binding sites present in the two high affinity M6P-binding domains [39]. Even though the binding sites of M6P and IGF-II are distinct, these two ligands can reciprocally inhibit each other [40]. Indeed, IGF-II has been shown to decrease binding of M6P-bearing lysosomal enzymes to CI-M6PR [41, 42]. Similarly, β-galactosidase and cathepsin D, two lysosomal enzymes containing M6P moieties, have been shown to decrease the binding of CI-M6PR to IGF-II [43]. These cross-interactions that partially shield other binding sites could be caused by a steric hindrance or a conformational change in the receptor. CI-M6PR has been also shown to bind with high affinity a third type of ligand, the retinoic acid at a site that is distinct from those of M6P and IGF-II, but the precise localization of the retinoic acid binding site remains unknown. Retinoic acid exerts diverse biological effects involved in the control of cell growth in embryogenesis and oncogenesis through its binding to the nuclear retinoid receptors. The interaction of retinoic acid with CI-M6PR may modulate a retinoic acid response pathway that is important for cell growth regulation [44]. Immuno-fluorescence techniques have allowed to show that retinoic acid can alter the intracellular distribution of both CI-M6PR and lysosomal enzymes [45].
Figure 1.
Besides IGF-II, retinoic acid and lysosomal enzymes carrying M6P, plasma membrane CI-M6PR has been shown to bind a number of other extracellular ligands bearing the M6P recognition marker (Table 1). Some multi-protein complexes, containing latent-TGFβ or urokinase-type plasminogen activator receptor (uPAR), bind to the membrane CI-M6PR to be activated extracellularly. Other proteins bind to CI-M6PR to be internalized or addressed to the lysosomes and then degraded. Since membrane CI-M6PR has an important role in some cancers and in sorting of lysosomal enzymes, this review will focus on the different functions of cellular CI-M6PR. According to the type of ligand, CI-M6PR may modulate a large panel of biological pathways, such as cell migration, wound healing, angiogenesis, cell growth inhibition, apoptosis or viral infection.
Table 1.
Multiple ligands and functions of CI-M6PR
| Sites and ligands | CI-M6PR functions | Associated Diseases | Ref. |
|---|---|---|---|
| M6P binding sites (repeats 1–3 and 9) | [38] | ||
| ~ 50 lysosomal enzymes (secreted cathepsins) | Lysosome biogenesis | Most of lysosomal diseases | [71] |
| Tumor dissemination | Cancer metastasis | [118] | |
| Latent TGFβ complex | TGFβ activation | Wound healing | [100] |
| Granzyme B | Cell entry and apoptosis | Immune cytotoxic cell response | [111] |
| Leukemia Inhibitory Factor, Macrophage-Colony-Stimulating Factor | Cell entry and degradation in lysosome | Haematopoietic disorders, multiple myeloma, Castelman disease, polycythemia vera | [76] |
| Proliferin | Cell growth, angiogenesis | Fibrosarcomas | [114] |
| gD, glycoprotein of herpes virus | Virus entry | Viral infection | [110] |
| Renin | Cardiac (pro)renin uptake | Cardiovascular disorders | [84] |
| IGFII binding site (repeat 11) | Cell entry and degradation in lysosome | Cell growth in embryogenesis and oncogenesis | [36] |
| uPAR/plasminogen binding site (repeat 1) | Regulation cell surface uPAR, activation uPA, activation of TGFβ | Fibrinolysis, cell adhesion and migration | [104] |
| Retinoic acid (unknown site) | Modulation of retinoic acid action | Cell growth in embryogenesis and oncogenesis | [44] |
Therefore, CI-M6PR potentially constitutes, by its multiple functions, a target for several clinical applications and justifies the development of synthetic M6P analogues to compensate for the poor stability of natural M6P in human serum and to increase the binding affinity with CI-M6PR.
II. SYNTHETIC ANALOGUES TO TARGET THE MEMBRANE CI-M6PR
1. RECOGNITION REQUIREMENTS
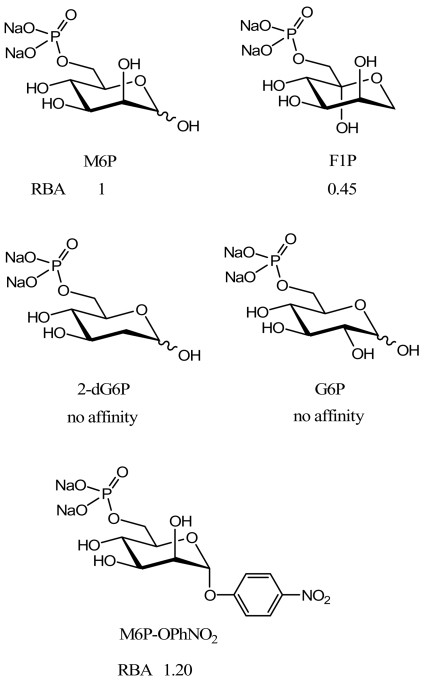
Some structural features have been shown to be crucial for the binding of M6P to the receptor. The hydroxyl group at the 2-position of the pyranose ring must be axial to allow a strong binding to the CI-M6PR. Both the absence of the 2-hydroxyl group (2-deoxy-glucose 6-phosphate (2dG6P)) and epimerization at the 2-position (glucose 6-phosphate (G6P)) abolish recognition of M6P by the receptor [16]. Structural modifications of the M6P anomeric centre do not impair the binding to the CI-M6PR since fructose 1-phosphate (F1P) is recognized as easily as M6P [16, 46]. The F1P representation given in Fig. (2) shows the high structural similarity between this compound and M6P. Moreover, the replacement of the hydroxyl group at the anomeric position by an aromatic aglycone, such as a para-nitrophenoxy group, slightly improves the recognition by CI-M6PR. This could be due to the lipophilic interactions between the aromatic moiety of the ligand and the binding pocket of CI-M6PR [46].
Figure 2.
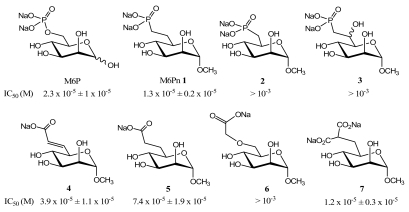
The distance between the negative charge and the pyranose ring plays also a key role in the recognition. We demonstrated that this distance must be in the same range as in M6P since the isosteric mannose 6-phosphonate (M6Pn) analogue 1 is well recognized by the CI-M6PR, whereas the non isosteric M6Pn analogue 2 displays a weak affinity [47] (Fig. (3)). On the other hand, the β-hydroxyphosphonate isosteric analogues 3 can not bind to CI-M6PR [48]. This means that the replacement of the phosphomonoester linkage by a methylene group does not affect the recognition phenomenon [47] but, further substitutions at the 6-position decrease the ligand-receptor interaction, probably by steric hindrance [48].
Figure 3.
It has been shown that a negatively charged phosphodiester analogue of M6P (with one methyl ester on the phosphate moiety) displays an affinity for the receptor similar to that of M6P [25]. This result demonstrates that only one negative charge is necessary for the binding to the CI-M6PR. This was confirmed by the synthesis and biological evaluation of carboxylate isosteric analogues of M6P. A specific receptor binding assay demonstrated that the unsaturated isosteric analogue 4 binds to CI-M6PR as well as M6P, while the saturated isosteric analogue 5 binds to the receptor with a slightly weaker affinity than M6P (Fig. (3)) [49]. These data confirmed previous results obtained by ligand-dependent elution of the receptor from pentamannose 6-phosphate affinity columns [50]. This difference in binding affinity can probably be explained by the geometry of the double bond in 4 which leads to greater interaction with the M6P-binding sites of CI-M6PR, in comparison to the single bond in 5. On the other hand, the non isosteric derivative 6 is weakly recognized by the CI-M6PR. We suppose that the analogue 6 does not match exactly the M6P-binding sites of the receptor because the distance between the negative charge and the pyranose ring in 6 is lengthened by comparison to that in M6P. The lengthening of the lateral chain probably impairs the binding affinity. A higher binding affinity was obtained with the malonate analogue 7 which is recognized by CI-M6PR as M6P itself. This last result does not correlate exactly with the IC50 values obtained by Berkowitz et al. [51]. However, their binding test followed a procedure in which CI-M6PR was covalently bound to a sepharose resin, while in our case CI-M6PR was not bound to a support. These procedures probably account for differences in affinity.
In summary, the analogues synthesized to target CI-M6PR must be isosteric to M6P to efficiently bind the receptor. Moreover, a single negative charge is sufficient to allow the binding to the receptor while the phosphorus atom is not necessary to ensure recognition. However, the best ligands for CI-M6PR contain two negative charges like in the malonate or phosphonate isosteric analogues of M6P.
Christensen et al. have synthesized M6P glycopeptides in order to determine their affinity for CI-M6PR and have shown that the invariant core of the oligosaccharide moiety of the lysosomal enzymes is not needed for their recognition by CI-M6PR. High affinity for CI-M6PR was maintained by keeping the terminal phosphorylated dimannoside on the antennary moieties whilst the invariant saccharidic core could be replaced by tri-, tetra-, or penta-peptides with no significant variations in the affinity [52, 53]. However, these data have to be put in perspective because the removal of the aminobenzoyl group, which is bound to the peptide lysine tail, caused a significant decrease in the affinity for CI-M6PR. This result demonstrates that a small change in the structure of the peptide can have a strong influence on the recognition process.
Roche et al. have functionalized bovine serum albumin (BSA) with M6P residues (i.e., M6P20-BSA) in order to target CI-M6PR [54] at the cell surface of monocytes and macrophages [54, 55]. The functionalization of a protein in order to target CI-M6PR has been developed with the aim of avoiding liver fibrosis by selective delivery of anti-fibrotic drugs to the hepatic stellate cells. They used human serum albumin (HSA), functionalized with M6P residues (i.e., M6PX-HSA) to cargo bioactive compounds, such as the antifibrotic drugs doxorubicin [56], pentoxifylline (PTX) [57], 18β-glycyrrhetinic acid (18β-GA) [58], mycophenolic acid [59, 60] or the apoptosis-inducing drug 15-deoxy-D12,14-prostaglandin J2 (15dPGJ2) [61], to the stellate cells which are implicated in the formation of fibrosis. They showed that with this approach they can efficiently target these cells via the CI-M6PR as the formation of fibrosis was significantly decreased. Moreover, Beljaars et al. demonstrated that the lower-derivatized albumins (i.e., M6P2-HSA, M6P4-HSA, and M6P10-HSA) remain in the blood for a long time, whereas the albumin preparations with the highest degree of M6P substitution (i.e., M6P28-HSA) selectively accumulate in the liver [62]. In order to improve the affinity for CI-M6PR, it would be interesting to use M6P analogues where the phosphate moiety has been replaced with other bioisosteric groups. For example, M6Pn 1 (Fig. (3)) has been used to improve the M6P treatment for wound healing [63] and was shown to reduce the size of scars as efficiently as M6P itself.
In order to study the interaction between M6P-bearing compounds and the CI-M6PR at the cell surface, an amphiphilic derivative, called M6Pn-CSA, was synthesized [64]. M6Pn and CSA stand for mannose 6-phosphonate and cholesteryl-succinyl-anilinyl, respectively. This amphiphilic steroidal glyco-conjugate was incorporated in the lipidic bilayer of liposomes and the resulting functionalized vesicles were incubated with MCF-7 breast cancer cells that over-express CI-M6PR. Strong liposome-cell interactions were observed demonstrating the high potential of such system for drug delivery applications. Then, another M6Pn-cholesteryl conjugate was synthesized to circumvent the synthetic difficulties encountered during the preparation of the M6Pn-CSA derivative [65, 66]. Hamdaoui et al. studied how M6P2-Pepstatine targeted tumour cells via CI-M6PR and showed that the bifunctional M6P2-Pepstatine has an antiproliferative activity against breast cancer cells [67]. Also in this case, it would be important to replace the phosphate moiety of M6P with a bioisosteric group to improve the affinity and the targeting of cancer cells that over-express CI-M6PR.
Thus, proteins, vesicles, nano-particles or other polymers could be functionalized with mannose 6-phosphate or its analogues in order to be used as carriers of bioactive molecules for the treatments of different diseases.
2. DOCKING M6P ANALOGUES IN THE CI-M6PR LIGAND BINDING POCKET
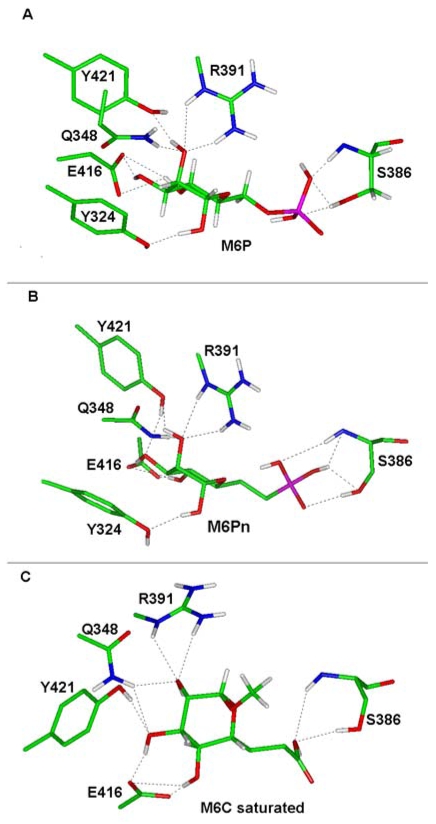
The crystal structure of domains 1–3 of CI-M6PR has been described [68, 69]. In this structure, the ligand binding pocket (LBP) is outlined by the β-strands 2, 3, 7, 8, 9, and loop C. LBP is also lined by 50% of uncharged amino acids Tyr(Y) 324, Gln(Q) 348, Cys(C) 356, C385, Ser(S) 386, Y421 and 50% of charged residues Glu(E) 323, Lys(K) 350, K358, E383, Arg(R) 391, E416. In order to understand the variations in the binding affinity of our analogues, we have analysed how the binding affinity changes in function of the structure modifications of each analogue. Molecular dynamic calculations of the analogues were performed using the InsightII software 2.7 (Accelrys, San Diego, CA). Energy was minimized using the Discover module until the deviation root mean square was below 0.01 kcal/mol and a consistent force field and the conjugate gradient algorithm were applied together with a cut-off distance of 25Ǻ. Ligand docking was initially carried out by superposition of the analogue structure to be tested onto the M6P agonist structure. Hydrogen bonds between residues Y324, Q348, S386, R391, E416, Y421 and the sugar were preserved. The ligand-receptor complex was then submitted to energy minimization until the maximum derivative was less than 0.2 kcal/Ǻ.
We found that the presence of S-S bonds (C385-C419, C339-C347) in the vicinity of the LBP within domain 3 (S386, E416, Q348) contributes to the conservation of the overall carbohydrate-binding cavity during our minimization. The ligand binding site (LBS), which gathers residues supporting direct interactions with the ligand, was not changed after minimization, even though moderate orientations of the side chains could occur, and was composed of residues Q348, R391, E416, Y324 and Y421 at one end of the pocket and, S386 at the opposite end. They stabilized with potential hydrogen bonds the hydroxyl groups of the ligand carried by C2, C3, C4, as well as the functional group carried by C8, i.e. M6 carboxylate saturated (M6C saturated).
However, discrete variations in the structure of the analogues, such as in the M6Pn, in which the oxygen atom on the side chain was substituted by a carbon atom, increase the relative bound affinity of the ligand (IC50 = 1.3 × 10−5 ± 0.2 × 10−5 M). As shown in the flattened view of the LBS (Fig. (4)) the reference M6P displays 11 potential hydrogen bonds and only 6 different anchorage points (Fig. (4A)) whereas the M6Pn analogue displays 12 potential hydrogen bonds with the essential residues in the LBS, and is anchored by 8 different positions to the CI-M6PR (Fig. (4B)). This increase in both the number of hydrogen bonds and the number of anchorage points, which stabilise the ligand within the LBS, might explain the better affinity of the M6Pn for the receptor as compared to that of M6P. We also observed that CI-M6PR exhibits a similar affinity for the M6-malonate analogue (IC50 = 1.2 × 10−5 ± 0.3 × 10−5 M), which is almost twice that of the reference M6P (IC50 = 2.3 × 10−5 ± 1.0 × 10−5 M). The malonate analogue is characterized by two carboxylate moieties instead of a phosphate moiety. This ligand allows the formation of 13 potential hydrogen bonds and 8 anchorage points (data not shown). The ratio between the number of anchorage points and the number of hydrogen bonds increases as the binding affinity increases (0.55 and 0.62 for M6P and the malonate analogue, respectively)
Figure 4.
In contrast, the M6C saturated ligand (methyl 6,7 dideoxy-α-D-manno-octopyranoside-uronate) displays a lower binding affinity (IC50 = 7.4 × 10−5 ± 1.9 × 10−5 M) than the M6P reference (Fig. (4C)). Moreover, the decrease in the binding affinity is correlated with a reduction in the ratio between the anchorage point number and the hydrogen bond number (0.55 and 0.50 for M6P and M6C saturated, respectively). In addition, the direct interaction of residue Y324 and the mannose was missing for that analogue, a residue known to be involved in the ligand stability as revealed by single amino acid substitutions [70].
III. ROLES OF CI-M6PR AND POTENTIAL THERAPEUTIC APPLICATIONS OF MANNOSE 6-PHOSPHATE ANALOGUES
1. CI-M6PR AS THE MAJOR TRANSPORTER TO THE LYSOSOME
An evident application of M6P derivatives is the transport of molecules to the lysosomes through the membrane CI-M6PR.
CI-M6PR AS A TOOL FOR LYSOSOMAL DISEASE TREATMENT
One of the major therapeutic applications of the M6P derivatives could be the enzyme replacement therapy (ERT) for lysosomal diseases. Each lysosomal disease is linked to the absence of a particular enzyme and the consequent lysosomal accumulation of a specific substrate. Soon after the discovery of the lysosome, de Duve [71] proposed that deficiencies of lysosomal enzymes could be rectified by the administration of exogenous enzymes. The treatment of patients with lysosomal storage diseases has been markedly improved in the past 10 years by the successful implementation of ERT. This therapy was first proposed for the treatment of Gaucher disease and, then, for other lysosomal storage diseases (Fabry, Pompe, MPS-I diseases). However, a major set-back is the enormous cost of such therapies and in many countries the health-care system cannot afford to pay for ERT. For example, the sale of the recombinant enzyme for the treatment of 4,500 Gaucher disease patients for a single year (2005) was estimated to at least 839 000,000 USD. This is merely the cost of the treatment of one of the 50 known lysosomal diseases. The treatments for other such rare diseases would take too many resources out of national health care budgets. The finding of new M6P analogues may help to decrease at least in part these costs. These analogues may improve both the affinity of the specific recombinant enzyme for CI-M6PR and the stability of the M6P moiety in the blood circulation. Consequently, they could significantly enhance ERT efficacy and reduce the active doses of enzyme that is required for an efficient treatment. Moreover, the successful application of these analogues, could promote their future utilisation in most of the 50 known lysosomal diseases. Several studies in mouse models, which were treated with high affinity ligands (i.e., phosphorylated oligomannose–containning poly-saccharides) conjugated to the oligosaccharide side-chain of the recombinant enzymes, demonstrated the role of CI-M6PR and the higher efficacy of high affinity M6P-glycoproteins in ERT. This approach allowed to decrease the effective dose of enzyme and to treat either less accessible tissues, such as skeletal muscles, or tissues with a relatively low abundance of CI-M6PR [72].
CI-M6PR AS A RECEPTOR FOR METABOLIC CLEARANCE OF EXTRACELLULAR LIGANDS
CI-M6PR binds and internalizes leukemia inhibitory factor
Leukemia inhibitory factor (LIF) is a multifunctional, highly glycosylated soluble protein belonging to the interleukin-6 (IL-6) subfamily of helical cytokines [73]. LIF exerts an important role in neuronal, platelet and bone formation. It also induces embryonic stem cells to retain their totipotency. Indeed, this multifunctional protein stimulates adipocyte lipid transport, adreno-corticotropic hormone production, proliferation of some hematopoeitic and muscle cells and inflammation [74]. Blanchard and co-workers have reported that glycosylated LIF, produced by transfected Chinese hamster ovary cells, binds to CI-M6PR [75]. Their subsequent study showed that M6P-containing LIF is naturally produced by a number of normal and tumour cell lines, and that another unrelated cytokine, the macrophage-colony-stimulating factor, was also able to bind to CI-M6PR in a M6P-sensitive manner [76]. The M6P-containing cytokine LIF is rapidly internalized and degraded by cells expressing CI-M6PR [76]. In addition, soluble CI-M6PR inhibits interleukin-6-type cytokine-dependent proliferation by neutralizing IGF-II [77]. Soluble CI-M6PR is a candidate natural molecule, presumably as other inhibitors of the IGF-II pathway, for the development of novel therapeutic strategies in the treatment of haematopoietic disorders in which IL-6-type cytokines play a role, particularly multiple myeloma.
CI-M6PR regulates renin and pro-renin clearance
The pool of cardiac renin comes mostly from the circulation. Renin and its inactive form, pro-renin, whose circulating level is much higher than that of renin, diffuse into the cardiac interstitial space [78, 79] or bind to receptors [80, 81]. One of these receptors is the (pro)renin receptor which binds equally well to renin and pro-renin. This receptor induces the activation of pro-renin without internalizing it [82]. CI-M6PR can also bind and internalize renin and pro-renin [83, 84]. After internalization pro-renin is swiftly activated by proteolytic cleavage [84, 85]. This mechanism leading to the production of renin is specific and saturable and can be antagonized by M6P [84]. It seems that this mechanism also participates to the renin and pro-renin clearance [86]. However, only a small percentage of renin and pro-renin carries an M6P signal and the amount captured by cardiomyocytes via CI-M6PR represents only 5–6% of the total circulating level [84, 87]. In conclusion, CI-M6PR in myocytes may participate in the cardiac (pro)renin uptake by binding to the fraction of human (pro)renin, that is characterized by the presence of the M6P recognition marker [86].
LYSOSOMAL TARGETING FOR CANCER THERAPY
CI-M6PR has been reported to be a potential tumour suppressor in 70% of hepatocarcinomas [88] and 15–30% of breast cancers [89–91]. The gene was deleted in one allele and mutated at the other one in these cancers. Moreover, over-expression of CI-M6PR induced regression of tumours in mice and growth inhibition in cancer cells. These effects are probably due to the different functions of this receptor that indirectly controls cell growth: (i) internalization and degradation of various growth-promoting factors, such as IGF-II [92], glycosylated LIF, and other M6P-containing cytokines, such as the macrophage colony-stimulating factor [76]; (ii) binding and uptake of granzyme B, an essential factor for T cell-mediated apoptosis [93]; (iii) regulation of the level of secreted lysosomal enzymes that are responsible for extracellular matrix degradation and tumour dissemination. Even though the CI-M6PR expression is decreased in some malignancies such as hepatocarcinoma, its over-expression in the majority of solid tumours, particularly in breast cancers [94], indicate that this receptor could be considered as a mean to address cytotoxic drugs to lysosomes. The routing of cytotoxic drugs via CI-M6PR may induce the lysis of lysosomes and then cell death. The higher specificity of CI-M6PR targeting in cancer cells versus normal cells could be due to the higher expression of CI-M6PR and its higher affinity at slightly acidic pH (pH 6–6.5) as found in the extracellular environment of solid tumors. This approach could be complementary to the classical chemotherapy. In fact, most anti-neoplastic drugs, such as anthracyclines (e.g., doxorubicin) or Vinca alkaloids, are weakly basic molecules which target the neutral pH compartment of tumours, but become inactive in a more acidic environment [95, 96].
Attempts to target the acidic extracellular compartment of solid tumors have been done by loading doxorubicin into pH-sensitive poly/PEG/folate micelles [97] or liposomes [98]. This conferred a longer half life to the drug and a higher efficacy probably by achieving higher local dose in the tumour. However, a major obstacle to successful cancer chemotherapy is the emergence of tumour chemo-resistance, which is due to different mechanisms, including over-expression of the multidrug resistance 1 gene product and decrease in the net intracellular accumulation of the drug [99]. A chemotherapy targeting CI-M6PR will also probably circumvent these chemo-resistance problems.
2. CI-M6PR AS A CRUCIAL PARTNER FOR THE ACTIVATION OF MULTIPLE PATHWAYS
CI-M6PR could activate several biological pathways either by acting at the cell membrane (TGFβ) or by internalizing extracellular ligands (glycoprotein D of the herpes virus, granzyme B and proliferin).
CI-M6PR AS AN EXTRACELLULAR ACTIVATOR OF LATENT TGFβ
The activation of latent transforming growth factor β (LTGFβ) to active TGFβ is a critical step in the healing process. LTGF-ss that comprises TGFβ bound to the latency associated peptide (LAP), which in turn may be bound to the LTGF-ss binding protein (LTBP), binds to cell-surface CI-M6PR via M6P-containing carbohydrates in the LAP [100–103]. The increase of M6P at a wounding site reduces the binding of LTGFβ to CI-M6PR and, consequently, affects the formation of fibrotic TGFβ.
In addition to its binding to LTGFβ CI-M6PR plays also a role in another process involved in LTGFβ activation, the plasminogen/plasmin conversion by a serine protease, the urokinase-type plasminogen activator (uPA). The pro-uPA is proteolytically cleaved and thereby activated, when bound to a specific uPAR at the cell surface. This uPAR has been reported to be a low affinity ligand for CI-M6PR [12, 104]. The full length uPAR and plasminogen appear both to interact with the N-terminus of repeat 1 of the CI-M6PR in a M6P independent manner and are situated on the opposite face of the molecule than the carbohydrate-binding sites [69] (Fig. (1)). Thus, it is possible that CI-M6PR binds simultaneously both uPAR/plasminogen and an M6P-oligosaccharide. Different roles for the binding of uPAR to CI-M6PR have been proposed, including the modulation of TGFβ activation, control of cell surface uPAR expression, or uPAR-mediated functions, such as fibrinolysis, cell adhesion, and migration [12, 104–108].
CI-M6PR AS A TRANSPORTER FOR INTRACELLULAR ACTIVATION
Herpes virus entry through CI-M6PR
The herpes simplex virus (HSV) glycoprotein D (gD) is essential for HSV entry into cells, it contains M6P on its Asn-linked oligosaccharides, and binds to CI-M6PR. It was supposed that the interaction between gD and CI-M6PR could play a role in some aspect of virus entry or egress [109]. In fact, CI-M6PR could serve as HSV receptors during virus entry into cells. Injection of soluble CI-M6PR and treatment with pentamannose-phosphate inhibit HSV plaque formation on monkey Vero cells, as do polyclonal rabbit anti-M6PR antibodies. It appears that M6P residues and M6P receptors are required for efficient transmission of HSV between cells, a process which differs in some respects from the entry of exogenous virus particles [110].
CI-M6PR binds granzyme B and induces cell death
CI-M6PR has been proposed as an alternative pathway to induce cell death through the delivery of lymphocyte granule toxins. Several studies indicate that CI-M6PR expressed at the cell membrane binds to granzyme B, a serine protease, secreted by cytotoxic T lymphocytes and natural killer cells, via its M6P moiety and induces endocytosis [93]. In the target cell, granzyme B activates an apoptotic pathway by cleaving key substrates in the cytoplasm, though these events are critically co-dependent on the pore-forming protein, perforin. Since granzyme B must access the substrates within the cell, the endocytosis through CI-M6PR binding is a key step in the granule-mediated killing process [111].
CI-M6PR regulates proliferin-induced angiogenesis
CI-M6PR is thought to be involved in the regulation of cell motility and angiogenesis through its binding to proliferin, a member of the prolactin/growth hormone family that is expressed in placenta [112–114]. Proliferin stimulates capillary endothelial cell migration, in culture, and induces neo-vascularisation, in vivo [115]. The angiogenic activity of proliferin depends on its interaction with CI-M6PR. This recognition involves at least in part the carbohydrate moiety of proliferin which competes with M6P [114]. Tumour growth depends on its vascularisation and needs micro-vessels development to expand its cell population and to become invasive [116]. Jackson et al. have described the expression profile of the proliferin gene in a progressive fibrosarcoma mouse tumour cell model. The expression of proliferin is detectable in cell lines derived from the mild, non-invasive tumour, is increased in cell lines from tumours at the aggressively invasive stage, and its expression remains high in cell lines from end stage fibrosarcomas [117]. Thus, the expression of proliferin seems to increase when a tumour becomes more invasive and exhibits a strong angiogenic activity.
IV. CONCLUSION AND PERSPECTIVES
The important diversity of ligands bearing an M6P signal suggests numerous potential therapeutic applications offered by CI-M6PR, which could be targeted by synthetic M6P analogues carried by lysosomal proteins or drugs. We and other groups have synthesized M6P analogues by changing the phosphate moiety into carboxylate, malonate or phosphonate moieties. Some of these modifications induced a gain of efficiency resulting in a better binding affinity for CI-M6PR and, importantly, in a stronger stability of these compounds in human serum. Indeed M6P analogues are not sensitive to blood phosphatases contrary to natural M6P.
Their application in lysosomal diseases is evident due to the 3 major limits of ERT in these diseases: (i) the replacing, recombinant enzyme needs an M6P signal, (ii) M6P signal is only produced by mammalian cells, (iii) ERT needs a large amount of recombinant enzymes. A new therapeutic approach, in which M6P analogues, selected for their affinity and their stability in circulation, are chemically coupled to the recombinant enzymes, could significantly improve ERT. This new lysosomal targeting could increase therapy efficiency at least by 10 fold and could allow: (i) to decrease the dosage needed for the treatment and, as a consequence, the side effects and (ii) to facilitate access to ERT to many underprivileged countries and (iii) to envisage a similar treatment of other lysosomal diseases. The fact that CI-M6PR was considered as a tumour suppressor does not appear as a contra-indication to future therapies using M6P analogues since these molecules will partially occupy plasma membrane receptors, corresponding to a minimal fraction of the total cellular receptor.
Other applications of the M6P analogues, which concern the competitive inhibition of different CI-M6PR extracellular ligands, have been already proposed depending on the patho-physiological condition. However, additional experiments on the physiological relevance of these unconventional M6P-based ligands and initial clinical trials should be performed before to consider the use of M6P analogues in the clinic. For example, the inhibition of the activation of latent TGF-β by M6P was envisaged as an anti-scarring therapy in adult wound healing. The prevention of activation of TGFβ1 and TGFβ2 by competition with M6P at the wound site results in markedly improved healing and less scarring. This type of competitive inhibition could be applied to other pathological processes, which are mediated through CI-M6PR interactions, such as the activation of complex structures in plasminogen activation or the internalization of growth-regulating proteins (i.e., LIF, granzyme B) or viral proteins.
Finally, the M6P analogues could also be used to target drugs or nano-particles to lysosomes where they will be activated by the large panel of acid protease activities of these structures. This approach could be used in the case of photo-sensitive, or pH-sensitive, drugs for imaging or therapy in cancers that express high levels of CI-M6PR.
Acknowledgments
This work was supported by grants from VML (Vaincre les Maladies Lysosomales) and ARC n° 3953 (Association pour la Recherche sur le Cancer).
References
- 1.Castino R, Demoz M, Isidoro C. J Mol Recognit. 2003;16:337–48. doi: 10.1002/jmr.643. [DOI] [PubMed] [Google Scholar]
- 2.Reitman ML, Kornfeld S. J Biol Chem. 1981;256:11977–80. [PubMed] [Google Scholar]
- 3.Waheed A, Hasilik A, von Figura K. J Biol Chem. 1981;256:5717–21. [PubMed] [Google Scholar]
- 4.Varki A, Kornfeld S. J Biol Chem. 1980;255:10847–58. [PubMed] [Google Scholar]
- 5.Reitman ML, Kornfeld S. J Biol Chem. 1981;256:4275–81. [PubMed] [Google Scholar]
- 6.Waheed A, Hasilik A, von Figura K. J Biol Chem. 1982;257:12322–31. [PubMed] [Google Scholar]
- 7.Kaplan A, Achord DT, Sly WS. Proc Natl Acad Sci U S A. 1977;74:2026–30. doi: 10.1073/pnas.74.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornfeld S. Annu Rev Biochem. 1992;61:307–30. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 9.Munier-Lehmann H, Mauxion F, Hoflack B. Biochem Soc Trans. 1996;24:133–6. doi: 10.1042/bst0240133. [DOI] [PubMed] [Google Scholar]
- 10.Dahms NM. Biochem Soc Trans. 1996;24:136–41. doi: 10.1042/bst0240136. [DOI] [PubMed] [Google Scholar]
- 11.Ni X, Canuel M, Morales CR. Histol Histopathol. 2006;21:899–913. doi: 10.14670/HH-21.899. [DOI] [PubMed] [Google Scholar]
- 12.Nykjaer A, Christensen EI, Vorum H, Hager H, Petersen CM, Roigaard H, Min HY, Vilhardt F, Moller LB, Kornfeld S, Gliemann J. J Cell Biol. 1998;141:815–28. doi: 10.1083/jcb.141.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahms NM, Lobel P, Breitmeyer J, Chirgwin JM, Kornfeld S. Cell. 1987;50:181–92. doi: 10.1016/0092-8674(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 14.Klier HJ, von Figura K, Pohlmann R. Eur J Biochem. 1991;197:23–8. doi: 10.1111/j.1432-1033.1991.tb15877.x. [DOI] [PubMed] [Google Scholar]
- 15.Stein M, Braulke T, Krentler C, Hasilik A, von Figura K. Biol Chem Hoppe Seyler. 1987;368:937–47. doi: 10.1515/bchm3.1987.368.2.937. [DOI] [PubMed] [Google Scholar]
- 16.Tong PY, Kornfeld S. J Biol Chem. 1989;264:7970–5. [PubMed] [Google Scholar]
- 17.Stein M, Meyer HE, Hasilik A, von Figura K. Biol Chem Hoppe Seyler. 1987;368:927–36. doi: 10.1515/bchm3.1987.368.2.927. [DOI] [PubMed] [Google Scholar]
- 18.Oshima A, Nolan CM, Kyle JW, Grubb JH, Sly WS. J Biol Chem. 1988;263:2553–62. [PubMed] [Google Scholar]
- 19.Lobel P, Dahms NM, Breitmeyer J, Chirgwin JM, Kornfeld S. Proc Natl Acad Sci U S A. 1987;84:2233–7. doi: 10.1073/pnas.84.8.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahagian GG, Steer CJ. J Biol Chem. 1985;260:9838–42. [PubMed] [Google Scholar]
- 21.von Figura K, Gieselmann V, Hasilik A. Biochem J. 1985;225:543–7. doi: 10.1042/bj2250543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.York SJ, Arneson LS, Gregory WT, Dahms NM, Kornfeld S. J Biol Chem. 1999;274:1164–71. doi: 10.1074/jbc.274.2.1164. [DOI] [PubMed] [Google Scholar]
- 23.Morgan DO, Edman JC, Standring DN, Fried VA, Smith MC, Roth RA, Rutter WJ. Nature. 1987;329:301–7. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- 24.Dahms NM, Wick DA, Brzycki-Wessell MA. J Biol Chem. 1994;269:3802–9. [PubMed] [Google Scholar]
- 25.Tong PY, Gregory W, Kornfeld S. J Biol Chem. 1989;264:7962–9. [PubMed] [Google Scholar]
- 26.Tong PY, Tollefsen SE, Kornfeld S. J Biol Chem. 1988;263:2585–8. [PubMed] [Google Scholar]
- 27.Han VK, D’Ercole AJ, Lund PK. Science. 1987;236:193–7. doi: 10.1126/science.3563497. [DOI] [PubMed] [Google Scholar]
- 28.Kandror KV, Pilch PF. Biochem J. 1998;331(Pt 3):829–35. doi: 10.1042/bj3310829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka Y, Rozek LM, Czech MP. J Biol Chem. 1985;260:9435–42. [PubMed] [Google Scholar]
- 30.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Nature. 1994;372:464–7. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 31.Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, Stewart CL. Genes Dev. 1994;8:2953–63. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig T, Eggenschwiler J, Fisher P, D’Ercole AJ, Davenport ML, Efstratiadis A. Dev Biol. 1996;177:517–35. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- 33.Costello M, Baxter RC, Scott CD. J Clin Endocrinol Metab. 1999;84:611–7. doi: 10.1210/jcem.84.2.5488. [DOI] [PubMed] [Google Scholar]
- 34.Bobek G, Scott CD, Baxter RC. Endocrinology. 1992;130:3387–94. doi: 10.1210/endo.130.6.1317782. [DOI] [PubMed] [Google Scholar]
- 35.Scott CD, Weiss J. J Cell Physiol. 2000;182:62–8. doi: 10.1002/(SICI)1097-4652(200001)182:1<62::AID-JCP7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt B, Kiecke-Siemsen C, Waheed A, Braulke T, von Figura K. J Biol Chem. 1995;270:14975–82. doi: 10.1074/jbc.270.25.14975. [DOI] [PubMed] [Google Scholar]
- 37.Devi GR, Byrd JC, Slentz DH, MacDonald RG. Mol Endocrinol. 1998;12:1661–72. doi: 10.1210/mend.12.11.0192. [DOI] [PubMed] [Google Scholar]
- 38.Dahms NM, Rose PA, Molkentin JD, Zhang Y, Brzycki MA. J Biol Chem. 1993;268:5457–63. [PubMed] [Google Scholar]
- 39.Reddy ST, Chai W, Childs RA, Page JD, Feizi T, Dahms NM. J Biol Chem. 2004;279:38658–67. doi: 10.1074/jbc.M407474200. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald RG, Pfeffer SR, Coussens L, Tepper MA, Brocklebank CM, Mole JE, Anderson JK, Chen E, Czech MP, Ullrich A. Science. 1988;239:1134–7. doi: 10.1126/science.2964083. [DOI] [PubMed] [Google Scholar]
- 41.Kiess W, Thomas CL, Greenstein LA, Lee L, Sklar MM, Rechler MM, Sahagian GG, Nissley SP. J Biol Chem. 1989;264:4710–4. [PubMed] [Google Scholar]
- 42.De Leon DD, Terry C, Asmerom Y, Nissley P. Endocrinology. 1996;137:1851–9. doi: 10.1210/endo.137.5.8612524. [DOI] [PubMed] [Google Scholar]
- 43.Kiess W, Thomas CL, Sklar MM, Nissley SP. Eur J Biochem. 1990;190:71–7. doi: 10.1111/j.1432-1033.1990.tb15547.x. [DOI] [PubMed] [Google Scholar]
- 44.Kang JX, Li Y, Leaf A. Proc Natl Acad Sci U S A. 1997;94:13671–6. doi: 10.1073/pnas.94.25.13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang JX, Bell J, Leaf A, Beard RL, Chandraratna RA. Proc Natl Acad Sci U S A. 1998;95:13687–91. doi: 10.1073/pnas.95.23.13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Distler JJ, Guo JF, Jourdian GW, Srivastava OP, Hindsgaul O. J Biol Chem. 1991;266:21687–92. [PubMed] [Google Scholar]
- 47.Vidil C, Morère A, Garcia M, Barragan V, Hamdaoui B, Rochefort H, Montero JL. Eur J Org Chem. 1999:447–450. [Google Scholar]
- 48.Vidal S, Vidil C, Barragan V, Morère A, Garcia M, Montero JL. Eur J Org Chem. 2000:3433–3437. [Google Scholar]
- 49.Jeanjean A, Garcia M, Leydet A, Montero JL, Morère A. Bioorg Med Chem. 2006;14:3575–82. doi: 10.1016/j.bmc.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 50.Vidal S, Garcia M, Montero JL, Morère A. Bioorg Med Chem. 2002;10:4051–6. doi: 10.1016/s0968-0896(02)00264-x. [DOI] [PubMed] [Google Scholar]
- 51.Berkowitz DB, Maiti G, Charette BD, Dreis CD, MacDonald RG. Org Lett. 2004;6:4921–4. doi: 10.1021/ol0479444. [DOI] [PubMed] [Google Scholar]
- 52.Christensen MK, Meldal M, Bock K, Cordes H, Mouritsen S, Elsner H. J Chem Soc Perkin Trans I. 1994:1299–1310. [Google Scholar]
- 53.Franzyk H, Christensen MK, Jorgensen RM, Meldal M, Cordes H, Mouritsen S, Bock K. Bioorg Med Chem. 1997;5:21–40. doi: 10.1016/s0968-0896(96)00194-0. [DOI] [PubMed] [Google Scholar]
- 54.Roche AC, Midoux P, Bouchard P, Monsigny M. FEBS Lett. 1985;193:63–8. doi: 10.1016/0014-5793(85)80080-6. [DOI] [PubMed] [Google Scholar]
- 55.Roche AC, Midoux P, Pimpaneau V, Negre E, Mayer R, Monsigny M. Res Virol. 1990;141:243–9. doi: 10.1016/0923-2516(90)90028-h. [DOI] [PubMed] [Google Scholar]
- 56.Greupink R, Reker-Smit C, Proost JH, van Loenen Weemaes AM, de Hooge M, Poelstra K, Beljaars L. Biochem Pharmacol. 2007;73:1455–62. doi: 10.1016/j.bcp.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalo T, Talman EG, van de Ven A, Temming K, Greupink R, Beljaars L, Reker-Smit C, Meijer DK, Molema G, Poelstra K, Kok RJ. J Control Release. 2006;111:193–203. doi: 10.1016/j.jconrel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 58.Luk JM, Zhang QS, Lee NP, Wo JY, Leung PP, Liu LX, Hu MY, Cheung KF, Hui CK, Lau GK, Fan ST. Liver Int. 2007;27:548–57. doi: 10.1111/j.1478-3231.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- 59.Greupink R, Bakker HI, van Goor H, de Borst MH, Beljaars L, Poelstra K. Pharm Res. 2006;23:1827–34. doi: 10.1007/s11095-006-9025-2. [DOI] [PubMed] [Google Scholar]
- 60.Greupink R, Bakker HI, Reker-Smit C, van Loenen-Weemaes AM, Kok RJ, Meijer DK, Beljaars L, Poelstra K. J Hepatol. 2005;43:884–92. doi: 10.1016/j.jhep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Hagens WI, Mattos A, Greupink R, de Jager-Krikken A, Reker-Smit C, van Loenen-Weemaes A, Gouw AS, Poelstra K, Beljaars L. Pharm Res. 2007 doi: 10.1007/s11095-006-9175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beljaars L, Molema G, Weert B, Bonnema H, Olinga P, Groothuis GM, Meijer DK, Poelstra K. Hepatology. 1999;29:1486–93. doi: 10.1002/hep.510290526. [DOI] [PubMed] [Google Scholar]
- 63.O’Kane S, Ferguson MW. Int J Biochem Cell Biol. 1997;29:63–78. doi: 10.1016/s1357-2725(96)00120-3. [DOI] [PubMed] [Google Scholar]
- 64.Barragan V, Menger F, Caran KL, Vidil C, Morère A, Montero JL. Chem Commun. 2001:85–86. [Google Scholar]
- 65.Vidal S, Montero JL, Leydet A, Morère A. Phosphorus Sulfur. 2002;177:2363–2377. [Google Scholar]
- 66.Vidal S, Morère A, Montero JL. Heteroat Chem. 2003:241–246. [Google Scholar]
- 67.Hamdaoui B, Dewynter G, Capony F, Montero JL, Toiron C, Hnach M, Rochefort H. Bull Soc Chim Fr. 1994;131:854–864. [Google Scholar]
- 68.Olson LJ, Dahms NM, Kim JJ. J Biol Chem. 2004;279:34000–9. doi: 10.1074/jbc.M404588200. [DOI] [PubMed] [Google Scholar]
- 69.Olson LJ, Yammani RD, Dahms NM, Kim JJ. Embo J. 2004;23:2019–28. doi: 10.1038/sj.emboj.7600215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hancock MK, Haskins DJ, Sun G, Dahms NM. J Biol Chem. 2002;277:11255–64. doi: 10.1074/jbc.M109855200. [DOI] [PubMed] [Google Scholar]
- 71.Deduve C. Fed Proc. 1964;23:1045–9. [PubMed] [Google Scholar]
- 72.Zhu Y, Li X, Kyazike J, Zhou Q, Thurberg BL, Raben N, Mattaliano RJ, Cheng SH. J Biol Chem. 2004;279:50336–41. doi: 10.1074/jbc.M409676200. [DOI] [PubMed] [Google Scholar]
- 73.Gearing DP. Adv Immunol. 1993;53:31–58. doi: 10.1016/s0065-2776(08)60497-6. [DOI] [PubMed] [Google Scholar]
- 74.Metcalf D. Stem Cells. 2003;21:5–14. doi: 10.1634/stemcells.21-1-5. [DOI] [PubMed] [Google Scholar]
- 75.Blanchard F, Raher S, Duplomb L, Vusio P, Pitard V, Taupin JL, Moreau JF, Hoflack B, Minvielle S, Jacques Y, Godard A. J Biol Chem. 1998;273:20886–93. doi: 10.1074/jbc.273.33.20886. [DOI] [PubMed] [Google Scholar]
- 76.Blanchard F, Duplomb L, Raher S, Vusio P, Hoflack B, Jacques Y, Godard A. J Biol Chem. 1999;274:24685–93. doi: 10.1074/jbc.274.35.24685. [DOI] [PubMed] [Google Scholar]
- 77.Duplomb L, Chaigne-Delalande B, Vusio P, Raher S, Jacques Y, Godard A, Blanchard F. Endocrinology. 2003;144:5381–9. doi: 10.1210/en.2003-0607. [DOI] [PubMed] [Google Scholar]
- 78.de Lannoy LM, Danser AH, van Kats JP, Schoemaker RG, Saxena PR, Schalekamp MA. Hypertension. 1997;29:1240–51. doi: 10.1161/01.hyp.29.6.1240. [DOI] [PubMed] [Google Scholar]
- 79.Heller LJ, Opsahl JA, Wernsing SE, Saxena R, Katz SA. Am J Physiol. 1998;274:R849–56. doi: 10.1152/ajpregu.1998.274.3.R849. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen G, Delarue F, Berrou J, Rondeau E, Sraer JD. Kidney Int. 1996;50:1897–903. doi: 10.1038/ki.1996.511. [DOI] [PubMed] [Google Scholar]
- 81.Sealey JE, Catanzaro DF, Lavin TN, Gahnem F, Pitarresi T, Hu LF, Laragh JH. Am J Hypertens. 1996;9:491–502. doi: 10.1016/0895-7061(96)00092-1. [DOI] [PubMed] [Google Scholar]
- 82.Nguyen G, Delarue F, Burckle C, Bouzhir L, Giller T, Sraer JD. J Clin Invest. 2002;109:1417–27. doi: 10.1172/JCI14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Admiraal PJ, van Kesteren CA, Danser AH, Derkx FH, Sluiter W, Schalekamp MA. J Hypertens. 1999;17:621–9. doi: 10.1097/00004872-199917050-00005. [DOI] [PubMed] [Google Scholar]
- 84.van Kesteren CA, Danser AH, Derkx FH, Dekkers DH, Lamers JM, Saxena PR, Schalekamp MA. Hypertension. 1997;30:1389–96. doi: 10.1161/01.hyp.30.6.1389. [DOI] [PubMed] [Google Scholar]
- 85.Saris JJ, Derkx FH, De Bruin RJ, Dekkers DH, Lamers JM, Saxena PR, Schalekamp MA, Jan Danser AH. Am J Physiol Heart Circ Physiol. 2001;280:H1706–15. doi: 10.1152/ajpheart.2001.280.4.H1706. [DOI] [PubMed] [Google Scholar]
- 86.Saris JJ, van den Eijnden MM, Lamers JM, Saxena PR, Schalekamp MA, Danser AH. Hypertension. 2002;39:573–7. doi: 10.1161/hy0202.103002. [DOI] [PubMed] [Google Scholar]
- 87.Faust PL, Chirgwin JM, Kornfeld S. J Cell Biol. 1987;105:1947–55. doi: 10.1083/jcb.105.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Souza AT, Hankins GR, Washington MK, Fine RL, Orton TC, Jirtle RL. Oncogene. 1995;10:1725–9. [PubMed] [Google Scholar]
- 89.Hankins GR, De Souza AT, Bentley RC, Patel MR, Marks JR, Iglehart JD, Jirtle RL. Oncogene. 1996;12:2003–9. [PubMed] [Google Scholar]
- 90.Chappell SA, Walsh T, Walker RA, Shaw JA. Br J Cancer. 1997;76:1558–61. doi: 10.1038/bjc.1997.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oates AJ, Schumaker LM, Jenkins SB, Pearce AA, DaCosta SA, Arun B, Ellis MJ. Breast Cancer Res Treat. 1998;47:269–81. doi: 10.1023/a:1005959218524. [DOI] [PubMed] [Google Scholar]
- 92.Kiess W, Blickenstaff GD, Sklar MM, Thomas CL, Nissley SP, Sahagian GG. J Biol Chem. 1988;263:9339–44. [PubMed] [Google Scholar]
- 93.Motyka B, Korbutt G, Pinkoski MJ, Heibein JA, Caputo A, Hobman M, Barry M, Shostak I, Sawchuk T, Holmes CF, Gauldie J, Bleackley RC. Cell. 2000;103:491–500. doi: 10.1016/s0092-8674(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 94.Berthe ML, Esslimani Sahla M, Roger P, Gleizes M, Lemamy GJ, Brouillet JP, Rochefort H. Eur J Cancer. 2003;39:635–42. doi: 10.1016/s0959-8049(02)00627-5. [DOI] [PubMed] [Google Scholar]
- 95.Kleeberger L, Rottinger EM. Cancer Chemother Pharmacol. 1993;33:144–8. doi: 10.1007/BF00685332. [DOI] [PubMed] [Google Scholar]
- 96.Gerweck LE, Kozin SV, Stocks SJ. Br J Cancer. 1999;79:838–42. doi: 10.1038/sj.bjc.6690134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee ES, Na K, Bae YH. J Control Release. 2005;103:405–18. doi: 10.1016/j.jconrel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 98.Storm G, Roerdink FH, Steerenberg PA, de Jong WH, Crommelin DJ. Cancer Res. 1987;47:3366–72. [PubMed] [Google Scholar]
- 99.Baldini N, Scotlandi K, Barbanti-Brodano G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S, Campanacci M, et al. N Engl J Med. 1995;333:1380–5. doi: 10.1056/NEJM199511233332103. [DOI] [PubMed] [Google Scholar]
- 100.Purchio AF, Cooper JA, Brunner AM, Lioubin MN, Gentry LE, Kovacina KS, Roth RA, Marquardt H. J Biol Chem. 1988;263:14211–5. [PubMed] [Google Scholar]
- 101.Dennis PA, Rifkin DB. Proc Natl Acad Sci U S A. 1991;88:580–4. doi: 10.1073/pnas.88.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shah M, Foreman DM, Ferguson MW. Lancet. 1992;339:213–4. doi: 10.1016/0140-6736(92)90009-r. [DOI] [PubMed] [Google Scholar]
- 103.Shah M, Foreman DM, Ferguson MW. J Cell Sci. 1994;107(Pt 5):1137–57. doi: 10.1242/jcs.107.5.1137. [DOI] [PubMed] [Google Scholar]
- 104.Godar S, Horejsi V, Weidle UH, Binder BR, Hansmann C, Stockinger H. Eur J Immunol. 1999;29:1004–13. doi: 10.1002/(SICI)1521-4141(199903)29:03<1004::AID-IMMU1004>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 105.Leksa V, Godar S, Cebecauer M, Hilgert I, Breuss J, Weidle UH, Horejsi V, Binder BR, Stockinger H. J Biol Chem. 2002;277:40575–82. doi: 10.1074/jbc.M207979200. [DOI] [PubMed] [Google Scholar]
- 106.Behrendt N, Ploug M, Patthy L, Houen G, Blasi F, Dano K. J Biol Chem. 1991;266:7842–7. [PubMed] [Google Scholar]
- 107.Ronne E, Behrendt N, Ellis V, Ploug M, Dano K, Hoyer-Hansen G. FEBS Lett. 1991;288:233–6. doi: 10.1016/0014-5793(91)81042-7. [DOI] [PubMed] [Google Scholar]
- 108.Blasi F, Carmeliet P. Nat Rev Mol Cell Biol. 2002;3:932–43. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 109.Brunetti CR, Burke RL, Kornfeld S, Gregory W, Masiarz FR, Dingwell KS, Johnson DC. J Biol Chem. 1994;269:17067–74. [PubMed] [Google Scholar]
- 110.Brunetti CR, Burke RL, Hoflack B, Ludwig T, Dingwell KS, Johnson DC. J Virol. 1995;69:3517–28. doi: 10.1128/jvi.69.6.3517-3528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Veugelers K, Motyka B, Goping IS, Shostak I, Sawchuk T, Bleackley RC. Mol Biol Cell. 2006;17:623–33. doi: 10.1091/mbc.E05-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Linzer DI, Nathans D. Proc Natl Acad Sci U S A. 1984;81:4255–9. doi: 10.1073/pnas.81.14.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Linzer DI, Lee SJ, Ogren L, Talamantes F, Nathans D. Proc Natl Acad Sci U S A. 1985;82:4356–9. doi: 10.1073/pnas.82.13.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee SJ, Nathans D. J Biol Chem. 1988;263:3521–7. [PubMed] [Google Scholar]
- 115.Volpert O, Jackson D, Bouck N, Linzer DI. Endocrinology. 1996;137:3871–6. doi: 10.1210/endo.137.9.8756559. [DOI] [PubMed] [Google Scholar]
- 116.Folkman J. Semin Oncol. 2002;29:15–8. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 117.Jackson D, Volpert OV, Bouck N, Linzer DI. Science. 1994;266:1581–4. doi: 10.1126/science.7527157. [DOI] [PubMed] [Google Scholar]
- 118.Garcia M, Platet N, Liaudet E, Laurent V, Derocq D, Brouillet JP, Rochefort H. Stem Cells. 1996;14:642–50. doi: 10.1002/stem.140642. [DOI] [PubMed] [Google Scholar]