Abstract
Activation of the high-osmolarity glycerol (HOG) pathway for osmoregulation in the yeast Saccharomyces cerevisiae involves interaction of the adaptor Ste50p with the cytoplasmic tail of single-transmembrane protein Opy2p. We have determined the solution structure of the Ste50p-RA (Ras association) domain, and it shows an atypical RA fold lacking the β1 and β2 strands of the canonical motif. Although the core of the RA domain is fully functional in the pheromone response, an additional region is required for the HOG pathway activation. Two peptide motifs within the intrinsically disordered cytoplasmic tail of Opy2p defined by NMR spectroscopy physically interact with the Step50p-RA domain. These Opy2p-derived peptides bind overlapping regions of the Step50p-RA domain with similarly weak affinities, suggesting a multivalent interaction of these proteins as a crucial point of control of the HOG pathway. As well, overall selection of signaling pathways depends on functionally distinct regions of the Ste50p-RA domain, implicating this element in the control of global regulatory decisions.
INTRODUCTION
The ability to generate adaptive responses to various environmental stresses is crucial for the survival of an organism. High osmolarity represents one such stress and the development of the capability of osmosensing and osmoregulation is critical for organisms to detect and respond to this challenge. The yeast Saccharomyces cerevisiae adapts to extracellular hyperosmolarity stress by activating its high-osmolarity glycerol (HOG) response pathway to direct the accumulation of intracellular glycerol as a compatible osmolyte (Brewster et al., 1993; Hohmann, 2002). In the HOG pathway, two upstream osmosensing branches, containing either the Sln1p or the Sho1p membrane protein, share a redundant function in the activation of a common downstream mitogen-activated protein kinase (MAPK) cascade consisting of the Pbs2p MAPKK and the Hog1p MAPK (Posas and Saito, 1997; O'Rourke and Herskowitz, 1998; Posas et al., 1998b; Wu et al., 1999). The activation of the HOG pathway by the SLN1 branch requires the action of two functionally redundant MAPKKKs, Ssk2p and Ssk22p, and their activator Ssk1p; the SHO1 branch uses the Ste11p MAPKKK to activate the pathway.
Although the mechanisms by which the MAP kinase cascade transmits signals to activate the HOG pathway are generally well understood, detailed knowledge of the mechanisms by which the upstream branches sense and respond to osmotic stresses remains elusive. Using systematic genetic array (SGA) analysis to identify gene(s) whose deletion caused synthetic osmosensitivity when the SLN1 branch was absent, we established that Opy2p is an essential component in the Sho1p/Ste11p/Ste50p part of the HOG pathway. Opy2p, a 360-amino acid–long protein with a single transmembrane domain, interacts with Ste50p through its cytoplasmic C-terminal region (Wu et al., 2006). More recently, two membrane mucin-like molecules, Msb2p and Hkr1p, have been implicated as osmosensors that also function in the SHO1 branch of the HOG pathway (Tatebayashi et al., 2007). Genetically, Opy2p functions at or above the level of Ste50p/Ste11p at or downstream of Sho1p (Wu et al., 2006) and downstream of Msb2p and Hkr1p (Tatebayashi et al., 2007).
During activation of the HOG pathway, Opy2p serves as a plasma membrane attachment site for the adaptor protein Ste50p (Wu et al., 2006). In addition to the HOG pathway, Ste50p also plays a critical role in modulating at least two other of the five distinct MAP kinase pathways in S. cerevisiae: the pheromone response and pseudohyphal growth control pathways (Gustin et al., 1998; Posas et al., 1998a; Wu et al., 1999; Jansen et al., 2001; O'Rourke et al., 2002). The main function of Ste50p in the activation of each of these MAP kinase pathways is in plasma membrane localization of the Ste11p MAPKKK (Posas et al., 1998b; Wu et al., 1999, 2006; Jansen et al., 2001). The Ste50p adaptor achieves this membrane association through the interaction of its RA (Ras association) domain with Opy2p (Wu et al., 2006) or with other membrane-anchored proteins, such as the small GTPase Cdc42p (Tatebayashi et al., 2006; Truckses et al., 2006). In each case it appears this membrane localization positions Ste11p for its activation by the PAK kinase member Ste20p, itself membrane-associated through interaction with Cdc42p (Peter et al., 1996; Leberer et al., 1997; Lamson et al., 2002; Ash et al., 2003). The interactions of the Ste50p-RA domain with different binding partners thus play a critical role in directing the specificity of pathway responses.
RA domains were first identified as effectors of small GTPases of the Ras family and have been found to play pivotal roles in transmitting signals to control many cellular processes (Ponting and Benjamin, 1996; Herrmann, 2003). Structurally, these conserved RA domains belong to the ubiquitin fold superfamily (Kiel and Serrano, 2006). Increasing evidence shows that RA domains have a much wider spectrum of binding partners than just Ras-like small GTPases. Examples include the RA domain from the tumor suppressor Nore 1, which binds intramolecularly its C1 domain (Harjes et al., 2006); the Ras-binding domain (RBD) domain of plexin B1 receptor, which associates with Rho GTPases in an atypical way (Tong et al., 2007); and the RBD of Raf, which binds the SH2 domain of the Grb10 adaptor protein (Nantel et al., 1998). The Ste50p-RA domain has been previously shown to interact with the small GTPase Cdc42p, a member of Rho type of the Ras superfamily. This interaction is implicated as a critical part of the activation of the Ste11p/Ste7p/Kss1pMAP kinase cascade controlling filamentous growth (Truckses et al., 2006) The interaction of the Ste50p-RA domain with Opy2p activating the HOG pathway is likely to be qualitatively different from that seen for Cdc42p since, in contrast to the GTPase, the associating region of Opy2p is predicted to be largely unstructured.
Using NMR spectroscopy we have determined the solution structure of the Ste50p-RA domain and show that it has an atypical fold that replaces the first two β-strands essential for the stability and the functionality of the canonical RA domain with a more unstructured region. Therefore, this Ste50p-RA domain defines a new subfamily within the ubiquitin-fold superfamily. Deletion mutants that retain only the core of this atypical domain are capable of signaling through the pheromone response pathway, but are totally defective in signaling through the HOG pathway. Two genetically and structurally identified regions within the cytoplasmic tail of Opy2p are critical for the interaction with the Ste50p-RA domain. Within each of these regions we have identified short peptides that interact with overlapping regions of the Ste50p-RA domain, albeit with a relatively low affinities. We propose that Opy2p anchors the Ste50p/Ste11p module to the membrane through multivalent binding to Ste50p dimers/oligomers and acts together with the Sho1p adaptor to activate the HOG pathway. Because mutations of Ste50p-RA domain have been obtained that affect interaction with Opy2p and have no affect on the pheromone pathway, it is clear that the RA domain can have different modes of interaction with the effectors of these signaling pathways. The structure of the Ste50p-RA domain, along with the interaction modes with its partners, provides an example of the structural and functional versatility of the RA fold.
MATERIALS AND METHODS
Materials
Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs (Beverly, MA) and GE Healthcare (Baie d'Urfé, QC, Canada). High-fidelity Expand thermostable DNA polymerase and tablet protease inhibitors were purchased from Roche Molecular Biochemicals (Laval, QC, Canada). Acid-washed glass beads (450–600 μm), synthetic α-mating factor, protease inhibitors, and bovine serum albumin were purchased from Sigma (Oakville, ON, Canada). Geneticin was purchased from Invitrogen (Burlington, ON, Canada), and nourseothricin (clonNAT) from Werner BioAgents (Jena-Cospeda, Germany). Plasmids pGEX-4T-3 and pGEX-2TK, glutathione-Sepharose beads, glutathione, NiNTA resin, and protein A/G Sepharose beads were obtained from GE Healthcare. The antibody against glutathione S-transferase (GST) was described previously (Wu et al., 1999), and horseradish peroxidase–conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Nitrocellulose and FVDF membranes were purchased from Millipore (Bedford, MA). The enhanced chemiluminescence (ECL) assay system was purchased from Roche Molecular Biochemicals.
Yeast Strains and Manipulations
Yeast media, culture conditions, and manipulations of yeast strains were as described (Rose et al., 1990). Yeast transformations with circular or linearized plasmid DNA were carried out after treatment of yeast cells with lithium acetate (Rose et al., 1990; Gietz et al., 1992). The yeast GST-ORF library was purchased from Invitrogen (Carlsbad, CA), and the yeast deletion strain collection was purchased from ATCC (Manassas, VA). The yeast strains used in this study are listed in Supplemental Table S2.
Construction of Plasmids
Both the wild-type and various deletion mutant alleles of OPY2 were constructed with either conventional cloning method or by mutagenic sewing PCR (Ho et al., 1989) followed by in vivo recombination (IVR) in yeast. Site-directed mutagenesis of STE50 was performed with QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to manufacturer's protocol. Mutations were confirmed by sequencing. All constructs were in low-copy centromere plasmids, and the expression was under the control of OPY2 promoter. The Escherichia coli expression constructs used in this study generated either His-tagged proteins or GST fusion proteins. The His-tagged Ste50p or Opy2p constructs were made using a derivative of the pET15b vector to express N-terminally tagged Ste50p and Opy2p fragments of various lengths. The GST-tagged Ste50p and Opy2p constructs were created using the pGEX vector.
Yeast HOG, Pheromone Response, and Other Assays
Halo assays to test cell growth inhibition in response to α-mating factor, assays for the ability of cells to grow on hyperosmotic media to test the function of the HOG pathway, and yeast extract preparation and Western blot analyses were performed as described previously (Wu et al., 1999). Quantitative β-galactosidase reporter assays for the pheromone response and HOG pathways were performed as described (Wu et al., 1999; Tatebayashi et al., 2006).
Protein Expression and Purification for NMR Studies
Ste50p fragments of various lengths (aa 187-327, 237-327, 231-336, and 250-327) were cloned using a pGEX or pET15b vector and expressed in the BL21 or Rosetta pLys strains of E. coli. The cytoplasmic fragment of Opy2p (aa 194-333) was expressed fused with maltose-binding protein with a thrombin cleavage site and the SID fragment (aa 266-345) was expressed using the pGEX vector. The s-peptide of Opy2 was produced as GST fusion using the pGEX vector and the Gold Magic strain of E. coli.
All proteins were purified by affinity chromatography using NiNTA resin (GE Healthcare) using standard protocols. The recombinant peptide was further purified by HPLC (Vydac C18 semipreparative column, 10 × 250 mm), using a linear gradient of 0–80% acetonitrile in 0.1% trifluoroacetic acid. Protein concentrations were estimated by the Bradford method and the BCA (Pierce, Rockford, IL) method following the protocols supplied by the manufacturers. The s- and f-peptides were synthesized using standard solid-phase protocols and purified by HPLC.
NMR Spectroscopy
NMR samples for structural analysis were prepared in 50 mM phosphate buffer, pH 6.4, 200 mM NaCl, 5 mM DTT, and 0.02% (wt/vol) sodium azide at concentrations 1.2–1.5 mM. All NMR data were collected at 300K on Bruker Avance500 (Billerica, MA) and Varian INOVA 800 (Sunnyvale, CA) spectrometers equipped with triple-resonance cryoprobes and with z-gradient pulse field gradient accessories.
Sequence-specific backbone and aliphatic side chain chemical shift assignments for the RA domain were obtained using HNCA, CBCA(CO)NH, HNCACA, H(CCCO)NH, HBHA(CO)NH, and (H)CC(CO)NH experiments using standard protocols (Cavanagh et al., 2007). 1H chemical shifts for aromatic side chains were primarily based on 2D DQF COSY, TOCSY, and nuclear Overhauser effect spectroscopy (NOESY) experiments. For the Ste50p/Opy2p complexes the experiments were carried out with one partner 15N/13C-labeled and the second one unlabeled in 25–50% molar excess. 1H chemical shifts were referenced directly to internal DSS (2, 2-dimethyl-silapentane-5-sulfonate) at 0 ppm, and the 13C and 15N chemical shifts were referenced indirectly to DSS using the method of Wishart et al. (Wishart et al., 1995). NMR spectra were processed using XWINNMR (Bruker Biospin) and analyzed using NMR-Pipe (Delaglio et al., 1995), and CARA and XEASY (Bartels et al., 1995) programs.
Structure Calculations
The 3D 1H-15N NOESY-HSQC, 3D 1H-13C NOESY-HSQC (in D2O), and 2D homonuclear NOESY experiments were used to collect NOE-restraints for structure calculations. A mixing time of 100 ms was used in all experiments. Dihedral constrains ϕ and ψ torsion angles were derived from 13Cα, 13Cβ, 13C′, 1Hα, and 15N chemical shifts using TALOS (Cornilescu et al., 1999). The Ste50-RA domain structure was calculated with the CANDID/CYANA 2.1 (Herrmann et al., 2002) torsion angle dynamic simulation algorithm using 200 starting conformers. NOEs upper limit distances were used together with dihedral angles in the standard CYANA protocol of seven interactive cycles of calculations. Automatic NOE assignments from CYANA were manually verified and corrected as needed. Twenty final structures with lowest energy (least structure violations) were retained for refinement. Hydrogen bond constrains derived from 1H-15N HSQC exchange experiment were added at the last stage of calculations. The summary of results is given in Supplemental Table S1. The structure was refined using Xplor-NIH NMR (Schwieters et al., 2003, 2006).
Preparation of RA Domain/Peptide Complexes for NMR Studies
The titrations of the Ste50p-RA domain with Opy2p peptides were carried out by adding lyophilized unlabeled peptide directly to the 15N-labeled RA sample in the NMR buffer. The peptide was added in several steps to 0.5 mM Ste50p-RA domain up to a final molar ratio of peptide to RA of 1:1.5. Because in the 1H-15N HSQC spectra for the complex the signals were shifted gradually titration curves were used to reassign backbone resonances.
Bioinformatic Analysis of Ste50p-RA Domain and the FID Region of Opy2p
Using the full-length sequences of S. cerevisiae Opy2p and Ste50p, we assembled a set of 30 Opy2p-like protein sequences and a set of 28 Ste50p-like protein sequences from fungal species by performing PSI-Blast searches (Altschul et al., 1997) against the NCBI RefSeq database (Pruitt et al., 2007). Multiple sequence alignments within each set were derived with the L-INS-i iterative refinement method of the MAFFT 6 program (Katoh and Toh, 2008), and analyzed with Jalview 2.4 (Waterhouse et al., 2009).
RESULTS
The RA Domain of Ste50p Adopts an Atypical Structure
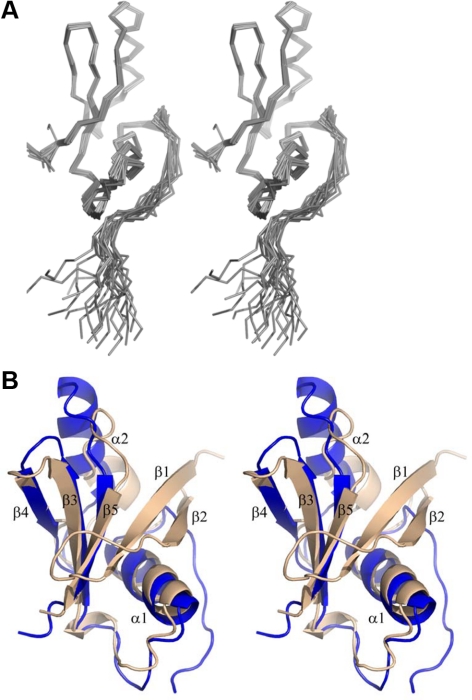
To provide a detailed description of the interaction between Ste50p and its binding partners, we first determined the structure of the Ste50p C-terminal region. This region contains a predicted domain (aa 235-327; Schultz et al., 1998; Kiel and Serrano, 2006; Letunic et al., 2009). This Ste50p-RA domain has been shown by in vitro binding assays to interact with specific Rho-type small GTPases rather than the classically expected Ras family members (Truckses et al., 2006; Annan et al., 2008). The initial construct corresponding to the predicted Ste50p-RA domain showed a well-dispersed 1H-15N HSQC spectrum indicative of a folded protein (data not shown), with the exception of the N-terminal region that was unfolded. Further analyses using heteronuclear NOE experiments confirmed this finding and showed that a shorter construct, Ste50p(250-327), behaved as folded. Thus, the N-terminal boundary of the structured Ste50p-RA domain was located near Asn250. With respect to the canonical RA fold this was unexpected, because the region 235-249 was predicted to contain the first strand of a β-sheet considered critical for the structural integrity of this fold. Together with the second β-strand of the canonical RA domain, it has been shown to form a highly stable hairpin that is important for folding of this domain, and which, from a functional perspective, is also important for the binding of effectors and regulators (Hilgenfeld,1995). To verify that the “missing strands” are not located in more distal regions of the protein, we analyzed a longer Ste50p fragment corresponding to aa187-346 was by NMR, and no additional folded elements were detected. A more detailed NMR study revealed that residues 250-261 corresponding to the expected second strand of the standard RA fold show evidence for interconverting heterogeneous states in the absence of a binding partner and are more mobile than the region encompassing aa 262-326 that forms a stable, well-folded core (Figure 1A). As predicted from the sequence alignment, the topology of the structured core corresponds to the canonical RA domain, excluding the first two β-strands. The central β-sheet of the Ste50p-RA domain thus has only three β-strands: β5↓-β3↑-β4↓ (with β2-β1 missing; Figure 1, A and B). Helix α1, which follows strand β2 in the canonical fold, and helix α2, located between strands β4 and β5, are present in the Ste50p-RA domain. Thus the Ste50p-RA domain indicates that the first two strands are not essential for the formation of a compact globular structure. The amino acid sequence corresponding to the folded core and the 250-261 region (second β-strand in the canonical RA domain) is highly conserved among Ste50p orthologues in fungi. By contrast, the region predicted as the first β-strand (235-249) of the canonical RA domain is highly variable (Supplemental Figure S1).
Figure 1.
The Ste50p-RA domain corresponding to aa 250-327 adopts an atypical RA domain fold. (A) The stereo view of the ensemble of 20 lowest-energy NMR structures of the Ste50p-RA domain. The first 10 residues were omitted from the figure for clarity. (B) Superposition of the Ste50p-RA domain (blue) with a canonical RA domain of RalGDS (tan; PDB code 1LXD).
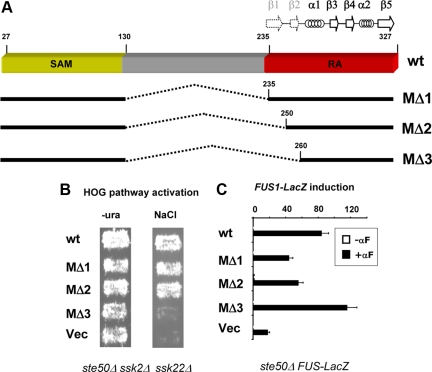
This somewhat atypical Ste50p-RA domain structure prompted us to probe the functional importance of the sequences corresponding to the canonical β1 and β2 strands. We took advantage of our previous finding that the N-terminal 26 residues and the central 131-218 residues of Ste50p are functionally dispensable (Wu et al., 1999). We constructed a series of in-frame internal deletion mutants that excluded increasing N-terminal regions of the predicted RA domain (Figure 2A). We found that Ste50p(27-130)::(235-327) and Ste50p(27-130)::(250-327) complemented Ste50p function in the activation of the HOG pathway, whereas Ste50p (27-130)::(260-327) did not. This showed that residues 235-249 (the predicted β1-strand) are not only structurally but also functionally dispensable, whereas residues 250-259 are critical for the function of Ste50p in this pathway (Figure 2B). Together, these experiments established the functional region of the Ste50p-RA domain in the HOG pathway as including residues 250-327. We have used this RA domain construct for further analysis of the interaction with Opy2p.
Figure 2.
Functional analysis of internal deletion mutants of Ste50p. (A) Schematic representation of internal deletion mutants of Ste50p. Dashed lines indicate deletions. The locations of β-strands and α-helices are indicated by arrows and coils. The dotted arrows mark the predicted but unobserved first two β-strands. (B) Yeast cells (ste50Δ ssk2Δ ssk22Δ) bearing different Ste50p mutants were assayed for their ability to grow on hyperosmolarity media. (C) Yeast cells (ste50Δ FUS1-LacZ) bearing different Ste50p mutants were assayed for their ability to activate the pheromone response pathway using a β-galactosidase reporter assay.
To investigate the importance of residues 250-259 in other pathways we interrogated the ability of Ste50p(27-130)::(260-327) to activate the pheromone response pathway. The construct retained fully functionality (Figure 2C), indicating that aa 250-259 are dispensable in this pathway, and thus the Ste50p-RA domain can utilize different interaction modes in different signaling pathways.
Two Functionally Important Regions of the Opy2 Cytoplasmic Tail
The protein Opy2p, an essential component of the SHO1 branch of the HOG pathway in the yeast S. cerevisiae, contains a single 93-115-aa transmembrane region. Sequence analysis predicted that the cytoplasmic tail of Opy2p is intrinsically unstructured (Supplemental Figure S2A). This prediction was confirmed by NMR spectroscopy of a recombinant 15N-enriched fragment of Opy2p encompassing amino acid residues 194-333, which showed a 1H-15N HSQC spectrum indicative of an unstructured polypeptide (Supplemental Figure S2B). We have previously shown that the region of the C-terminal tail of Opy2p consisting of amino acids residues 267-345 physically interacts with Ste50p, and we have termed this region SID (for Ste50p interacting domain; Wu et al., 2006).
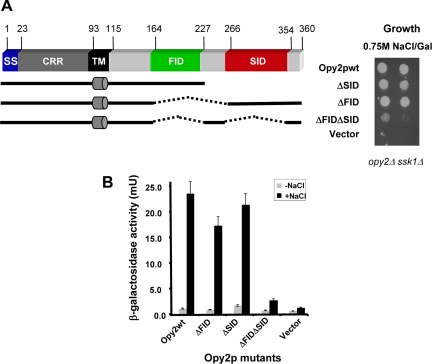
To further delineate region(s) important for Opy2p function in the activation of the HOG pathway, we generated a series of C-terminal deletions and assayed the ability of these proteins to complement Opy2p function in activating the HOG pathway in an opy2Δ strain. We found that Opy2p(1-227) was sufficient to complement the Opy2p function in the HOG pathway. This result indicates that the previously identified Ste50p interacting domain, SIDOpy2p (Wu et al., 2006) is not in itself absolutely essential for activation of the HOG pathway. A more extensive C-terminal deletion construct, Opy2p(1-164), was unable to complement the Opy2p function and thus a region corresponding to amino acid residues 164-227 constitutes another functionally important domain (FID) of Opy2p (Figure 3A). To further assess the contribution of either FIDOpy2p or SIDOpy2p to the function of Opy2p, we created internal deletions of each region separately and a deletion of both, and then assayed the ability of these deletion mutants to complement the function of Opy2p. Deletion of either FIDOpy2p or SIDOpy2p alone weakened but did not abolish the ability of these mutants to complement the function of Opy2p, as judged by the ability of cells lacking these regions to grow under hyperosmotic conditions (Figure 3A, right panel). Quantitative assays with a transcriptional reporter whose expression reflects Hog1p activity indicated that both mutants functioned somewhat less efficiently than the wild-type Opy2p. However, a mutant with both the FIDOpy2p and SIDOpy2p regions deleted had completely lost the ability to complement Opy2p function (Figure 3B). These results indicate that FIDOpy2p and SIDOpy2p are functionally partially redundant, with either one being sufficient for basic Opy2p function in the activation of the HOG pathway.
Figure 3.
Two functional regions of the Opy2p cytoplasmic tail. (A) Schematic representation of deletion constructs of the Opy2p cytoplasmic tail (left), assayed in yeast cells (opy2Δ ssk1Δ) by serial dilution spotting assay for their ability to grow on hyperosmolarity media (right). (B) The capacity of activating the HOG pathway by yeast cells (opy2Δ ssk1Δ) bearing different mutants assayed using a transcriptional reporter (8xCRE-CYC1-LacZ) assay.
The Ste50p-RA Domain Interacts with Peptides from Both SID and FID of Opy2p
The intrinsically unstructured nature of the cytoplasmic tail of Opy2p prompted us to consider that the Ste50p-RA domain may interact with short linear peptides within the FIDOpy2p and SIDOpy2p segments. To accurately delineate these critical regions we combined bioinformatics, NMR spectroscopy, genetic manipulations, and functional analysis.
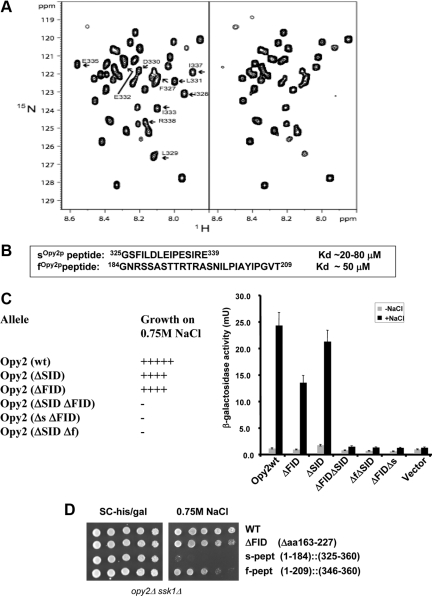
To determine the residues within the SIDOpy2p that are in physical contact with the Ste50p-RA domain, we expressed and purified the recombinant 15N-labeled SIDOpy2p fragment (aa 267-345). The HSQC spectra of this fragment titrated with the unlabeled Ste50p-RA domain revealed that only signals corresponding to residues 325-339 of SIDOpy2p showed significant changes in the presence of the Ste50p-RA domain (Figure 4A), suggesting that these residues contribute to the interaction. We termed this Ste50p-binding Opy2p peptide encompassing aa 325-339 the s-peptide (sOpy2p; Figure 4B).
Figure 4.
Two peptide motifs of Opy2p interact with the Ste50p-RA domain. (A) The HSQC spectra of 15N-labeled Opy2p SID (aa 267-345) in the absence (left) or presence (right) of the Ste50p-RA domain. Arrows indicate Opy2p residues involved in Ste50p-RA domain binding. (B) The affinity of synthesized s- and f-peptides for the Ste50p-RA domain. (C) Ability of yeast cells (opy2Δ ssk1Δ) transformed with OPY2 plasmids with indicated deletions to grow (+) or not (−) on hyperosmolarity media (left) and the capacity to activate a transcriptional reporter (8xCRE-CYC1-LacZ) for the HOG pathway (right). Δf (=Δaa185-209), Δs (=Δaa325-339). (D) The f-peptide but not the s-peptide is sufficient to complement the function of the Opy2p cytoplasmic tail. Yeast cells of opy2Δ ssk1Δ transformed with the OPY2 plasmids indicated were assayed for their ability to grow on hyperosmolarity media.
Next, we synthesized the 15-mer s-peptide of Opy2p to examine if it is on its own able to bind the Ste50p-RA domain. We analyzed the HSQC spectra of the 15N-labeled Ste50p-RA domain in the absence and presence of the unlabeled s-peptide. The changes in the HSQC spectra confirmed that the synthetic s-peptide interacts with the Ste50p-RA domain (Supplemental Figure S3). To quantify the strength of this interaction, we used fluorescence polarization and isothermal titration calorimetry (ITC) measurements and determined that the affinity (apparent Kd) is in the range of 20–80 μM (Figure 4B).
Because FIDOpy2p is also able to activate the HOG pathway in an Ste50p-dependent manner, we next examined if the FIDOpy2p segment also physically interacts with the Ste50p-RA domain. The sequence analysis of FIDOpy2p led to the identification of a highly conserved stretch of ∼25 amino acid residues within the FIDOpy2p region (Supplemental Figure S2C). A corresponding 26-mer peptide derived from S. cerevisiae Opy2p, 184GNRSSASTTRTRASNILPIAYIPGVT209, named the f-peptide (fOpy2p), was synthesized, purified, and tested for interaction with the 15N-labeled Ste50p RA domain. The HSQC spectra clearly showed that the synthetic f-peptide also binds the Ste50p-RA domain. ITC measurements indicated the apparent affinity of this interaction (Kd) to be ∼50 μM (Figure 4B). Intriguingly, the HSQC spectra of the 15N-labeled Ste50p-RA domain with the fOpy2p peptide indicated that similar regions of the Ste50p-RA domain are involved in the interaction with both the fOpy2p and sOpy2p peptides (see Figure 6, A and B). Taken together, these results suggest that the f-peptide and s-peptide bind to partially overlapping regions of Ste50p-RA domain, and each individual interaction has a low, but similar, affinity.
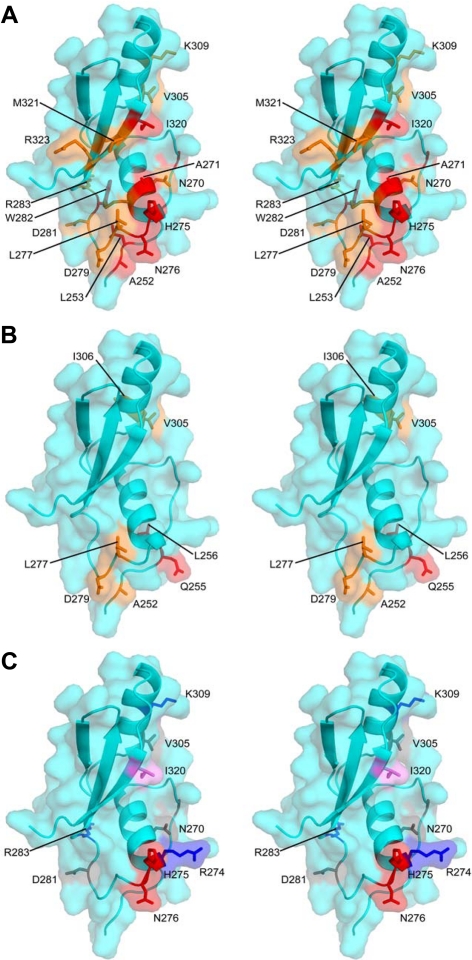
Figure 6.
Mapping Ste50p-RA domain surface residues for Opy2p interaction and their function in HOG pathway regulation described in Table 1 onto the structure of the Ste50p-RA domain. (A and B) Residues most affected in NMR experiments by binding of the sOPY2p and fOPY2p peptides, respectively. The most affected residues are in red, moderately affected in orange. (C) Residues mutated and functionally characterized as summarized in Table 1 are displayed as gray (mutation with no defect in the HOG pathway as compared with the wt Ste50p); red (mutations with defect in the HOG pathway signaling specifically through the Opy2p-FID (=ΔSID); blue (mutations with defect in the HOG pathway signaling specifically through the Opy2p-SID (=ΔFID); and violet (mutations defective in the HOG pathway signaling even with the wt Opy2p).
The s- and f-Peptides Are Key Functional Elements of SIDOpy2p and FIDOpy2p
To examine whether the sOpy2p and fOpy2p peptides are essential for the respective function of the SIDOpy2p and FIDOpy2p regions, we created a series of internal in-frame deletion mutant alleles of Opy2p and assessed their ability to complement the function of Opy2p to activate the HOG pathway under hyperosmotic stress. Like the Opy2p-ΔFID-ΔSID, which is unable to complement Opy2p, neither Opy2p-ΔFID-Δs nor Opy2p-Δf-ΔSID was able to complement the function of Opy2p required to survive hyperosmotic stress conditions (Figure 4C). These results indicated that deleting the peptide motifs has similar consequences to the deletion of their corresponding functional regions, suggesting that fOpy2p and sOpy2p peptides are essential for the function of the entire corresponding FIDOpy2p and SIDOpy2p regions, respectively.
Next, we addressed the question whether the fOpy2p and sOpy2p peptides are sufficient on their own to complement the Opy2p function in HOG pathway or whether there are other parts of the FIDOpy2p and SIDOpy2p segments that are also required for their function. We first created an Opy2p(1-184)::(346-360) mutant that was lacking both the FID and SID regions and was thus nonfunctional. We used this construct as a starting point for IVR to create Opy2p mutants carrying only the fOpy2p or sOpy2p peptide, designated Opy2p(1-209)::(346-360) or Opy2p(1-184)::(325-360), respectively. When these Opy2p alleles were assayed for their ability to activate the HOG pathway, we found that only the fOpy2p peptide was able to significantly activate the HOG pathway (Figure 4D). Although the fOpy2p peptide alone was functional, further analysis indicated that it functioned less efficiently than the FIDOpy2p or the wild type, as judged by the inability of mutant with only the fOpy2p peptide to survive elevated osmotic stress conditions under which the wild-type Opy2p and Opy2p-FID-ΔSID could still survive (data not shown). On the other hand, the sOpy2p peptide alone, in the context of this construct, was not sufficient to complement Opy2p (Figure 4D), indicating that some additional parts located between residues 227 and 325 within the SIDOpy2p also contribute to its function.
Both FIDOpy2p and SIDOpy2p Are Required for Full Response to Hyperosmotic Stress
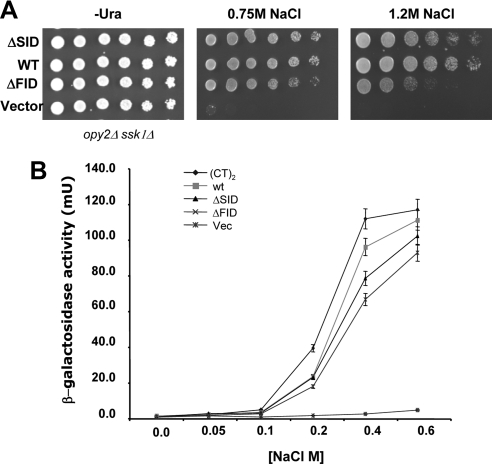
The FIDOpy2p and SIDOpy2p showed redundancy in the activation of the HOG pathway in qualitative assays such as cell growth on hyperosmotic media. However, semiquantitative assays of HOG pathway activity in terms of hyperosmotic-stress–dependent transcriptional activation showed significant differences between mutant Opy2p alleles and wild-type alleles. To analyze the Opy2p-SID (=ΔFID) and Opy2p-FID (=ΔSID) in a more quantitative manner, we performed spotting assays of the yeast strains bearing different alleles of Opy2p onto media with different levels of hyperosmolarity. Opy2p with deletion of either SIDOpy2p or FIDOpy2p survived 0.75 M NaCl almost as well as the wild type, but showed obvious differences in the ability to survive the more severe osmotic stress of 1.2 M NaCl (Figure 5A). Dose–response curves of HOG-pathway–dependent transcriptional activity as a function of salt concentration showed that the Opy2p mutant alleles have significantly lower slopes compared with that of the wild type (Figure 5B), indicating that the mutants are less responsive compared with the wild type. That is, Opy2p mutants require higher doses of stress/stimulation for a given amount of HOG pathway activity compared with the wild type. These data suggest that both FIDOpy2p and SIDOpy2p regions, interacting with the Ste50p-RA domain, are required for quantitatively proper signaling in the HOG pathway.
Figure 5.
Both the FID and SID regions of Opy2p are required for normal response to hyperosmolarity. (A) Yeast cells (opy2Δ ssk1Δ) transformed with OPY2 plasmids with indicated deletions were assayed for the ability to grow on the hyperosmolarity media indicated. (B) Dose–response curves of Opy2p alleles. Yeast cells as in A) bearing the Opy2p alleles indicated were assayed for transcriptional reporter (8xCRE-CYC1-LacZ) activity under indicated osmotic stress conditions for 1 h before the β-galactosidase activity was measured. (CT)2 indicates an Opy2p construct having a duplicated cytoplasmic tail (see text for details).
The interaction between the isolated Ste50p-RA domain and separate fOpy2p or sOpy2p peptides is of low affinity, with a Kd in the range of 20–80 μM. Such relatively weak binding suggested that avidity effect may play a functional role in HOG pathway activation. Observation of dimers/oligomers of Ste50p both in vivo (Slaughter et al., 2008) and in vitro (Ekiel and Wu, unpublished data) provided further support for this hypothesis. We therefore examined the effect of increased copy number of the Ste50p-RA domain–interacting segments of Opy2p (FID and SID) on the overall responsiveness to hyperosmotic stress. We created an Opy2p variant with a duplicated cytoplasmic tail (aa 163-360) of Opy2p with two copies of FID and SID each and measured the dosage response curves. In contrast to Opy2p with only one copy of either FID or SID, Opy2p with more copies of the Ste50p-RA domain–interacting segments showed a dose–response curve with steeper slope, or hypersensitivity to osmotic stresses, compared with that of the wild type (Figure 5B).
Mapping the Interface of the Ste50p-RA Domain Interacting with Opy2p
The NMR titration experiments of the Ste50p-RA domain with fOpy2p and sOpy2p peptides indicated that these two peptides bound to overlapping regions of the RA domain. A subset of residues of the Ste50p-RA domain that displayed chemical shifts in the presence of the sOpy2p were picked up for mutagenesis to confirm their functional importance as the Ste50p-RA domain–interacting interface with Opy2p. A total of 10 residues were mutated and assayed: S259A, N270A, R274A, H275A/N276A, D281A, R283A, V305T, K309A, and I320K. Because of the apparent functional redundancy of the FIDOpy2p and SIDOpy2p regions of Opy2p, the Ste50p mutants were assayed in combination with each of three Opy2p alleles: Opy2p-SID (= ΔFID), Opy2p-FID (=ΔSID), and wild type, in order to identify Ste50p mutants specifically defective in signaling in combination with particular Opy2p alleles.
The yeast strains (ste50Δ opy2Δ ssk1Δ) bearing all these combinations of Ste50p mutants and Opy2p alleles were assayed for their ability to activate the HOG pathway as indicated by their ability to grow on hyperosmolarity media. The results are summarized in Table 1. All the Ste50p-RA domain mutants, except for I320K, were able to survive the hyperosmotic insult when coupled with the wild-type Opy2p protein. In contrast, Ste50p R274A, K309A, and R283A mutants were found to be only defective in the HOG pathway signaling when combined with Opy2p-SID (=ΔFID), but were able to signal when Opy2p-FID (=ΔSID) was present, indicating that these Ste50p mutants are defective in signaling specifically through the SIDOpy2p. Similarly, the H275A/N276A double mutant was specifically defective in HOG pathway signaling through the FIDOpy2p (Table 1, Supplemental Figure S4A). These results suggest that the overlapping binding sites of the Ste50p-RA domain with FIDOpy2p and SIDOpy2p are genetically separable and that this approach can be used to further dissect the interaction between the Ste50p-RA domain with either FIDOpy2p or SIDOpy2p regions of Opy2p. The remaining mutants: S259A, N270A, D281A and V305T, showed no detectable defect with all Opy2p alleles tested (Table 1, Supplemental Figure S4A).
Table 1.
Ste50p mutants function differentially in HOG and pheromone response pathways
| STE50 alleles |
OPY2 alleles |
|||
|---|---|---|---|---|
| HOG pathway activity |
Pheromoneresponse | |||
| ΔFID (SID) | ΔSID (FID) | WT | WT | |
| Vector | − | − | − | + |
| WT | + | + | + | +++ |
| S259A | + | + | + | ND |
| R274A | − | + | + | +++ |
| H275A/N276A | + | − | + | +++ |
| N270A | + | + | + | ND |
| D281A | + | + | + | ND |
| R283A | − | + | + | +++ |
| V305T | + | + | + | ND |
| K309A | − | + | + | +++ |
| I320K | − | − | − | + |
| R274A/H275A/N276A | ND | ND | − | +++ |
Note that ste50Δ causes a severe defect in pheromone response, but is not completely sterile. Wild-type Ste50p is required for normal pheromone response (+++) as measured by halo assay and transcriptional reporter assay (see Supplemental Figure S4 for details). In the assay for HOG pathway activity, the ability to grow on hyperosmolarity media is indicated by positive sign (+) and inability to do so by negative sign (−). ND, not determined.
Because we identified two classes of Ste50p mutations, one defective in signaling through SIDOpy2p and other through FIDOpy2p, we reasoned that a Ste50p mutant combining these changes should fail to interact even with the wild-type Opy2p. Indeed, a strain carrying the triple Ste50p mutant R274A/H275A/N276A was severely defective in surviving hyperosmotic stress even in the presence of wild-type Opy2p (Table 1, Supplemental Figure S4, A and C). These residues are clustered at the beginning of the loop that follows helix α1, at the end of a long groove in the Ste50p-RA domain structure (Figure 6C). Although these residues appear to harbor two preferential interaction sites for distinct Opy2-peptide motifs, the close spatial proximity of these sites suggests the likelihood that Ste50p-RA domain interactions with these Opy2-peptide regions are mutually exclusive.
Because Ste50p is also required for proper signal transduction in the pheromone response pathway (Rad et al., 1992; Wu et al., 1999), we examined whether the Ste50p mutants defective in HOG pathway activation are also defective in pheromone response. They were subjected to both halo assays measuring pheromone-induced cell cycle arrest and reporter assays monitoring pheromone-induced gene expression (Wu et al., 1999). All the Ste50p mutants described above, except I320K, were fully able to elicit the pheromone response (Table 1, Supplemental Figure S4, B and C), indicating that these mutations have no adverse effect in the pheromone response pathway and that the Ste50p interactions regulating the pheromone pathway occur through a different interface than that with Opy2p. These results are consistent with the previous observation that Opy2p is not required in the pheromone response (Wu et al., 2006).
DISCUSSION
Ste50p is a key element of the MAP kinase pathways governing the hyperosmotic response, the pheromone response, and pseudohyphal growth in S. cerevisiae. A structural element within Ste50p that responds to the upstream signals of these various pathways is located in the C-terminal segment and has been predicted to have an RA domain fold. This element's interaction with several partners raises the question of the mode of their binding. We have determined the solution structure of the Ste50p-RA domain and characterized its interaction with Opy2p, a plasma-membrane–localized protein that is essential for the SHO1 branch of the HOG MAP kinase pathway. Further, we showed that mutations of the Ste50p-RA domain that block binding of Opy2p and shut down the HOG response have no effect on pheromone response, establishing that the Ste50p-RA domain interacts with at least some of its various partners through different surfaces.
The predicted Ste50p-RA domain extends between residues 235-327. However, the aa 235-249 corresponding to the β1-strand of the canonical RA domain lacks sequence conservation among fungal Ste50p orthologues, and none of the canonical RA domain typical sequence patterns can be found in this region (Kiel and Serrano, 2006). Further, our sequence analysis, deletion mutagenesis, and structural characterization of this domain showed that this “β1-strand” is functionally dispensable in both the HOG and pheromone pathways and is structurally not integral to the Ste50p-RA domain. Indeed, the Ste50p-RA domain adopts an atypical fold that lacks not only the β1- but also the β2-strand of the canonical fold and thus forms a separate subfamily within the ubiquitin fold superfamily. The peptide corresponding to the β2-strand is required for activation of the HOG pathway but shows more flexibility than the rest of the structure and is loosely associated with the core. Significantly, this β2-strand is dispensable for pheromone response, supporting its nonessentiality for the structural integrity of the Ste50p-RA domain.
In the canonical RA domain the strands β1 and β2 and helix α1 have been shown to mediate interactions with the effectors from the Ras family (Hilgenfeld, 1995; Wohlgemuth et al., 2005), The Ste50p-RA domain interacts with its partner(s) in a different way; indeed the mutants defective in Opy2p signaling are located mainly within α1, loop 3, α2, and β5 (Figure 6), but also include the “β2-strand” replacement region. The I320K mutation (in β5) showing signaling defects in both the pheromone response and the HOG pathway (Table 1, Supplemental Figure S4, A and B) may be involved in the interactions with different partners required for these two pathways. Alternatively, the observed defects in multiple signaling pathways may indicate a general role of I320 in maintaining the structural integrity of the RA domain. However, the first possibility is more likely because I320 is solvent-exposed and clearly involved in the interaction with Opy2p (Supplemental Figure S3). Further studies will be required to identify the partner involved in the pheromone response pathway and its mode of interaction with the Ste50p-RA domain. Our findings demonstrate that the Ste50p-RA domain interacts not only with Rho GTPases (Cdc42p) as previously shown (Truckses et al., 2006) but also with two regions within an intrinsically disordered C-terminal cytoplasmic tail of Opy2p. Thus it appears that RA domains have a binding capacity not limited to the Ras family members as implied by their name.
We have further identified the structural mode of the interaction of the Ste50p-RA domain with the cytoplasmic tail of Opy2p. Using genetic approaches we have mapped two regions in Opy2p that interact with the Ste50p-RA domain and thus provide the specific connection between Ste50p and the plasma membrane critical for the activation of Ste11p MAPKKK in the osmotic response. Analysis of the Opy2p by deletion mutagenesis defined two physically separate and functionally partially redundant regions essential for activating the HOG pathway; these regions have been designated FIDOpy2p and SIDOpy2p. Dose–response analysis of cells with Opy2p mutants bearing different numbers of Ste50p-RA domain–interacting regions revealed that both SIDOpy2p and FIDOpy2p were required for the normal response to hyperosmotic stress. Cells with an Opy2p with only one region, either SIDOpy2p or FIDOpy2p, were hyporesponsive to osmotic stress, as these mutants needed higher osmotic stress compared with the wild type to produce the same amount of reporter activity. On the other hand, constructs containing extra copies of SIDOpy2p and FIDOpy2p were more responsive to osmotic signals.
Within each of these two regions a short ∼15–25 amino acid long linear peptide was identified by NMR that interacts physically with the Ste50p-RA domain. The fOpy2p peptide is highly conserved in fungi, whereas the sOpy2p is more variable. Such a different conservation pattern for the two functionally redundant motifs may indicate that they have other functions in addition to interacting with the Ste50p-RA domain. The affinity of sOpy2p or fOpy2ppeptides toward the isolated Ste50p-RA domain is weak, with the Kd in the range of 20–80 μM. NMR analysis of 15N-labeled Ste50p-RA domain with either sOpy2p or fOpy2p peptide indicated that the peptides bind in the same general region of the RA domain. Nevertheless, their binding sites are functionally only partially overlapping because the R274ASte50-RA mutation affects only signaling through the SIDOpy2p, whereas the H275A/N276ASte50-RA mutant affects only signaling through FIDOpy2p. As expected, the combined triple mutant R274A/H275A/N276ASte50-RA is also defective in signaling through the wild-type Opy2p.
Such adjacent location of residues affecting f- and s-peptide binding to Ste50p-RA domain would argue against the possibility of a simultaneous binding of two distinct Opy2 interaction motifs to the same Ste50p-RA domain. On the other hand, the weak binding affinity of individual peptides and deleterious effects on HOG pathway activation by deleting either one of them strongly supports the notion that both peptides are involved in the interaction with Ste50p simultaneously. These conflicting concepts could be reconciled by taking into account that Ste50p forms dimers or higher oligomers (Slaughter et al., 2008). We propose a model in which the Opy2p tail interacts simultaneously with two/multiple Ste50-RA domains within the Ste50p oligomer. If this model is correct then an Opy2p with multiple SID and FID regions should show more sensitivity to osmotic stress than the wild type. Indeed, we have shown that Opy2p mutants with a duplicated cytoplasmic tail (containing two copies of SIDOpy2p and FIDOpy2p) require lower osmotic stress to produce the same amount of reporter activity than does the wild type. It is conceivable that the extent of multivalent interaction between Ste50p and Opy2p depends on hyperosmotic stress conditions, which cause significant shrinkage of cells, and thus increase the local concentration of Opy2p and Ste50p and may promote multivalent interactions.
Ste50p has also been shown to be involved in the activation of the pseudohyphal growth through the interaction of the Ste50p-RA domain with the Rho GTPase Cdc42p (Truckses et al., 2006). Several Ste50p-RA domain mutations affecting pseudohyphal growth have been previously identified, namely K254A, I267A/L268A, and R274A/N276A, which implicated these residues in binding Cdc42p (Truckses et al., 2006). Of these only R274 and N276 are involved in the Opy2p binding, thus indicating a potentially partial overlap of the Cdc42p and Opy2-binding sites over the loop 3 of the RA domain.
Overall, we have shown that the Ste50p-RA domain lacks the β1- and β2-strands of the canonical RA domain fold and that it is involved in interactions with components of various pathways through different mechanisms leading to differentiation of downstream effects. We have identified the region of Ste50p-RA domain surface that interacts with Opy2p and have shown that two short peptides of Opy2p convey the majority of the contacts. The weak binding of individual peptide motifs suggests that avidity effects play a crucial role in Opy2p-Ste50p interactions and that oligomerization of Ste50p plays a functional role in the cellular response to osmotic stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jason Baardsnes and Mr. Patrice Bouchard for performing SPR and fluorescence polarization analyses, and Dr. H. Saito (University of Tokyo, Tokyo, Japan) for plasmids. This work is partially supported by the Genomic Health Initiative (GHI) of National Research Council of Canada. The National Research Council of Canada publication number for this work is 50653.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0645) on October 21, 2009.
REFERENCES
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan R. B., Wu C., Waller D. D., Whiteway M., Thomas D. Y. Rho5p is involved in mediating the osmotic stress response in Saccharomyces cerevisiae, and its activity is regulated via Msi1p and Npr1p by phosphorylation and ubiquitination. Eukaryot. Cell. 2008;7:1441–1449. doi: 10.1128/EC.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash J., Wu C., Larocque R., Jamal M., Stevens W., Osborne M., Thomas D. Y., Whiteway M. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics. 2003;163:9–20. doi: 10.1093/genetics/163.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., X. T.-h., Billeter M., Güntert P., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules J. Biomol. NMR. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother W. J., Palmer A. G., III, Rance M., Skelton N. J. Protein NMR Spectroscopy: Principles and Practice. 2nd ed. San Diego, CA: Academic Press; 2007. [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes E., Harjes S., Wohlgemuth S., Muller K. H., Krieger E., Herrmann C., Bayer P. GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1. Structure. 2006;14:881–888. doi: 10.1016/j.str.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Herrmann C. Ras-effector interactions: after one decade. Curr. Opin. Struct. Biol. 2003;13:122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert P., Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. Regulatory GTPases. Curr. Opin. Struct. Biol. 1995;5:810–817. doi: 10.1016/0959-440x(95)80015-8. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Buhring F., Hollenberg C. P., Ramezani Rad M. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genom. 2001;265:102–117. doi: 10.1007/s004380000394. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kiel C., Serrano L. The ubiquitin domain superfold: structure-based sequence alignments and characterization of binding epitopes. J. Mol. Biol. 2006;355:821–844. doi: 10.1016/j.jmb.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Wu C., Leeuw T., Fourest-Lieuvin A., Segall J. E., Thomas D. Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. SMART6, recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A., Mohammad-Ali K., Sherk J., Posner B. I., Thomas D. Y. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J. Biol. Chem. 1998;273:10475–10484. doi: 10.1074/jbc.273.17.10475. [DOI] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I., O'Shea E. K. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman A. M., Park H. O., van Lohuizen M., Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Benjamin D. R. A novel family of Ras-binding domains. Trends Biochem. Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- Posas F., Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F., Takekawa M., Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1998a;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- Posas F., Witten E. A., Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1998b;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Maglott D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad M. R., Xu G., Hollenberg C. P. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol. Gen. Genet. 1992;236:145–154. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters C. D., Kuszewski J. J., Clore G. M. Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectrosc. 2006;48:47–62. [Google Scholar]
- Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Slaughter B. D., Huff J. M., Wiegraebe W., Schwartz J. W., Li R. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS ONE. 2008;3:e1931. doi: 10.1371/journal.pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K., Tanaka K., Yang H. Y., Yamamoto K., Matsushita Y., Tomida T., Imai M., Saito H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K., Yamamoto K., Tanaka K., Tomida T., Maruoka T., Kasukawa E., Saito H. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006;25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Chugha P., Hota P. K., Alviani R. S., Li M., Tempel W., Shen L., Park H. W., Buck M. Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain. J. Biol. Chem. 2007;282:37215–37224. doi: 10.1074/jbc.M703800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckses D. M., Bloomekatz J. E., Thorner J. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:912–928. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S., Bigham C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth S., Kiel C., Kramer A., Serrano L., Wittinghofer F., Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J. Mol. Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Wu C., Jansen G., Zhang J., Thomas D. Y., Whiteway M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006;20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Leberer E., Thomas D. Y., Whiteway M. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.