Abstract
The actin microstructure in dendritic spines is involved in synaptic plasticity. Inositol trisphosphate 3-kinase A (ITPKA) terminates Ins(1,4,5)P3 signals emanating from spines and also binds filamentous actin (F-actin) through its amino terminal region (amino acids 1-66, N66). Here we investigated how ITPKA, independent of its kinase activity, regulates dendritic spine F-actin microstructure. We show that the N66 region of the protein mediates F-actin bundling. An N66 fusion protein bundled F-actin in vitro, and the bundling involved N66 dimerization. By mutagenesis we identified a point mutation in a predicted helical region that eliminated both F-actin binding and bundling, rendering the enzyme cytosolic. A fusion protein containing a minimal helical region (amino acids 9-52, N9-52) bound F-actin in vitro and in cells, but had lower affinity. In hippocampal neurons, GFP-tagged N66 expression was highly polarized, with targeting of the enzyme predominantly to spines. By contrast, N9-52-GFP expression occurred in actin-rich structures in dendrites and growth cones. Expression of N66-GFP tripled the length of dendritic protrusions, induced longer dendritic spine necks, and induced polarized actin motility in time-lapse assays. These results suggest that, in addition to its ability to regulate intracellular Ca2+ via Ins(1,4,5)P3 metabolism, ITPKA regulates structural plasticity.
INTRODUCTION
Dendritic spines, the tiny knob-like protrusions that cover the dendrites of excitatory neurons, receive synaptic signals from the axon terminals of other neurons and participate in signal integration. Within spine heads, the second messenger Ins(1,4,5)P3 [phosphatidylinositol (1,4,5)-trisphosphate; IP3] is generated near the plasma membrane and diffuses to the endoplasmic reticulum to trigger intracellular Ca2+ release in the main dendrite (Berridge, 1998; Ross et al., 2005). The neuronal enzyme inositol trisphosphate 3-kinase A (ITPKA) is highly enriched in spines, where it selectively phosphorylates IP3 at the 3-hydroxyl position to produce Ins(1,3,4,5)P4 (IP4; Irvine et al., 2006). A number of studies implicate IPTKA as a regulator of synaptic plasticity, but how ITPKA regulates synaptic signals remains poorly understood (Irvine et al., 1999). ITPKA terminates IP3 signals that release intracellular Ca2+, and it also generates the messenger IP4, whose physiological role and precise mechanism(s) of action remain unclear (Irvine and Schell, 2001; Miller et al., 2008). Disruption of ITPKA in mice increases long-term potentiation (Jun et al., 1998), yet infusion of IP4 into hippocampal neurons via pipette enhances long-term potentiation by increasing Ca2+ entry (Szinyei et al., 1999). ITPKA expression in the hippocampus increases after spatial learning (Kim et al., 2004).
Spines also respond to synaptic stimuli by changing shape and size, a process controlled largely by the actin cytoskeleton (Bramham, 2008; Honkura et al., 2008). Independent of its enzymatic activity, ITPKA binds filamentous actin (F-actin), via a domain that resides in the 66 most N-terminal amino acids (N66; Schell et al., 2001). This targets ITPKA onto F-actin inside spines, positioning the enzyme to control local IP3 and IP4 lifetimes near the postsynapse. The catalytic activity of ITPK resides in the C-terminal half of the protein and is conserved in all isoforms and among all metazoans (Irvine and Schell, 2001). By contrast, the N-terminal F-actin interaction domain occurs only in birds and mammals, and its structure and function are much less understood (Irvine et al., 2006). We previously demonstrated that targeting ITPKs to F-actin enhances their ability to terminate IP3 signals, presumably by localizing the enzyme near sites of IP3 production (Lloyd-Burton et al., 2007). Furthermore, the localization of ITPKA and F-actin in spines is regulated by synaptic activity (Schell and Irvine, 2006).
In our earlier work, we had noted that expression of the ITPKA N-terminal F-actin–binding domain (N66) in cells can modify F-actin structure (Schell et al., 2001). Furthermore, a recent study using nonneuronal cells found that ITPKA expression modifies F-actin structure in a manner that is independent of enzymatic activity (Windhorst et al., 2008). Here, we tested the hypothesis that ITPKA not only binds F-actin, but also cross-links the filaments into bundles. These are filament superstructures in which the sides of the filaments are cross-linked closely together (reviewed in (Bartles, 2000). Using in vitro biochemical assays combined with fixed and live imaging of hippocampal neurons, we delineated the molecular interaction between the amino terminus of ITPKA and F-actin and then determined its role in targeting IPTKA in neurons. Our data suggest that the F-actin–binding domain of IPTKA induces bundles of F-actin in dendritic spines, causing an increase in the average length of dendritic protrusions. We also demonstrate that the full-length 66-amino acid F-actin–binding domain (N66), but not the minimal F-actin–binding region (N9-52), selectively targets the enzyme to dendrites. In mature synapses, F-actin bundled by the IPTKA amino terminus occurred preferentially near the motile tips of spine heads. We propose that, in addition to its ability to regulate IP3 Ca2+ signals, ITPKA also regulates structural plasticity in spines by modifying F-actin microstructure.
MATERIALS AND METHODS
Cell Culture
The rat glioma cell line C6 was grown in DMEM containing 10% fetal bovine serum, penicillin/streptomycin, and high glucose (Invitrogen 11965). C6 cells were transfected with Fugene HD (Roche, Indianapolis, IN), according to manufacturer's instructions. All procedures involving animals were carried out according to protocols approved by the USU Animal Use and Care Committee. Primary hippocampal mixed neuron-glial cultures were isolated from neonatal (postnatal day 0) rat pups, digested in papain, and maintained in serum-free Neurobasal A medium supplemented with B27 (Invitrogen, Carlsbad, CA). Neurons were transfected by the Ca2+ phosphate method at days 7–10 in vitro (DIV) and maintained for up to 5 wk, as previously described (Schell and Irvine, 2006).
F-Actin Binding and Bundling In Vitro
The coding regions for rat N66, N66(L34P), and N9-52 were cloned downstream of the 58-kDa solubility-enhancing bacterial protein NusA with hexahistidine in vector pET43A (Novagen, Madison, WI), as described previously (Schell et al., 2001). Compared with our previous study, the efficiency and quality of expression was improved by the use of the Overnight Express System (Novagen). Nevertheless, we were unable to obtain the fusion protein NusA-N9-52 in more than ∼50% full-length form, and this hindered our ability to reach saturating conditions in the binding assays (see Supplementary Figure S1 and Figure 5). Moreover, we were unable to express any soluble, full-length ITPTKA, even when fused to NusA. Fusion proteins were extracted in B-PER (Pierce, Rockford, IL) and purified using metal affinity chromatography on HisPur cobalt resin (Pierce). Buffer was exchanged, and protein was concentrated, by repeated concentration/dilution cycles (Ultra-15, Amicon, Beverly, MA) of eluted, pooled fractions using 5 mM Tris, pH 8.0; aliquots were snap-frozen and stored at −80°C. Rabbit skeletal muscle actin (Cytoskeleton, Denver, CO) was polymerized at a concentration of 121 μM. Binding assays were done using modifications of our previous methods (Schell et al., 2001). F-actin–binding experiments were done in actin-binding buffer (ABB; 20 mM Tris-HCl, 10 mM NaCl, 1 mM MgCl2, 1 mM ATP, and 1 mM dithiothreitol, pH 8.0) at an actin concentration of 10 μM, with NusA or NusA-N66 concentrations between 0.1 and 100 μM. After incubation for 30 min at room temperature, samples were centrifuged at 430,000 × g for 30 min at 4°C to pellet F-actin. Supernatants and pellets were dissolved in SDS-PAGE sample buffer, separated on 8% denaturing gels, stained with Coomassie blue, and dried. F-actin–bundling assays were done similar to binding assays, except that samples were centrifuged at only 7000 × g for 15 min to pellet F-actin bundles. Intensity of the Coomassie-stained bands was determined by scanning the dried gels followed by digital densitometry of the images using ImageJ (http://rsb.info.nih.gov/ij/; Wayne Rasband, NIH). Curve fitting and calculation of affinity were determined with Prism software (Graphpad, San Diego, CA). For binding, a single binding site equation (Y = Bmax * x/(Kd + x) produced the best curve fit; for bundling, a single-site Hill slope (Y = Bmax * X/Kdh + Xh), with a Hill coefficient (h) of 3 was used to produce the best-fit curve. For microscopic analysis of the bundles produced in vitro, F-actin was mixed with NusA-N66 or NusA to give a final concentration of 10 μM F-actin and 25 μM ligand in ABB and incubated 1 h at room temperature. Fluorescent phalloidin (Alexa 488; Molecular Probes, Eugene, OR) was then added to a final concentration of 8.3 nM and a drop of this mixture was placed between a glass slide and a coverslip and then viewed immediately using a 63× objective.
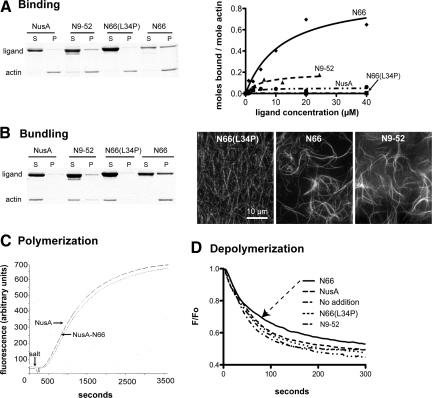
Figure 5.
Biochemical comparison of fusion proteins containing the full, truncated, and mutated ITPKA F-actin–binding domain. NusA fusion proteins were purified and binding (A) or bundling assays (B) were performed as described in Materials and Methods. (A) N9-52 binds F-actin, but N66(L34P) does not. Coomassie-stained gels (left) show soluble (S) and pelleted (P) fractions in representative F-actin–binding assays. Curves (right) show binding curves for NusA, N9-52, N66(L34P), and N66. N9-52 shows F-actin–binding activity, but we were not able to achieve saturation (see text and Supplementary Figure S1). (B) N9-52 bundles F-actin, but N66(L34P) does not. Gel (left) depicts representative results from F-actin–bundling assays. Micrographs depict F-actin filaments (10 μM) stained with phalloidin after the addition of 25 μM of various fusion proteins. (C) Effect of NusA or NusA-N66 on actin polymerization. Actin (5.8 μM) comprised of 18% pyrene-labeled monomers was polymerized by the addition of salt in the presence of 10 μM NusA or NusA-N66. Relative fluorescence indicates actin polymerization, and no differences in the rates were observed between NusA alone and NusA-N66. (D) Actin comprised of 70% pyrene-labeled monomers was polymerized for 1 h at a concentration of 1 μM and then mixed with buffer (no addition) or with 20 μM of various fusion proteins and allowed to incubate for 30 min to facilitate binding. Samples were then rapidly diluted 1:10 in 1× polymerization buffer to bring the actin concentration below the critical concentration, and the decrease in fluorescence was recorded. The presence of NusA, N9-52, or N66(L34P) had no effect on F-actin depolymerization, whereas N66 (arrow) showed a modest propensity to reduce the rate and extent of polymerization. Data are the mean of three determinations per condition after normalization of data to F0 = 1.
F-Actin Polymerization In Vitro
Lyophilized rabbit skeletal muscle actin and pyrene-labeled actin were each dissolved in general actin buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.5 mM dithiothreitol, and 0.2 mM ATP), and diluted to 0.45 mg/ml. After 1 h on ice, each was centrifuged for 1 h at 200,000 × g at 4°C to deplete residual actin polymers. Supernatants were removed to fresh tubes and stored on ice. Polymerization assays were performed with 5.8 μM actin in a volume of 750 μl in quartz cuvettes at room temperature in the presence of 10 μM bacterially expressed NusA or NusA-N66, purified as described above. The actin used was 18% pyrene-actin labeled. Assays were initiated by the addition of a polymerization buffer stock to produce a final concentration of 12.5 mM KCl, 0.5 mM MgCl2, and 0.25 mM ATP. Fluorescence measurements (excitation 365 nm, emission 407 nm; 2.5-nm slit widths) were collected every 10 s in a Perkin Elmer-Cetus fluorescence spectrophotometer (Norwalk, CT). Because our polymerization assays were not “seeded” with actin polymers, they exhibited a lag phase and proceeded for ∼1 h before approaching steady-state levels of polymerization. In preliminary experiments, we determined that the rate of actin polymerization was identical in the absence or presence of 10 μM NusA protein, indicating that NusA itself does not affect polymerization.
F-Actin Depolymerization In Vitro
F-actin (1 μM) composed of 70% pyrene-labeled monomers was allowed to polymerize in 1× polymerization buffer for 1 h at room temperature. Aliquots of F-actin were then brought to 20 μM final concentration of various fusion proteins [NusA, NusA-N66, NusA-N66(L34P), and NusA-9-52] and incubated for at least 30 min. Depolymerization assays were initiated by rapid 1:10 dilutions of F-actin/fusion protein mixtures into 1× polymerization buffer, bringing the final F-actin concentration below the critical concentration (0.1 μM) and the final fusion protein concentration to 2 μM. Fluorescence readings were collected every 2 s for 5 min. Data shown in Figure 5 depict the mean of triplicate determinations normalized to initial fluorescence values (F0 = 1.0).
N66 Pulldown Assay
NusA-N66 and NusA were each expressed in bacteria and purified as described above. To immobilize NusA-N66 to beads, buffer was exchanged into 0.2 mM NaHCO3, pH 8.9, 0.5 mM NaCl by repeated concentration/dilution through a centrifugal filter device (Amicon, Ultra-15). Purified protein was coupled overnight at 4°C to CNBr-activated Sepharose 4 fast flow beads (GE Life Sciences, Piscataway, NJ), at a coupling concentration of 5 mg/ml mixed with an equal volume of packed, activated beads. After extensive washing and capping of remaining reactive groups with Tris/ethanolamine in TBS, beads were stored in Tris-buffered saline at 4°C. Pulldown assays were performed in microcentrifuge tubes in a volume of 100 μl. Each assay consisted of 50 μl NusA-N66 bead slurry and a final concentration of 50 μM purified, soluble NusA or NusA-N66. After incubation for 2 h at room temperature, beads were washed four times at 4°C in 1 ml ABB, followed by elution of bound protein with 6 M urea in TBS. Samples were analyzed on 8% SDS-PAGE gels. The presence of urea in the eluted samples caused the eluted proteins to migrate slightly slower through the gel, producing a slightly larger apparent molecular weight compared with the starting protein in Tris buffer.
Molecular Biology
All inserts for cloning were produced using PCR-based cloning with pfu Turbo polymerase (Stratagene, La Jolla, CA) in the presence of 10% DMSO. Site-directed mutagenesis and deletion was performed with the QuikChange method (Stratagene), as previously described (Lloyd-Burton et al., 2007). All bacterial expression constructs were designed using the strategy described in Schell et al. (2001). For the truncation analysis, unique restriction sites were designed for directional cloning at the HindIII/BamHI sites of vector pEGFPN1 (Clontech, Palo Alto, CA), in which the sequence for enhanced green fluorescent protein (EGFP) contained the A206K mutation to render it monomeric (pmEGFP; Zacharias et al., 2002). Thus, all mEGFP fusions of the ITPKA actin-binding domain incorporated a linker sequence of DPPVAT located between the cloned insert and mEGFP. To delete the coding region for amino acids 53–66 in full-length ITPKA-mEGFP, we used the following primer pair: sense: 5′-GCC GCA GCG GCC GCA CCT AAC GGG CTC CCG-3′; and antisense: 5′-CGG GAG CCC GTT AGG TGC GGC CGC TGC GGC-3′. To create the untagged, full-length ITPKA expression construct, the full-length sequence for rat ITPKA was cloned with its stop codon intact into vector pCMVTag3A, such that the open reading frame terminated after the last IPTKA codon, but before the FLAG tag sequence. Tandem tomato (tdTomato) cDNA was the gift of Dr. Roger Tsien (UCSF). The open reading frame of tdTomato was amplified by PCR and cloned into vector pCMVTag2A (Stratagene) at the XhoI/ApaI sites to create the mammalian expression vector used as a cytosolic fill protein in neuronal cotransfection experiments. All clones used in the study were confirmed by DNA digest, the presence of cellular fluorescence (when applicable), and sequencing. Sequence alignments were created using the default settings of ClustalX for the MAC OS (http://www.embl.de/∼chenna/clustal/darwin/; Chenna Ramu, EMBL) and presented as shaded alignments using McBoxshade (Michael Baron, BBSRC). Secondary structure prediction (see Figure 2) was based on the consensus prediction of the JPred3 server (http://www.compbio.dundee.ac.uk/www-jpred/; Barton Group, University of Dundee) (Cole et al., 2008).
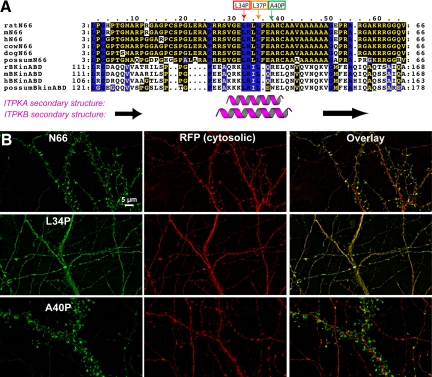
Figure 2.
The L34P point mutation within the ITPKA F-actin–binding domain destroys its localization to dendritic spines. (A) Alignment of F-actin–binding domains of ITPKA (top six sequences) and ITPKB (bottom four sequences) from various mammals. Residues shaded in yellow with a black background are identical; black residues with a blue background are conserved among all sequences shown. The cartoons under the alignment depict the consensus of a protein secondary structure prediction program (JPred3). The purple helices and black arrows demarcate predicted α-helices and β-strands, respectively. The small boxes and arrows above the alignment indicate three residues within the predicted, conserved helix. These residues were each singly mutated to proline to test for their effects on F-actin localization in cells. (B) Effects of point mutations on localization of N66 in hippocampal neurons. Plasmids coding for N66, N66(L34P), or N66 (A40P) fused at their C-terminus to mEGFP were each were cotransfected with the soluble marker tdTomato into primary hippocampal neurons at 8 DIV and fixed at 12 DIV. Nonmutated N66 is highly concentrated in F-actin–rich dendritic spines (top). The L34P mutation rendered the GFP fluorescence coincident with the soluble marker (middle). By contrast, the A40P mutation (bottom) had no obvious effect on the targeting, as GFP fluorescence remained highly associated with dendritic spines, similar to the unmutated N66 region. The L37P mutation produced a phenotype intermediate between the two other mutants (not shown). Scale bar, 5 μm.
Immunocytochemistry and Fluorescence Microscopy
Cells grown on coverslips were washed twice in warm PBS containing Ca2+ and Mg2+ and then fixed in 37°C 4% formaldehyde in 0.1 M sodium phosphate, pH 7.4, for 15 min. After permeabilization in 0.1% TX-100 for 10 min, cells were blocked and stained by indirect immunofluorescence, as previously described (Schell and Irvine, 2006). Secondary antibodies were species-appropriate Alexa 488, 568, or 647–labeled whole IgG (Invitrogen). In some cases, cells were labeled with fluorophore coupled to phalloidin during the incubation with secondary antibody. Coverslips were mounted onto slides in Prolong gold antifade plus DAPI (Invitrogen) and air-dried for 48 h. Images were collected with a Zeiss Axoivert 200M wide-field microscope (Thornwood, NY) equipped with a CCD camera (Orca ER; Hamamatsu, Bridgewater, NJ) and 63× or 100× NA 1.4 objectives. In some experiments, live cells were extracted at room temperature in BRB cytoskeleton stabilization buffer (80 mM Pipes/KOH, pH 6.8, 4% polyethylene glycol 8000, 1 mM MgCl2, and 1 mM EGTA), containing 1% TX-100 for 5 min before fixation, as previously described (Schell et al., 2001). We also tried an alternative extraction condition using 1% TX-100 in PEM-GA buffer (100 mM Pipes, pH 6.9, 1 mM MgCl2, 1 mM EGTA, 4.2% sucrose, and 0.05% glutaraldehyde). For semiquantitative analysis of resistance to extraction with 1% TX-100, transfected C6 cells on pairs of coverslips were washed with BRB buffer with or without 1% TX-100 for 5 min and then fixed. The number of transfected cells were counted inside a 2.8-mm2 region, and then this value was multiplied by 16 to estimate the number of transfected cells per 18-mm round coverslip. For neurons, all transfected cells on a coverslip were counted. Percent resistant cells is expressed as the number of cells on the extracted coverslip divided by the number on the control coverslip. Data for the extraction experiments are shown in Supplementary Figure S2.
Quantification of Neuronal Protrusion Length and Density
Hippocampal neurons were cultured for 8 d and then cotransfected with soluble tdTomato plus mEGFP alone or mEGFP fused to N66, N66 (L34P), or N9-52. At 14 DIV, cells were fixed and double-stained to enhance fluorescence signals by labeling with antibodies to red fluorescent protein (rabbit polyclonal anti-RFP, red channel; Rockland, Gilbertsville, PA) or GFP (monoclonal 3E6, green channel; Invitrogen). For the comparison of protrusion length and density, the investigator (H.W.J.) was blinded to the cotransfection condition, and only the red channel (soluble protein) was used for protrusion measurements so as not to bias the assay toward F-actin–bound fluorescence. For each condition, three-image Z-stacks covering a distance of 3 μm were collected from the first major branch off of the apical dendrite. Data were collected from at least six different neurons per condition. Stacks were exported to ImageJ where they were flattened as maximal projections and then analyzed using the NeuronJ plug-in (http://www.imagescience.org/meijering/software/neuronj/) (Meijering et al., 2004). For each neuron, the number of protrusions measured varied between 30 and 74, so that the protrusion length measurements for each condition were compiled from between 180 and 750 protrusions. Statistical testing was by one-way ANOVA and Bartlett's statistic.
Deconvolution Confocal Microscopy
To generate the high-resolution images of growth cones depicted in Figure 9, we used a microscopy technique that we described previously to improve signal-to-noise ratio and resolution (Schell and Irvine, 2006). Briefly, hippocampal neurons cotransfected on 8 DIV with the tdTomao red fluorescent protein (RFP) to mark the cytosolic compartment, along with various GFP-tagged versions of the ITPKA amino terminal region. On 12 DIV, cells were fixed and immunostained for RFP (red channel) and GFP (green channel) to enhance the signals. Cells were also labeled for F-actin using Alexa 647-phalloidin (Molecular Probes). Stacks of images consisting of 25–30 slices through 5–7 μm in the Z-plane were collected on a Zeiss LSM 710 inverted confocal microscope using a 100× 1.4 NA oil objective and an optical zoom setting of 2.6. The pinhole was set to 1 Airy unit for the Alexa 647 channel, which produced Airy unit values of 1.2 and 1.3 for the red and green channels, respectively. Stacks of images were subjected to 15 rounds of constrained reiterative deconvolution using the Autoquant blind three-dimensional (3D) algorithm (Media Cybernetics, Silver Spring, MD). Data are presented as maximal projections of the deconvolved Z-stacks.
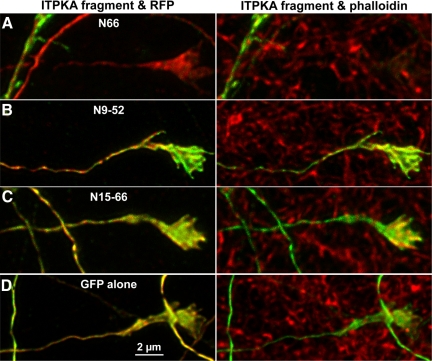
Figure 9.
N66 is largely excluded from axons and growth cones, whereas N9-52 occurs on F-actin–rich structures throughout the neuron. Hippocampal neurons were cotransfected at 8 DIV with various parts of the ITPKA F-actin–binding domain fused to GFP, and tdTomato (RFP) as a cytosolic marker. At 12 DIV, cells were fixed and triple-labeled for GFP (green), RFP (red), and phalloidin (far-red). Confocal stacks of growth cone images were and subjected to blind reiterative 3D deconvolution to enhance the signal-to-noise ratio. Images depicted are maximal projections of deconvolved Z-stacks. Left, overlays of green fluorescence with cytosolic RFP (red); right, overlays of GFP fluorescence and phalloidin labeling of F-actin (red). Note that phalloidin labels both transfected and untransfected cells. (A) N66 localizes mainly to F-actin–rich dendrites and is largely excluded from F-actin–rich growth cones. (B) By contrast, N9-52 localizes to F-actin—rich domains in growth cones and also identifies regions of axons which are more coincident with phalloidin (left) than with the cytosolic marker (right). (C) The N15-66 fragment is largely coincident with the cytosolic marker, as is D, the GFP alone negative control. Scale bar, 2 μm.
Time-Lapse Imaging and Creation of “Motility-Grams”
Hippocampal neurons grown on 18-mm round no. 1.5 coverslips (Warner Instruments, Hamden, CT) were transfected on DIV 7 or 8 and then grown until 18 DIV. Coverslips were placed into Ludin chambers (Life Imaging Services, Basel, Switzerland) in the open configuration. Cells were imaged in normal conditioned growth medium supplemented with 10 mM HEPES, pH 7.2. Ludin chambers with coverslips and medium were placed into a custom-fit adaptor and then into the Zeiss heating insert P, which was then sealed with a clear plastic lid to allow environmental control of humidity and CO2. Images were collected using a 63× 1.4 NA oil objective on a Zeiss Axiovert inverted microscope equipped with an Exfo X-cite 120 light source with liquid light guide and a Hamamatsu Orca ER CCD camera. The microscope was enclosed inside a plastic heating chamber to maintain a temperature of 37°C (Pecon, Erbach, Germany). Acquisition was controlled by Volocity software (Improvision, Lexington, MA), and typical time-lapse conditions used a neutral density filter (uvnd 1.3, 5% transmission; Chroma, Brattleboro, VT), 2× camera binning, and frame rates of three or six frames/min. Camera exposure times ranged between 50 and 250 ms. Under these conditions, one region of interest could typically be imaged for at least 90 min, and cells on each coverslip remained viable for at least 6 h. After acquisition, image stacks were exported to ImageJ (version 1.43f) and subjected to the delta F down time function from the McMaster Biophotonics plug-in bundle (Tony Collins, McMaster University). This plug-in subtracts successive frames in the series and stores the pixel values that have changed intensity values between frames. The difference stack was flattened as a projection of SD to create 2D images of motile regions such as those shown in the red channel in Figure 10. Total neuronal outlines (the green channel) were created by making averaged projections of the original image stacks, and these were merged with the motility images to create the final motility-grams. For final time-lapse movies, images were autocontrasted in ImageJ before exporting in the QuickTime MP4 format.
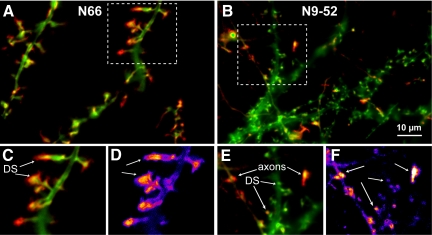
Figure 10.
N66-dependent F-actin bundling regulates the location of F-actin dynamics at synapses. Hippocampal neurons were transfected with GFP-tagged N66 or N9-52 on 8 DIV and imaged at 18 DIV. Time-lapse images of live cells were collected at a rate of three frames/min for 47 min (movies can be viewed as Supplementary Movies 4 and 5). The motility during the time-lapse experiment is depicted in red, representing a comparison of the SD of pixels between frames, and the green channel shows an averaged projection of fluorescence during the experiment. (A) Motility of the N66 F-actin–binding/bundling domain. Dynamic regions are polarized toward the spine tips and only rarely are motile regions observed in axons. (B) Motility of the N9-52 F-actin–binding domain. Motile regions occur in dendritic spines, similar to N66, but are much less pronounced. Instead, vivid N9-52-GFP dynamics could be observed in thin-caliber axons and in growth cones, demonstrating that the full-length amino terminal F-actin–binding domain (N66) is necessary to target the EGFP reporter selectively to spine heads. (C) Magnification of boxed region shown in A. Arrows point to tips of dendritic spines (DS), which the regions of highest N66-GFP motility. (D) The relative intensity of motility is indicated in pseudocolor, showing regions of high motility in hot colors and low motility in cool colors. (E) Detail of boxed region shown in B. Highly labile axonal regions of N9-52-GFP motility are indicated by the top set of arrows, whereas motility in dendritic spines (DS) is indicated by the bottom set. (F). Relative motility is shown in pseudocolor, as described in D. Scale bar, 10 μm (top panels).
Fluorescence Recovery after Photobleaching
C6 glioma cells grown on glass coverslips were transfected with ITPKA-mEGFP, ITPKA-mEGFP(delta 52-66), N66-mEGFP, or N9-52-mEGFP, N15-66-mEGFP, N66(L34P)-mEGFP, or mEGFP alone. Fluorescence recovery after photobleaching (FRAP) was performed 48 h after transfection at room temperature (∼22°C) in imaging buffer containing (in mM) NaCl, 145; KCl, 5; CaCl2, 3; MgSO4, 1; NaH2PO4, 1.2; glucose, 10; and HEPES, 20, pH 7.4. Coverslips were placed into Ludin Chambers (open configuration) and then mounted onto the stage of a Zeiss LSM710 confocal microscope. Because our aim was to estimate the relative binding affinity of various EGFP-tagged constructs for F-actin, we focused on relatively stable F-actin structures in cells (stress fibers; see Figure 6). To further increase F-actin stability, cells were pretreated with 5 μM jasplakinolide (Molecular Probes; diluted 1:1000 from a 5 mM DMSO stock) for 15 min before imaging, and the drug was present throughout the FRAP experiment. During the experiments, the pinhole was set to 3 Airy units, and the laser power was 0.2% during frame acquisitions. For constructs showing recovery rates significantly slower than a soluble protein (the four leftmost constructs depicted in Figure 6C), a small rectangular region of interest of approximate dimensions 0.5 × 0.2 μm located on a stress fiber was bleached by two iterations (pixel dwell time of 25 μs/pixel) of 100% power of the 458/488 laser lines. Data were collected at a 256 × 256-pixel resolution, at a frame rate of 6 Hz. The above method did not, however, produce sufficiently high frame rates to allow measurement of soluble proteins, which recover at diffusion-controlled rates (Sprague and McNally, 2005). Thus, for the four fusion proteins depicted on the rightmost part of Figure 6C, we set the microscope zoom to a 1.4 × 1.1-μm rectangle (46 × 34 pixels) before bleaching the same 0.5 × 0.2-μm region similar to the other FRAP experiments. This maneuver allowed us to obtain frame rates of 91 Hz (11 ms/frame). The recovery rate of one construct (N9-52-mEGFP) was compared using the two methods of acquisition (Figure 6B), and the kinetics obtained were not significantly different. Bleach recovery data were automatically normalized and then fit to a single exponential, using the FRAP analysis module of the Zeiss ZEN software. For each condition, data shown are the mean of nine independent trials of bleaching of nine different stress fibers, from three different cells per condition. For proteins appearing to be cytosolic, there were no labeled stress fibers, so we bleached an equivalent region outside of the nucleus. Statistical testing of differences in recovery half-times used an upaired Student's two-tailed t test (Graphpad Prism).
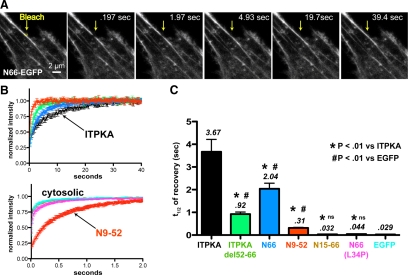
Figure 6.
FRAP reveals relative affinities for F-actin in cells. mEGFP fusions of ITPKA, ITPKA (residues 52-66 deleted), N66, N9-52, N15-66, N66(L34P), or mEGFP alone, were transfected into C6 glioma cells. Before the FRAP experiment, cells were treated with jasplakinolide for >15 min to stabilize F-actin. (A) Representative experiment depicting a peripheral region of one cell transfected with N66-mEGFP. The arrow points to a small region of interest (yellow box) located on a stress fiber; this was bleached and recovery was recorded over time. Scale bar, 2 μm. (B) Recovery was normalized to prebleach values and fit to an exponential recovery curve. Shown are the mean recovery for nine trials (±SEM). The top set of curves, with an x-axis spanning 40 s, depicts slower recovering constructs, which show substantial F-actin binding. The bottom set of curves, with an x-axis spanning 2 s, depicts three constructs recovering at or near diffusion-controlled (cytosolic) rates and the N9-52 fragment, which recovered at an intermediate rate, which was about ten times slower than cytosolic proteins. (C) Half-times of recovery for each EGFP construct tested. Data are the mean of nine determinations per condition (three cells per condition). *Statistically significant difference from full-length IPTKA and statistically significant difference from EGFP, the cytosolic control; #p > 0.01, Student's two-tailed t test.
RESULTS
In our preliminary experiments we observed that the expression of the N-terminus of ITPKA (N66) affected actin structure in a variety of cell types. When N66 was fused C-terminally to EGFP and expressed in neurons, glia, C6 cells, COS7 cells, and HeLa cells, it induced the formation of long protrusions and thick stress fibers, which stained intensely for the F-actin–binding toxin phalloidin. The formation of protrusions was not simply a consequence of the weak propensity of EGFP to dimerize, because the A206K mutation that renders EGFP monomeric (Zacharias et al., 2002) had no effect on the formation of protrusions (not shown). The protrusion-inducing effect occurred to various extents in different cell types, but appeared to be a universal property of N66 expression in cells. The extent of protrusions was positively correlated to the extent of overexpression, as judged by the intensity of green fluorescence. Because the morphology of the most intensely labeled F-actin structures resembled stress fibers and filopodia, which are composed of filament bundles, we hypothesized N66 could bundle or cross-link F-actin in cells.
N66 Binds and Bundles F-Actin In Vitro with Similar Affinity
To test biochemically for bundling activity, we expressed the putative bundling domain (N66) as a fusion protein in bacteria (see Supplementary Figure S1). In previous studies, we tested N66 F-actin binding in vitro by fusing it to a solubility-enhancing bacterial protein called NusA (Schell et al., 2001). However, our ability to perform in vitro studies was limited by the tendency of N66 fusions to remain in the inclusion body fraction of bacterial extracts and to express as a truncated protein. By adopting an improved bacterial expression system (see Materials and Methods), we achieved better yields of full-length fusion protein compared with our previous study. The improved expression made possible a more thorough in vitro analysis of the interactions between N66 and F-actin.
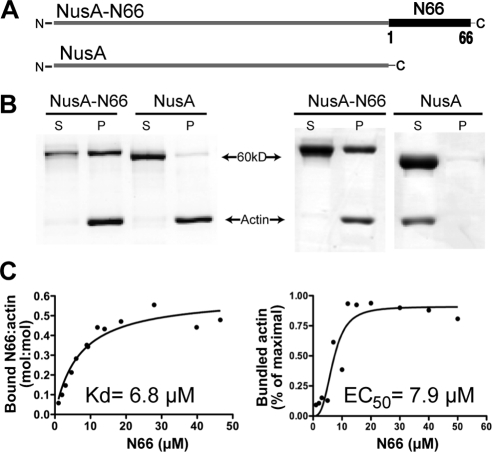
In centrifugation-based binding studies in which a putative F-actin–binding ligand is spun at 430,000 × g with purified skeletal muscle F-actin, we determined the affinity of N66 for F-actin to be 6.8 μM, with a binding curve that showed a best fit to a single binding site (Figure 1, B and C, left). Under saturation conditions, the stoichiometry of actin molecules to molecules of NusA-N66 was ∼2:1. The Kd value obtained (6.8 μM) is lower affinity than what we reported in a previous study (2.7 μM; Schell et al., 2001). In those earlier experiments, the bacterially expressed fusion protein was partly truncated, and a number of assumptions, corrections, and assay modifications were required to estimate the affinity. As the values obtained in the present study used full-length N66-NusA fusion protein and standard binding techniques, we consider the affinity and stoichiometry estimates obtained here to be more accurate. We should note, however, that the N66 region in our experiments has been fused at its N-terminus to the NusA protein, whereas in the physiological situation, the N-terminus is free, and lies upstream of the rest of ITPKA.
Figure 1.
Biochemical characterization of F-actin binding and bundling by the ITPKA N-terminus. (A) Schematic representation of the bacterially expressed fusion proteins. The 66-amino terminal resides of ITPKA (N66), which include the F-actin–binding domain, were fused to the solubility-enhancing protein NusA. The purified fusion proteins have a molecular weight of ∼60 kDa. (B) Coomassie-stained gels depicting results of F-actin binding (left) or bundling assays (right). For binding, fusion proteins were mixed with 10 μM F-actin and centrifuged at 430,000 × g. Bound protein and F-actin appear in the pellet (P), and unbound protein and G-actin (globular actin) appear in the supernatant (S). Gels depicted illustrate the result when the fusion protein concentration is 20 μm. For bundling, the mixture was spun at 7000 × g, which sediments filament bundles but not individual filaments. (C) Determinations of affinities for binding (left) and bundling (right).
We next tested F-actin bundling by modifying the binding assay to include a 7000 × g centrifugation step. During the lower-speed spin, individual F-actin filaments remain in the supernatant, whereas the denser bundles of F-actin appear in the pellet (Figure 1B, right). This assay provided clear evidence of F-actin bundling by N66 in vitro, with an EC50 for bundling of 7.9 μM—a value very similar to the Kd of N66 for F-actin binding (Figure 1C, right). In contrast to the binding curves, the bundling curves showed positive cooperativity (Hill constant of 3). These data indicate that N66 is sufficient to bundle F-actin in vitro and that the limiting step in bundling is likely to be F-actin binding.
A Point Mutation in the Predicted Helical Region of ITPKA Destroys F-Actin Binding in Cells
Actin bundling involves the cross-linking of filaments, and this can occur by at least three different mechanisms (Janmey, 2001). One mechanism involves the contribution of a third protein to mediate cross-linking, but our in vitro bundling assays using purified proteins (Figure 1, B and C) made this possibility unlikely. If an F-actin–binding protein possesses two distinct F-actin–binding sites, a single molecule can mediate cross-linking. Alternatively, two or more molecules of protein are required if the cross-linking involves the formation of homodimers, each possessing single F-actin–binding sites. In our first attempts to distinguish these possibilities we performed site-directed mutagenesis on the predicted alpha helix region of N66 (Figure 2). A previous report showed that the F-actin–binding region of the nonneuronal isoform ITPKB consisted of tandem (predicted) alpha helices, each capable of F-actin binding (Brehm et al., 2004). That study also suggested that there is weak sequence homology between the first F-actin–binding helix of ITPKB and the predicted helix in ITPKA. Figure 2A depicts an alignment of the F-actin–binding domains of ITPKA and the first F-actin–binding domain of ITPKB from various mammals. In the earlier report, two point mutations were identified in the actin-binding domain in the first predicted helix of human ITPKB (L139P and L143P), which disrupted F-actin binding (Brehm et al., 2004). We therefore made the analogous mutations in N66 (L34P and L37P), as well as in a nearby residue (A40P), which we also predicted to be part of the helix.
When the three different point mutations in N66 were expressed as EGFP fusion proteins in hippocampal neurons, they produced very different localizations (Figure 2B). The mutation L34P appeared to completely abolish all F-actin binding by N66. All colocalization with phalloidin was lost (not shown) and the GFP fluorescence instead colocalized with RFP, a cotransfected cytosolic marker (Figure 2B, middle row of images). Moreover, extraction of live neurons expressing the L34P mutant under conditions that preserve the binding of N66 to the cytoskeleton abolished cell-associated fluorescence of the N66(L34P)-GFP mutant (see Supplementary Figure S2). The N66(L37P) mutant also showed a greatly reduced colocalization with phalloidin; however, the morphological association with F-appeared partly retained (not shown). By contrast, the A40P mutant appeared to remain completely spine-associated and exhibited a localization that was indistinguishable from nonmutated N66 (Figure 2B, cf. top row and bottom row). These studies indicate that the ability of N66 to bind F-actin probably occurs via a single α-helix and that this binding appears to be completely eliminated by a helix-disrupting mutation at residue L34. These studies also support the likelihood of weakly conserved structural homology between the F-actin–binding domains of ITPKA and ITPKB, as previously suggested (Brehm et al., 2004).
The L34P Mutation Is Sufficient to Render Full-Length ITPKA Cytosolic
The experiments shown in Figure 2 used the N66 domain in isolation to test F-actin binding in neurons. Full-length IPTKA contains a number of structural elements that could conceivably contribute to its intense localization to dendritic spines in neurons. For example, the proline rich-region in the extreme amino terminus could bind SH3 domains within the synaptic scaffold, or a polybasic region located around amino acid 60 could participate in the interaction with the cytoskeleton. C-terminal to N66, the calmodulin binding domain and/or the putative protease substrate domain could regulate dendritic spine localization. Moreover, it is conceivable that the fusion of ITPKA to EGFP prevents protein–protein interactions through steric hindrance.
To test whether the ITPKA F-actin binding domain is necessary and sufficient for localizing the full-length untagged enzyme to spines, we cotransfected hippocampal neurons with soluble EGFP and the untagged full-length version of ITPKA or a corresponding version containing the L34P mutation (Figure 3). Cells were then fixed and ITPKA was visualized with a polyclonal antibody against rat brain ITPKA (Takazawa et al., 1990) and an mAb against GFP to enhance the signals. The ITPKA antibody was used at a high dilution (1:5000) such that it was only sensitive enough to detect overexpressed protein but not endogenous protein (which does not reach high levels until a week later in rat brain and in cell culture; Moon et al., 1989; Schell and Irvine, 2006). Although wild-type ITPKA was localized to spines and obviously different from the soluble EGFP marker (Figure 3A), the L34P mutant appeared identical to EGFP, except that it was too large to traverse the nuclear pore, unlike EGFP (Figure 3, D vs. E). Moreover, although about a third of the wild-type protein resisted extraction with TX-100 (similar to a GFP-tagged version; see Supplementary Figure S2), the L34P mutant appeared to be extracted completely because no antibody-stained neurons could be identified under this condition (not shown). The percent TX-100 resistance was consistently lower in neurons compared with C6 cells, perhaps reflecting a more dynamic actin pool in neurons (Supplementary Figure S2B).
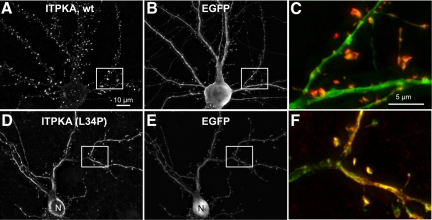
Figure 3.
The predicted helix in the F-actin–binding domain targets full-length ITPKA to spines. Full-length rat ITPKA or ITPKA(L34P) were coexpressed with the soluble marker EGFP in hippocampal neurons at 8 DIV and then fixed at 14 DIV and stained with a rabbit antibody made against rat brain ITPKA (left panels and red) or a mouse mAb against GFP (middle panels and green). Antibody to ITPKA was used at a high dilution so as to be below the detection limit for labeling endogenous ITPKA in the cultures at this age. (A) Full-length ITPKA is intensely localized to dendritic spines and is very different from the cytosolic marker EGFP shown in B. (C) Higher magnification overlay view of the boxed area illustrates the typically high concentration of ITPKA (red) in dendritic spines. (D) Full-length ITPKA containing the point mutation L34P appears cytosolic in neurons, as demonstrated by its similar distribution to EGFP shown in E. (F) Higher magnification view of the boxed area shows a high degree of coincidence (yellow) between EGFP and ITPKA(L34P), illustrating that ITPKA(L34P) is no more concentrated in spines than the cytosolic marker. Note that ITPKA(L34P), in D is excluded from the nucleus (N) because the full-length protein is too large (54 Kd) to traverse the nuclear pore, despite being cytosolic. In contrast, EGFP (28 kDa) enters the nucleus (cf. D and E, nucleus near bottom). Scale bar, 10 μm.
These results indicate that the predominant means by which ITPKA is localized to dendritic spines is through the ability of the predicted α-helical region in its N-terminus to bind F-actin–rich spines. Moreover, if other protein–protein interactions occur with ITPKA within neurons, they are insufficient to localize the enzymatic activity selectively to the dendritic spine compartment. Third, these studies demonstrate that full-length, untagged ITPKA (53 kDa) does not passively traverse the nuclear pore. This observation may be relevant to the highly active processes of IP3 metabolism that occur in the nucleus and suggests that the nuclear pool of IP3 is metabolized predominantly by other types of inositol polyphosphate kinases (Seeds et al., 2007). If ITPKA participates in nuclear IP3 metabolism, it would first have to undergo N-terminal proteolysis (Pattni and Banting, 2004).
Truncation Analysis of N66 Indicates the Minimal F-Actin–binding Domain to Be Residues 9-52
The observation that a single point mutation destroys F-actin localization in cells (Figure 2) supports the notion that F-actin bundling by N66 requires the association of two or more F-actin–binding domains located on different molecules of N66. To explore in more detail the minimal requirements of F-actin binding, we performed an extensive truncation analysis of N66 (Figure 4). We fused various fragments of N66 at their C-terminus with mEGFP, expressed them in the C6 glioma cell line, and costained for the F-actin marker phalloidin. Figure 4A depicts a graphic of the truncation scheme. We previously showed that neither residues 1-33 nor 33-66 possessed F-actin binding in cells (Schell et al., 2001). Likewise, residues 15-66 (Figure 4B, bottom left), 25-66 (Supplementary Movie 3), and 30-66 (not shown) appeared largely cytosolic in cells, as indicated by the lack of colocalization with phalloidin and by their ability to traverse the nuclear pore and label the nucleus. In the highest expressing cells, the 15-66 construct occasionally showed weak colocalization with phalloidin, as indicated by the arrows in Figure 4B (bottom panels). By contrast, residues 1-66 (Figure 4B, top) 9-66 (second row), 1-52 (not shown), and 9-52 (third row) appeared highly coincident with phalloidin (right panels). A weak nuclear fluorescence was sometimes observed for 9-52, indicating that a small fraction of this fragment is cytosolic. By these criteria, residues 9-52 comprise the minimal F-actin–binding domain in cells. In addition, the first two prolines (residues 4 and 7), which form a potential PxxP motif for binding SH3 domains, appear to be dispensable for F-actin binding.
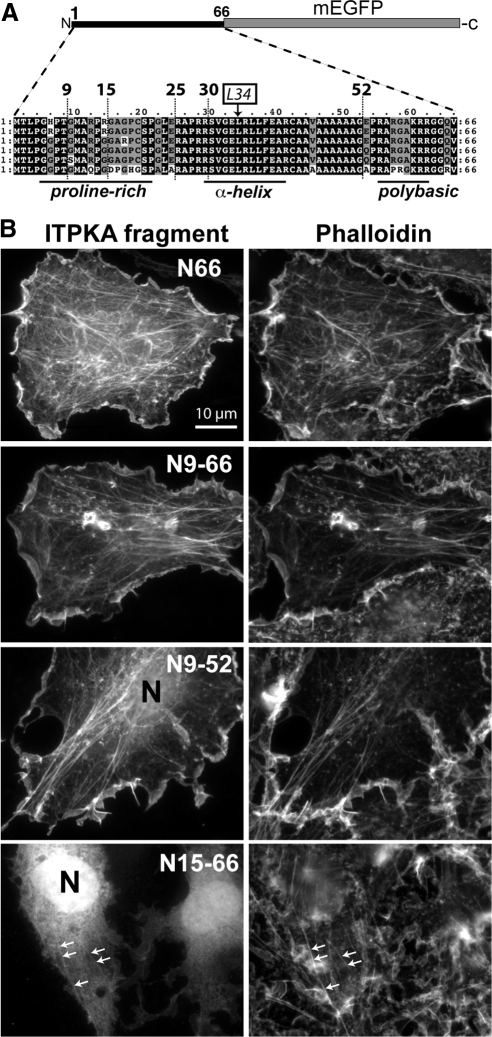
Figure 4.
Truncation analysis of ITPK F-actin–binding domain identifies the minimal F-actin–binding region. (A) Structural basis of the truncation strategy. Sequences from the N66 regions of ITPKA from mammals were aligned to identify regions of structural conservation, and then different stretches of amino acids were fused upstream at their C-termini to mEGFP for expression in cells. The ITPKA F-actin–binding domain consists of a proline-rich region, followed by a predicted α-helix, followed by a polybasic region. From top to bottom of the alignment, the species depicted are ITPKA from rat, mouse, human, cow, dog, and opossum. (B) When the fusion proteins were expressed in C6 cells (left) residues 1-66 (top row) 9-66 (second row), and 9-52 (third row) appeared coincident with staining for phalloidin (right). By contrast, residues 15-66 appeared cytosolic and entered the nucleus (N) freely (bottom row). Modest amounts of nuclear fluorescence (N) were sometimes observed for the 9-52 construct (third row), indicating that a minor fraction of this construct occurs in the cytosolic fraction in live cells at steady state. Some of the cells expressing the 15-66 construct showed a weak colocalization with phalloidin (bottom row, arrows), suggesting that a minor fraction of this construct occurs bound to F-actin in live cells at steady state. Scale bar, 10 μm. Supplementary Figure SS2A shows color overlays of the cells depicted here, along with results from detergent extraction experiments.
We also expressed the truncation series in C6 cells and neurons and extracted the live cells with 1% TX-100 under two different cytoskeleton-preserving conditions (Supplementary Figure S2). Consistent with their association with phalloidin in unextracted cells, GFP-tagged fragments 1-66 and 9-66 were more than 50% resistant to extraction (Supplementary Figure S2). Unexpectedly, two fragments that appeared cytosolic under physiological conditions (15-66 and 25-66; Supplementary Movie 1) became associated with F-actin during the live cell extraction with TX-100 (Supplementary Movie 2). The presence of the polybasic region (amino acids 52-66) was required to observe this apparent translocation—but its presence was not sufficient to mediate the interaction with F-actin by itself, because full-length N66 containing the helix-disrupting mutation L34P did not exhibit this behavior (Supplementary Figure S2). By contrast, construct N9-52, which comprised the minimal F-actin–binding domain by morphological criteria (colocalization with phalloidin Figure 4B, third row), was completely extracted from the cells under these conditions (Supplementary Figure S2), suggesting that its affinity for F-actin is lower than that of the full-length N66 domain.
Overall, our studies using live cell extraction as a measure of F-actin association suggest that the polybasic region (amino acids 52-66), although neither necessary nor sufficient to mediate an interaction with F-actin on its own, can participate in an interaction with F-actin if some of the helical character of the upstream domain is retained. By contrast, the minimal helical domain consisting of amino acids 9-52 is able to interact with F-actin by itself; however, its lower affinity for F-actin (compared with full-length N66) allows it to be extracted under cytoskeleton-preserving conditions, possibly because it becomes diluted to a concentration far below its affinity constant.
Amino Acids 9-52 Exhibit Reduced F-Actin Binding, Whereas the L34P Mutation Eliminates Binding
To provide biochemical evidence to complement the morphological data obtained in cell cultures, we performed a more thorough characterization of the interaction between F-actin and N66, N66(L34P), and N9-52 in vitro (Figure 5). These domains were fused at their N-termini to the NusA protein, expressed in bacteria, and purified as described in Materials and Methods. The four proteins tested are depicted diagrammatically in Supplementary Figure S1A, and their electrophoretic properties are shown in Supplementary Figure S1B. F-actin–binding assays demonstrated that, similar to N66, N9-52 could bind F-actin, but N66(L34P) and NusA could not (Figure 5A). A substantial portion of the N9-52 fusion protein (43%) expressed in bacteria as a C-terminally truncated protein, and only the full-length protein bound F-actin and thus appeared in the pellet after cosedimentation with F-actin (Supplementary Figure S1C). Although this supports our claim that N9-52 is a minimal F-actin–binding domain (which therefore loses its ability to bind F-actin when additional residues are lost), it also made it difficult to obtain sufficiently high concentrations of full-length N9-52 to reach saturation in binding assays, as shown in Figure 5A (right). The binding curves shown in Figure 5A show the estimated concentration of full-length N9-52 used in the assay after correction for 43% of it being truncated and thus unable to bind F-actin. We were unsuccessful using higher concentrations in the assay because this caused aggregation and precipitation of protein. Nevertheless, the data clearly indicate that the affinity of N9-52 for F-actin is considerably lower than N66, and this supports our conjecture based on live cell extractions. The data also indicate that the L34P mutation completely destroys F-actin binding in vitro, supporting the conclusions obtained in both C6 cells and in neurons.
To directly investigate bundling of F-actin by IPTKA domains, we subjected the four fusion proteins to in vitro bundling assays, similar to those shown in Figure 1. Qualitative experiments (Figure 5B) indicated that residues 9-52, when used at sufficiently high concentration (20 μM, corrected for percent truncated length protein), could bundle F-actin similar to 1-66. The N66(L34P) mutant showed no bundling activity. Microscopic analysis of F-actin mixed with the different fusion proteins confirmed these data, because both N66 and N9-52—but not N66(L34P)—produced prominent F-actin bundles (Figure 5B, right). Thus, similar to the full N66 domain, the F-actin–bundling activity of residues 9-52 appeared to be determined by its affinity for F-actin binding, supporting a model of bundling that involved homodimerization of residues N9-52, which comprise the putative helical region.
Our initial observation of increased F-actin–rich cellular protrusions in cells expressing N66 raised the possibility that the interaction with actin was somehow affecting the rates of actin polymerization or depolymerization. To test this, we measured the rate of F-actin polymerization based on the increased fluorescence of pyrene actin when it is incorporated into filaments (Figure 5C). NusA alone at a concentration of 10 μM had no effect on polymerization/depolymerization rates compared with samples containing pure actin only. The presence of N66 at concentrations (10 μM) that exceeded its affinity for F-actin had no measurable effect on the rate of F-actin polymerization.
Many proteins that bind to the sides of actin filaments provide stability to the filaments in a way that reduces the rate or extent of actin depolymerization. This stabilization can even occur with small side-binding molecules, such as phalloidin (Coluccio and Tilney, 1984). Thus, we tested whether any of our fusion proteins exhibited this property in depolymerization assays. Polymerized F-actin at a concentration of 1 μM was mixed with 20 μM final concentration of various fusion proteins to allow binding. Each mixture was then diluted below the critical concentration for polymerization, and the rate and extent of fluorescence decrease was recorded (Figure 5D). NusA, N9-52, and N66(L34P) showed no measurable effect on F-actin depolymerization compared with buffer alone. By contrast, N66 caused a modest reduction in the rate and extent depolymerization (Figure 5D, arrow). The effect was most apparent when the depolymerization reactions had progressed for 50 s or more. These data indicate that the N66 domain of ITPKA may be able to reduce the rate, and possibly the extent, of depolymerization. Because our biochemical experiments (Figure 1) showed that N66 binds approximately two actin monomers within the context of a filament, the depolymerization data are consistent with the idea that N66 stabilizes preformed F-actin oligomers or protofilaments comprised of a small number of monomers.
Photobleaching Reveals Relative Affinities of IPTKA Fragments for F-Actin in Cells
Our cellular and biochemical experiments provided qualitative support that N9-52 could bind F-actin, but we were unable raise the concentrations of NusA-N9-52 high enough to obtain saturation binding (Figure 5 A, right). As an alternative means of obtaining information about the relative affinities of ITPKA fragments for F-actin in live cells, we used FRAP (Sprague and McNally, 2005). C6 glioma cells were transfected with various ITPKA fragments fused at the C-terminus to mEGFP. For FRAP, we wanted to minimize effects caused by actin turnover and dynamics, so we focused on stress fibers, which are relatively stable F-actin structures composed of bundles. To stabilize the F-actin further, we pretreated the cells with jasplakinolide, a cell-permeable F-actin side-binding toxin. Small rectangular spots located on stress fibers were photobleached using maximal laser intensity and the recovery kinetics were recorded and then fit to single exponential curves. For constructs that did not visibly associate with stress fibers, FRAP was performed on regions of interest located outside the nucleus. Figure 6A shows a representative experiment using N66-mEGFP expressing cells, where a small region on a stress fiber is bleached and its fluorescence recovers; this experiment is also depicted in time-lapse in Supplementary Movie 3. Figure 6B shows the recovery curves for each of the seven constructs tested. The top panel shows the kinetics for four constructs that showed a significant association with F-actin in cells, as suggested in earlier experiments by colocalization with phalloidin. The bottom panel shows four constructs that recovered with more rapid kinetics, some of which showed the diffusion-controlled recovery expected for a cytosolic protein. Note that the N9-52-mEGFP construct is shown in both panels (red traces), to allow easier comparison of the different scales on the x-axes. For all constructs, fluorescence recovered to ∼100% of prebleach values, indicating that there was no significant immobile phase.
The half-times of recovery for the seven constructs showed highly significant differences from one another and varied among one another by more than 100-fold (Figure 6C). Full-length ITPKA-mEGFP exhibited a mean half-time of recovery of ∼3.5 s. Merely deleting the 14 amino acids containing the polybasic region C-terminal to the predicted alpha helix (residues 52-66) reduced the half-time more than threefold, suggesting that the presence of this region contributes to high-affinity F-actin binding in cells. The N66 construct showed a mean recovery half-time of ∼2 s, whereas the value for N9-52 was 0.3 s. The reported half-times of recovery for cytosolic proteins are ∼0.02 s (Sprague and McNally, 2005). To confirm this under our experimental conditions, we bleached soluble EGFP and measured a mean half-time of 0.029 s (Figure 6, B and C). Likewise, the N66(L34P) point mutation showed a fast, diffusion-controlled recovery, with a mean half-time of 0.044 s, and the 15-66 construct showed a mean half-time of 0.032 s. Thus, the N9-52 minimal F-actin–binding domain, despite having a recovery significantly faster than full-length ITPKA, still recovered more than ten times slower than soluble proteins (Figure 6B, bottom curves). These data provide further support that N9-52 comprises a minimal F-actin–binding domain that possesses a lower affinity for F-actin than the full N66 domain. These data also indicate that the L34P mutation is sufficient to render N66 a cytosolic protein and that the polybasic region (52-66) cannot sustain F-actin binding independent of the upstream helical region. Overall, these results are consistent with the results obtained from our other experiments and suggest that the relative affinities for F-actin measured biochemically are preserved in living cells.
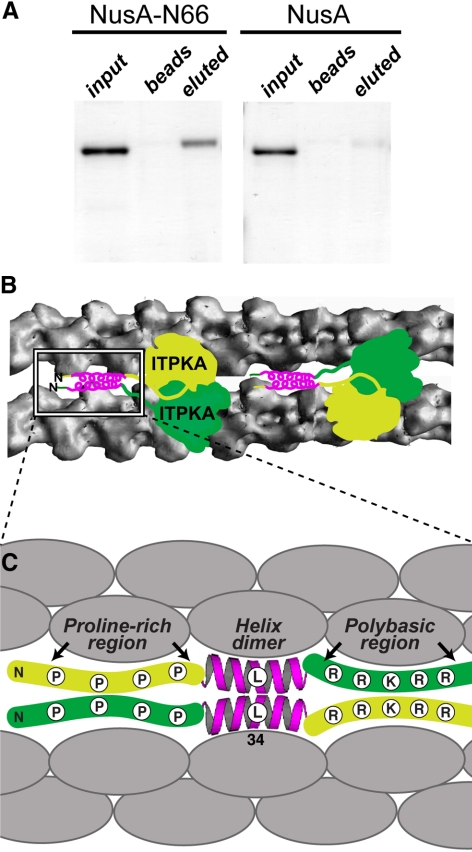
N66 Interacts with Itself In Vitro
To test directly whether N66 can associate with itself and thus bundle F-actin via homodimerization, we devised pulldown experiments in which NusA-N66 was covalently coupled to Sepharose beads and then incubated either with soluble NusA or soluble NusA-N66 to allow for protein–protein interaction with immobilized NusA-N66 (Figure 7A). After extensive washes, followed by elution from beads with urea, we separated the recovered fractions by SDS-PAGE. The results show that NusA-N66—but not NusA alone—binds to N66 immobilized on beads (Figure 7A). These studies provide evidence that the ITPKA-mediated bundling of actin involves the interaction of two or more molecules of N66. Taken together, our data suggest that the self-association domain includes the predicted α-helical region. Thus, both binding and bundling can be destroyed by the L34P mutation. A hypothetical model based on our data are presented in Figure 7, B and C. In this model, at least two actin monomers within the context of a filament (shaded gray) are necessary to interact with the helix-containing region 9-52 in N66. Two (or more) molecules of the helix-containing domain are predicted to mediate the multimerization, causing filament bundling. The polybasic regions immediately C-terminal to the helix can increase the affinity of the interaction with F-actin, but only in the context of the predicted upstream helix. We found no evidence that the polybasic region 52-66 can interact with F-actin under conditions where the helical region is truncated or if it is destroyed by the L34P point mutation.
Figure 7.
Self-association of N66. (A) Bacterially expressed NusA-N66 was coupled to Sepharose beads and then incubated with NusA-N66 or NusA. After extensive washing, bound proteins were eluted from the beads by incubation with 6 M urea. NusA-N66 was bound and eluted (left), but not the NusA control (right). Thus, N66 binds to itself in vitro. The presence of urea in the eluted samples causes the eluted proteins to migrate slightly slower through the gel, producing a slightly larger apparent molecular weight. (B) Proposed model for the bundling mechanism as suggested by our results. In this model, the ITPKA occurs as a dimer that binds the sides of actin filaments. On the basis of the solved crystal structure of IPTKA catalytic domain, we have drawn the ITPKA dimer (2 × 53 kDa) approximately to scale relative to the actin filaments (43 kDa per monomer). The diagram of the actin filaments is based on a recently published model (Oda et al., 2009). The sites on the filaments where ITPKA binds are not known. The boxed region highlights a hypothetical model of the F-actin–binding region at the N-terminus of ITPKA. (C) The N-terminal domain of IPTKA (N66) consists of a minimal F-actin binding comprised of an α-helix centered around residue 34. This helix is preceded by a proline-rich region and is followed by a polybasic region. Our data show that residues 9-15, which include proline 13, are necessary for the helix to bind and/or fold properly, whereas the polybasic region increases the affinity of the helix for F-actin, but cannot bind F-actin in isolation. Based on the stoichiometry of binding measured in biochemical assays, one ITPKA molecule binds two actin monomers in the context of the actin filament. Actin bundling occurs through homodimerization of the helical region, but may be enhanced by adjacent regions. The extent of bundling correlates with the affinity of ITPKA for F-actin, and thus all three structural elements in N66 contribute to both binding and bundling. Therefore the full N66 domain is required to mediate the correct targeting and function of ITPKA (see also Figures 8–10).
N66-induced Bundling Restricts ITPKA to Dendrites and Increases Protrusion Length
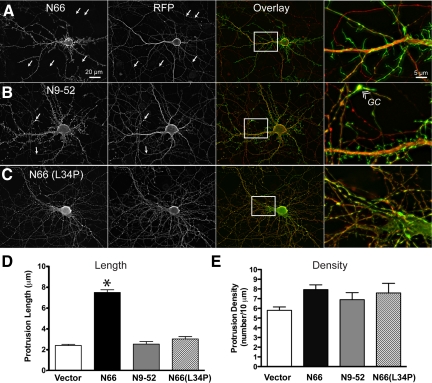
To explore the functional consequences of N66-induced F-actin bundling in neurons, we expressed either N66 or N9-52, or N66(L34P) together with soluble tdTomato (RFP) in hippocampal neurons (Figure 8). Both ITPKA fragments appeared to be highly enriched in spike-like protrusions that emanated from dendrites and were not strongly labeled with RFP. By contrast, the N66(L34P) mutant appeared 100% coincident with the cytosolic marker. Remarkably, N66—but not N9-52—was largely restricted to dendrites and appeared only weakly in the thin caliber neuronal processes indicative of axons (Figure 8A, arrows). Nor was N66 observed in structures having the morphology of (actin-rich) growth cones except in extreme cases of overexpression. By contrast, N9-52 labeled spiky, putative F-actin–rich structures in dendrites and also was found in axons (Figure 8B), including growth cones (GC, Figure 8B, far right). The L34P mutation in N66 rendered the fusion proteins completely cytosolic, similar to the RFP (Figure 8C).
Figure 8.
N66 regulates protein targeting and protrusion length in dendrites. Hippocampal neurons were cotransfected at 8 DIV with GFP-tagged fragments of the ITPKA F-actin–binding domain [N1-66, N9-52, and N66 (L34P)] or EGFP vector alone, and the cytosolic marker tdTomato (RFP). Cells were fixed at 12 DIV and visualized by fluorescence microscopy. (A) The full N66 domain (amino acids 1-66), which binds and bundles F-actin, preferentially localizes to dendritic protrusions and is excluded from axons (arrows). The magnified image (far right) shows clearly the preferential targeting of N66 (green) to dendritic protrusions and very low levels in axons, which exhibit a more constant, untapered caliber than dendrites and terminate in growth cones (see B, far left). (B) The minimal F-actin–binding domain (N9-52) also localizes to F-actin—rich protrusions; however, the green fluorescence is observed both in axons (arrows) and dendrites. Different from to N66, N9-52 occurs in F-actin–rich growth cones (GC, right panel) at the tips of axons. (C) The L34P mutation destroys the preferential targeting of N66 to dendrites. The green fluorescence of N66(L34P)-mEGFP appears similar to the cytosolic marker. (D) Protrusion lengths were determined in a blind manner from cytosolic fluorescence of the RFP channel. The ITPKA F-actin–bundling domain approximately triples the length of F-actin protrusions, whereas the F-actin–binding domain localizes to protrusions of a length similar to controls. The asterisk indicates statistical significance (p > 0.01, one-way ANOVA, with Bartlett's statistic). (E) Protrusion numbers per unit of dendritic length were determined from images of the red channel and were found to be similar for all constructs tested (no significant differences among groups, one-way ANOVA with Bartlett's statistic). Thus, The N66 domain has no effect on protrusion density. Scale bars, 20 μm (low-magnification images, left and middle panels) and 5 μm (high magnification, right panel).
The length of dendritic protrusions was also affected by the presence of full-length N66 (Figure 8D). When N66 was transfected into neurons at 8 DIV and fixed at 12 DIV, we observed an approximate tripling of the average length of dendritic protrusions (from 2.2 μm in controls to 7.5 μm in cells expressing N66; Figure 8D, left). This observation may be explained by a stabilizing effect of the F-actin–binding domain on maturing spines or possibly by bundle-dependent recruitment of accessory proteins. This effect was not observed in neurons transfected with the N9-52 fragment, despite its localization on F-actin. In N9-52–transfected cells, protrusion length did not differ from that in EGFP-expressing neurons (Figure 8D, left). We found no effect of N66 on the numbers of protrusions in dendrites (Figure 8D, right).
The wide-field microscope analysis shown in Figure 8 suggested that the full N66 fragment produced preferential targeting to F-actin located in dendrites and spines, whereas the N9-52 fragment also labeled axons and growth cones. To explore further this apparent F-actin dependent mechanism of polarized targeting of ITPKA in neurons, we performed deconvolution confocal microscopy on cells cotransfected with GFP-tagged N66, N9-52, N15-66 or GFP alone—together with RFP as a cytosolic marker. Cells were fixed and then additionally labeled with fluorescent phalloidin in the far-red channel to allow comparison of the constructs with a cytosolic marker and an F-actin marker simultaneously. We focused on axons and growth cones, because the F-actin microstructure is difficult to discern in these small compartments using a wide-field microscope. In confirmation of our preliminary impressions, N66 seldom labeled growth cones (defined morphologically), except rarely in very highly expressing cells (Figure 9A). By contrast, N9-52 prominently labeled F-actin in growth cones in almost every transfected neuron (Figure 9B). The N15-66-GFP and GFP alone were both colocalized with the soluble marker and not with phalloidin, consistent with our biochemical, morphological, and photobleaching studies. Although thin-caliber axons are not considered to be F-actin–rich structures, they do contain F-actin, which is important for regulating microtubule polarity (Hasaka et al., 2004). Our experiments suggest the presence of putative F-actin “hotspots” in the axon, which are labeled by N9-52. Additional live cell experiments below indicate that axons contain labile and dynamic regions, which can be visualized with N9-52-GFP (Figure 10).
ITPKA-mediated F-Actin Bundling Regulates F-Actin Motility in Dendritic Spines
To evaluate the consequences of the ITPKA interaction with F-actin, we examined the motility of ITPKA F-actin–binding fragments in live neurons. We collected time-lapse movies of 18 DIV neurons expressing either the full N66 binding/bundling domain or the N9-52 binding domain (Figure 10, and Supplementary Movies 4 and 5). By 18 DIV most of the neurons in the cultures display mature spines, which possess a discernable head and neck. To visualize motility, we created motility-grams, in which pixels that changed intensity between successive frames over the course of the time-lapse movie are shaded in red and then overlaid on the average neuronal fluorescence. F-actin motility visualized by N66 was largely constrained to the spine heads, and few regions of motility could be observed in axons (Figure 10, left side, and Supplementary Movie 4). By contrast, the 9-52 construct was a general marker of F-actin motility throughout the neurons and indicated extensive actin motility in spines and growth cones (Figure 10, right side, and Supplementary Movie 5).
High-magnification images of the spine morphologies and motile regions visualized by the two constructs (Figure 10, C–F) indicated that in both cases motility could be observed in dendritic spines (DS). However, the dendritic necks were obviously longer in the presence of N66 and the sites of motility were polarized toward the spine tips. Stubby spines, with small or absent dendritic necks, were rarely observed in the case of N66 expression, but were common when N9-52 was expressed. By contrast, the N9-52 construct showed the highest motility not in spines, but in axons. These “flares” of N9-52–labeled fluorescence would appear and disappear, as they transiently emanated from the axons and then retracted (Figure 10, E and F, and Supplementary Movie 5). In general, N66 expression and motility exhibited far more polarity than N9-52, both at the level of axons versus dendrites and also at the level of the spine head.
Thus, physiological F-actin–dependent targeting of IPTKA in neurons appears to depend on the presence of the full N66 domain. This domain is both necessary and sufficient for the restricting IPTKA to spines, where it associates with a bundled fraction of motile F-actin located in the spine head, near the postsynaptic membrane. The increase in the length of spine necks upon expression of N66 suggests that ITPKA participates in the modulation of spine structure. Taken together, our data suggest that ITPKA-mediated effects on the structure of dendritic spine actin are mediated by the actin binding and bundling properties of its N-terminus.
DISCUSSION
Unique Biochemical Properties of ITPKA Interaction with F-Actin
The F-actin–bundling domain (N66) occurs only in mammalian and bird varieties of ITPKA, and it shows weak sequence and (predicted) structural similarity to the first of two F-actin–binding domains in mammalian ITPKB, but not to other F-actin–binding proteins (Brehm et al., 2004). Our binding studies indicate that the molar ratio of N66 and actin monomers in F-actin is 1:2. On the basis of this finding, we propose that each molecule of N66 binds two molecules of actin within the context of a filament and that two molecules of N66 dimerize to bundle the filaments (Figure 7). Early gel filtration studies of ITPKA isolated from rat brain suggested that ITPKA occurs as a dimer (Johanson et al., 1988). Our in vitro actin polymerization data indicate that the N66 domain does not affect polymerization but may weakly stabilize filaments from depolymerization, perhaps at the dimer or trimer stage.
The core of the F-actin interaction region consists of a predicted α-helical region, which is both necessary and sufficient for binding and bundling. The interaction between full-length ITPKA and F-actin was destroyed by mutating a leucine at position 34. A stretch of proline-rich amino acids precedes the putative helix. Truncation of the first nine amino residues in this region does not destroy F-actin binding, but truncating the first 15 residues does. This indicates that the most N-terminal PxxP motif is dispensable for F-actin binding. We also delineated an accessory role in the F-actin interaction possessed by a polybasic region (amino acids 52-66) located C-terminally to the predicted helix. Although this region did not posses the ability to interact with F-actin on its own, it synergized with the helical region to increase the affinity for F-actin. Polybasic regions occur on other proteins known to regulate F-actin dynamics, such as some formins (Kanaya et al., 2005), Rac1 (Hajdo-Milasinovic et al., 2007), and N-WASP (Papayannopoulos et al., 2005). Whether the ITPKA polybasic region has an analogous function is not known.
Our observation that N66-decorated regions of F-actin motility show polarization in spines suggests a polarized orientation for ITPKA-induced bundles in spines. Previous studies that have examined the polarity of F-actin in spines have shown that the barbed ends localize near the postsynaptic membrane, suggesting that a parallel bundle orientation predominates in spines (Fifkova and Delay, 1982). Parallel bundling may determine the locus of F-actin turnover by focusing the motile regions near the plasma membrane (Bartles, 2000). This would concentrate the rapidly growing barbed ends near the postsynaptic density, creating a microdomain of rapid actin turnover near the synapse and away from the more stable pools of actin located in the spine neck (Honkura et al., 2008).
The minimal ITPKA F-actin–binding domain (N9-52) consists of 43 residues. It is one of the smallest F-actin-specific binding peptides yet described. Residues 9-52 fused to fluorescent proteins may have general utility as “live cell” F-actin reporters. Different from other actin reporters such as GFP-actin (Schell and Irvine, 2006), phalloidin (Mahaffy and Pollard, 2008), or Lifeact (Riedl et al., 2008; Munsie et al., 2009), fluorescent N9-52 does not modify F-actin structure or turnover significantly. We have tested N9-52-mEGFP in a range of cell types and have found it to be a suitable reporter of F-actin (H.W.J. and M.J.S., unpublished data). This property is likely due to its relatively low affinity for F-actin, as we demonstrated biochemically and with FRAP. A low-affinity F-actin–binding domain may be less prone to affecting F-actin structure and dynamics merely because of the high rate at which it dissociates from filaments. Indeed, our data suggest that a minority of N9-52 occurs in the cytosol in cells (Figure 4). The low affinity may prevent excessive stiffening of the filaments and/or may reduce the propensity of the domain to compete with other physiological side-binding proteins.
The F-Actin Interaction Is the Predominant Means of Localizing ITPKA to Dendritic Spines
When the L34P mutation is incorporated into full-length, untagged ITPKA (Figure 3), the enzyme is rendered cytosolic but does not enter the nucleus. This indicates that the intense concentration of ITPKA in dendritic spines (Go et al., 1993; Yamada et al., 1993) is explained fully by the F-actin–binding and –bundling properties of the ITPKA N-terminus. Although the full-length IPTKA contains a number of potential sites for protein–protein interaction, these appear to be insufficient to grossly affect enzyme targeting independently of F-actin binding. Our data indicate that high-affinity ITPKA F-actin binding and/or bundling mediates enzyme targeting to dendrites and a concomitant de-enrichment in axons and growth cones. Because high densities of F-actin occur in both neuronal compartments (Cingolani and Goda, 2008), it is not obvious how the ITPKA interaction with F-actin affects the differential targeting to neuronal subcompartments. The selective and polarized targeting may be a consequence of different affinities for actin in different cellular locations and contexts, or possibly due to the influence of bundle-selective myosin motors (Nagy et al., 2008).
The high concentration of ITPKA in dendritic spines suggests that IP3 metabolism is heterogeneous or polarized within subdomains of neurons. Molecular targets of the ITPKA product IP4 are likely to be dendritic spine-enriched proteins rather than presynaptic, despite the fact that IP3 signaling occurs in both locations (Takei et al., 1998). Additionally, our data show that ITPKA is excluded from the nucleus, suggesting that other enzymes preferentially metabolize the nuclear pool of IP3 (Seeds et al., 2007; Resnick and Saiardi, 2008). In preliminary studies, we have observed that the prenylation domain of the IP3 type 1 5-phosphatase (De Smedt et al., 1996) localizes this IP3 metabolizing enzyme preferentially to axons (H.W.J. and M.J.S., unpublished data), suggesting that metabolic pathways downstream of IP3 generation may be different in axons versus dendrites.
Implications for Spine Structural Plasticity
Expression of the ITPKA F-actin–bundling domain causes the length of dendritic protrusions to triple. In biochemical assays, N66 exhibited a weak ability to slow F-actin depolymerization, so the protrusion-inducing effect could be an inherent property of the domain. Interestingly, other spine-enriched F-actin–bundling proteins produce a similar effect in neurons. Drebrin (Shirao et al., 1994; Hayashi and Shirao, 1999), neruabin (Terry-Lorenzo et al., 2005), and CamKII beta (Ca2+/CaM-depenent kinase II beta; Okamoto et al., 2007) all affect dendritic spine morphology through actin bundling. Bundles may recruit additional proteins, and these could participate in spine and/or filopodial elongation. Increasing the distance between the spine head and the main dendrite through elongation of the spine neck (as we show in Figure 10) affects the diffusion of small molecules into the main dendrite (Grunditz et al., 2008). The bundling properties of ITPKA may therefore synergize with its enzymatic properties to further enhance the compartmentalization of IP3 metabolism in spines.
Actin bundling proteins are critical to the creation and maintenance of F-actin superstructure in cells, and different subsets of bundling proteins conceivably impart diverse and/or plastic characteristics on spines. We showed previously that ITPKA-enriched bundles form polarized Y-shaped structures in spines, which radiate between the endoplasmic reticulum and the postsynaptic membrane (Schell and Irvine, 2006). Such “bundle bridges” have also been depicted in a number of ultrastructural studies (Wilson et al., 1983; Morales and Fifkova, 1989; Capani et al., 2001; Rostaing et al., 2006), but their relationship to structural plasticity is unknown. We also showed previously that ITPKA-labeled actin filaments in dendritic spines undergo a rapid and reversible reorganization in response to Ca2+ influx (Schell and Irvine, 2006). One component of this reorganization involves bulk movements of actin filaments. We suggest that rapid reorganization of ITPKA-decorated F-actin bundles modifies spine actin microstructure in response to extracellular signals, and this in turn underlies some kinds of experience-dependent structural plasticity in dendritic spines.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roger Tsien (University of California, San Diego) for providing tdTomato cDNA; Meeta Desai for pioneering the protrusion analysis; Ikuko Fujiwara, Wolfgang Wagner, and John Hammer (NHLBI, Bethesda, MD) for helpful discussions; and Gudrun Ihrke (USU) for improving the logic, text, and figures. This work was supported by USU Grants RO75LN and RO70NX and a Research Starter Award from the PhRMA Foundation awarded to M.J.S. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the view of the Department of Defense or the Uniformed Services University.
Abbreviations used:
- CaM
calmodulin
- DIV
days in vitro
- F-actin
filamentous actin
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- IP3
inositol (1,4,5)-trisphosphate
- ITPKA
inositol trisphosphate 3-kinase isoform A
- ITPKB
inositol trisphosphate isoform B
- mEGFP
monomeric enhanced green fluorescent protein
- N66
amino terminal residues 1-66 of ITPKA
- N9-52
residues 9-52 of ITPKA
- RFP
red fluorescent protein
- tdTomato
tandem tomato red fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0083) on October 21, 2009.
REFERENCES
- Bartles J. R. Parallel actin bundles and their multiple actin-bundling proteins. Curr. Opin. Cell Biol. 2000;12:72–78. doi: 10.1016/s0955-0674(99)00059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J. Neuronal calcium signaling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- Bramham C. R. Local protein synthesis, actin dynamics, and LTP consolidation. Curr. Opin. Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Brehm M. A., Schreiber I., Bertsch U., Wegner A., Mayr G. W. Identification of the actin-binding domain of Ins(1,4,5)P3 3-kinase isoform B (IP3K-B) Biochem. J. 2004;382:353–362. doi: 10.1042/BJ20031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capani F., Martone M. E., Deerinck T. J., Ellisman M. H. Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study. J. Comp. Neurol. 2001;435:156–170. doi: 10.1002/cne.1199. [DOI] [PubMed] [Google Scholar]
- Cingolani L. A., Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio L. M., Tilney L. G. Phalloidin enhances actin assembly by preventing monomer dissociation. J. Cell Biol. 1984;99:529–535. doi: 10.1083/jcb.99.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smedt F., Boom A., Pesesse X., Schiffmann S. N., Erneux C. Post-translational modification of human brain type I inositol-1,4,5-trisphosphate 5-phosphatase by farnesylation. J. Biol. Chem. 1996;271:10419–10424. doi: 10.1074/jbc.271.17.10419. [DOI] [PubMed] [Google Scholar]
- Fifkova E., Delay R. J. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J. Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go M., Uchida T., Takazawa K., Endo T., Erneux C., Mailleux P., Onaya T. Inositol 1,4,5-trisphosphate 3-kinase highest levels in the dendritic spines of cerebellar Purkinje cells and hippocampal CA1 pyramidal cells. A pre- and post-embedding immunoelectron microscopic study. Neurosci. Lett. 1993;158:135–138. doi: 10.1016/0304-3940(93)90247-i. [DOI] [PubMed] [Google Scholar]
- Grunditz A., Holbro N., Tian L., Zuo Y., Oertner T. G. Spine neck plasticity controls postsynaptic calcium signals through electrical compartmentalization. J. Neurosci. 2008;28:13457–13466. doi: 10.1523/JNEUROSCI.2702-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdo-Milasinovic A., Ellenbroek S. I., van Es S., van der Vaart B., Collard J. G. Rac1 and Rac3 have opposing functions in cell adhesion and differentiation of neuronal cells. J. Cell Sci. 2007;120:555–566. doi: 10.1242/jcs.03364. [DOI] [PubMed] [Google Scholar]
- Hasaka T. P., Myers K. A., Baas P. W. Role of actin filaments in the axonal transport of microtubules. J. Neurosci. 2004;24:11291–11301. doi: 10.1523/JNEUROSCI.3443-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Shirao T. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J. Neurosci. 1999;19:3918–3925. doi: 10.1523/JNEUROSCI.19-10-03918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N., Matsuzaki M., Noguchi J., Ellis-Davies G. C., Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Irvine R. F., Lloyd-Burton S. M., Yu J. C., Letcher A. J., Schell M. J. The regulation and function of inositol 1,4,5-trisphosphate 3-kinases. Adv. Enzyme Regul. 2006;46:314–323. doi: 10.1016/j.advenzreg.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine R. F., McNulty T. J., Schell M. J. Inositol 1,3,4,5-tetrakisphosphate as a second messenger—a special role in neurones? Chem. Phys. Lipids. 1999;98:49–57. doi: 10.1016/s0009-3084(99)00017-1. [DOI] [PubMed] [Google Scholar]
- Irvine R. F., Schell M. J. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Janmey P. A. Creating a niche in the cytoskeleton: actin reorganization by a protein kinase. Proc. Natl. Acad. Sci. USA. 2001;98:14745–14747. doi: 10.1073/pnas.011601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson R. A., Hansen C. A., Williamson J. R. Purification of D-myo-inositol 1,4,5-trisphosphate 3-kinase from rat brain. J. Biol. Chem. 1988;263:7465–7471. [PubMed] [Google Scholar]
- Jun K., et al. Enhanced hippocampal CA1 LTP but normal spatial learning in inositol 1,4,5-trisphosphate 3-kinase(A)-deficient mice. Learn Mem. 1998;5:317–330. [PMC free article] [PubMed] [Google Scholar]
- Kanaya H., Takeya R., Takeuchi K., Watanabe N., Jing N., Sumimoto H. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 2005;10:665–678. doi: 10.1111/j.1365-2443.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- Kim I. H., Park S. K., Sun W., Kang Y., Kim H. T., Kim H. Spatial learning enhances the expression of inositol 1,4,5-trisphosphate 3-kinase A in the hippocampal formation of rat. Brain Res. Mol. Brain Res. 2004;124:12–19. doi: 10.1016/j.molbrainres.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Lloyd-Burton S. M., Yu J. C., Irvine R. F., Schell M. J. Regulation of inositol 1,4,5-trisphosphate 3-kinases by calcium and localization in cells. J. Biol. Chem. 2007;282:9526–9535. doi: 10.1074/jbc.M610253200. [DOI] [PubMed] [Google Scholar]
- Mahaffy R. E., Pollard T. D. Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry. 2008;47:6460–6467. doi: 10.1021/bi702484h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E., Jacob M., Sarria J. C., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miller A. T., Chamberlain P. P., Cooke M. P. Beyond IP 3, roles for higher order inositol phosphates in immune cell signaling. Cell Cycle. 2008;7:463–467. doi: 10.4161/cc.7.4.5518. [DOI] [PubMed] [Google Scholar]
- Moon K. H., Lee S. Y., Rhee S. G. Developmental changes in the activities of phospholipase c, 3-kinase, and 5-phosphatase in rat brain. Biochem. Biophys. Res. Commun. 1989;164:370–374. doi: 10.1016/0006-291x(89)91728-2. [DOI] [PubMed] [Google Scholar]
- Morales M., Fifkova E. In situ localization of myosin and actin in dendritic spines with the immunogold technique. J. Comp. Neurol. 1989;279:666–674. doi: 10.1002/cne.902790412. [DOI] [PubMed] [Google Scholar]
- Munsie L. N., Caron N., Desmond C. R., Truant R. Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat. Methods. 2009;6:317. doi: 10.1038/nmeth0509-317. [DOI] [PubMed] [Google Scholar]
- Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Iwasa M., Aihara T., Maeda Y., Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Narayanan R., Lee S. H., Murata K., Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc. Natl. Acad. Sci. USA. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V., Co C., Prehoda K. E., Snapper S., Taunton J., Lim W. A. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol. Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Pattni K., Banting G. Ins(1,4,5)P3 metabolism and the family of IP3–3Kinases. Cell Signal. 2004;16:643–654. doi: 10.1016/j.cellsig.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Resnick A. C., Saiardi A. Inositol polyphosphate multikinase: metabolic architect of nuclear inositides. Front. Biosci. 2008;13:856–866. doi: 10.2741/2726. [DOI] [PubMed] [Google Scholar]
- Riedl J., et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W. N., Nakamura T., Watanabe S., Larkum M., Lasser-Ross N. Synaptically activated Ca2+ release from internal stores in CNS neurons. Cell Mol. Neurobiol. 2005;25:283–295. doi: 10.1007/s10571-005-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing P., Real E., Siksou L., Lechaire J. P., Boudier T., Boeckers T. M., Gertler F., Gundelfinger E. D., Triller A., Marty S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur. J. Neurosci. 2006;24:3463–3474. doi: 10.1111/j.1460-9568.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- Schell M. J., Erneux C., Irvine R. F. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 2001;276:37537–37546. doi: 10.1074/jbc.M104101200. [DOI] [PubMed] [Google Scholar]
- Schell M. J., Irvine R. F. Calcium-triggered exit of F-actin and IP(3) 3-kinase A from dendritic spines is rapid and reversible. Eur. J. Neurosci. 2006;24:2491–2503. doi: 10.1111/j.1460-9568.2006.05125.x. [DOI] [PubMed] [Google Scholar]
- Seeds A. M., Frederick J. P., Tsui M. M., York J. D. Roles for inositol polyphosphate kinases in the regulation of nuclear processes and developmental biology. Adv. Enzyme Regul. 2007;47:10–25. doi: 10.1016/j.advenzreg.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirao T., Hayashi K., Ishikawa R., Isa K., Asada H., Ikeda K., Uyemura K. Formation of thick, curving bundles of actin by drebrin A expressed in fibroblasts. Exp. Cell Res. 1994;215:145–153. doi: 10.1006/excr.1994.1326. [DOI] [PubMed] [Google Scholar]
- Sprague B. L., McNally J. G. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Szinyei C., Behnisch T., Reiser G., Reymann K.G. Inositol 1,3,4,5-tetrakisphosphate enhances long-term potentiation by regulating Ca2+ entry in rat hippocampus. J. Physiol. 1999;516(Pt 3):855–868. doi: 10.1111/j.1469-7793.1999.0855u.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takazawa K., Lemos M., Delvaux A., Lejeune C., Dumont J. E., Erneux C. Rat brain inositol 1,4,5-trisphosphate 3-kinase. Ca2(+)-sensitivity, purification and antibody production. Biochem. J. 1990;268:213–217. doi: 10.1042/bj2680213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K., Shin R. M., Inoue T., Kato K., Mikoshiba K. Regulation of nerve growth mediated by inositol 1,4,5-trisphosphate receptors in growth cones. Science. 1998;282:1705–1708. doi: 10.1126/science.282.5394.1705. [DOI] [PubMed] [Google Scholar]
- Terry-Lorenzo R. T., Roadcap D. W., Otsuka T., Blanpied T. A., Zamorano P. L., Garner C. C., Shenolikar S., Ehlers M. D. Neurabin/protein phosphatase-1 complex regulates dendritic spine morphogenesis and maturation. Mol. Biol. Cell. 2005;16:2349–2362. doi: 10.1091/mbc.E04-12-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. J., Groves P. M., Kitai S. T., Linder J. C. Three-dimensional structure of dendritic spines in the rat neostriatum. J. Neurosci. 1983;3:383–388. doi: 10.1523/JNEUROSCI.03-02-00383.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhorst S., Blechner C., Lin H. Y., Elling C., Nalaskowski M., Kirchberger T., Guse A. H., Mayr G. W. Ins(1,4,5)P3 3-kinase-A overexpression induces cytoskeletal reorganization via a kinase-independent mechanism. Biochem. J. 2008;414:407–417. doi: 10.1042/BJ20080630. [DOI] [PubMed] [Google Scholar]
- Yamada M., Kakita A., Mizuguchi M., Rhee S. G., Kim S. U., Ikuta F. Specific expression of inositol 1,4,5-trisphosphate 3-kinase in dendritic spines. Brain Res. 1993;606:335–340. doi: 10.1016/0006-8993(93)91004-c. [DOI] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.