Abstract
More than half of the world's population uses rice as a source of carbon intake every day. Improving grain quality is thus essential to rice consumers. The three main properties that determine rice eating and cooking quality—amylose content, gel consistency, and gelatinization temperature—correlate with one another, but the underlying mechanism of these properties remains unclear. Through an association analysis approach, we found that genes related to starch synthesis cooperate with each other to form a fine regulating network that controls the eating and cooking quality and defines the correlation among these three properties. Genetic transformation results verified the association findings and also suggested the possibility of developing elite cultivars through modification with selected major and/or minor starch synthesis-related genes.
Keywords: association analysis, grain quality, starch synthesis
Rice (Oryza sativa L.) is a pivotal cereal crop that provides the staple food for more than half of the world's population. Successful application of hybrid production technology has greatly increased grain yield, but grain quality of hybrid rice remains to be improved. Grain quality has now become the primary consideration of rice customers and breeding programs. Cultivars with different grain qualities are also required for medicinal, ceremonial, or special production purposes. Tremendous efforts have been made to understand the genetic basis of rice eating and cooking quality (ECQ) (1–6), which is mainly affected by three physicochemical properties: amylose content (AC), gel consistency (GC), and gelatinization temperature (GT) (7–9), but so far only the Waxy gene (10) has been found to affect AC and ALK (4, 11) to GT. We are therefore still facing many challenging questions. For example, how many major and minor genes control grain ECQs? Are AC, GC, and/or GT controlled by one or multiple genes? What is the relationship among these genes? Is it possible to breed elite varieties with desired ECQ properties by genetic manipulation of AC, GC, and/or GT?
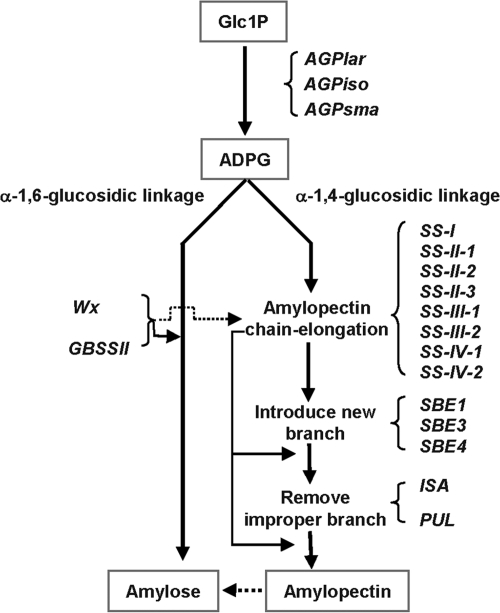
Given that starch comprises approximately 90% of the rice grain, genes involved in starch biosynthesis are naturally expected to affect ECQs. Starch biosynthesis is a complex system composed of multiple subunits or isoforms of four classes of enzymes: ADP-glucose pyrophosphorylase (AGP), starch synthase (SS), starch branching enzyme (SBE), and starch debranching enzyme (DBE) (12–14) (Fig. 1). Each enzyme plays a distinct role (15, 16), but presumably functions as part of a network. For example, the rice sugary mutant is defective in ISA (Isoamylase-type DBE), but the expression levels of several other starch synthesis-related genes (SSRGs) were also affected (17). As parts of this complex synthesis pathway, genes controlling amylose synthesis also affect amylopectin formation (18), and amylopectin can give rise to amylose (19). These relationships jointly make the genetic dissection of ECQs difficult.
Fig. 1.
A simplified starch synthesis system in cereal. Eighteen genes are involved in or play distinct roles in different steps of starch synthesis. AGP, ADP-glucose pyrophosphorylase; AGPlar, AGP large subunit; AGPiso, AGP large subunit isoform; AGPsma, AGP small subunit; GBSS, granule-bound starch synthase; SS, soluble starch synthase; SBE, starch branching enzyme; ISA, isoamylase; PUL, pullulanase; ISA and PUL belong to starch debranching enzyme (DBE).
QTL mapping and cloning provides useful information of genetic loci, but usually it is difficult to isolate genes of minor effects that play a minor role or to elucidate a complex network due to narrow germplasms used in single experiments. Association mapping is a powerful tool for studying genetic loci involved in the inheritance of complex traits (20–22), and an initial analysis of kernel composition traits has revealed promising associations with starch biosynthesis genes in maize (23, 24). To gain a broader understanding of the functions of these SSRGs on rice grain ECQs, we carried out a candidate-gene association mapping study and gene transformation verification, demonstrating that SSRGs form a fine network to control ECQs by regulating AC, GC, and/or GT and that rice with a desired ECQ can be achieved through genetic modification of one or more SSRGs in the regulating network.
Results
Diverse Panel of Rice Varieties Used for Analysis.
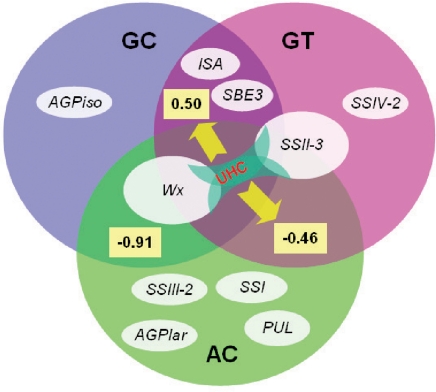
To have enough statistical power to test an association, it is critical to choose various germplasms that span a full range of phenotypic variations. A diverse panel of rice varieties, including 33 indica and 37 japonica, were measured for AC, GC, and GT in 2 successive years with three replications each year, and various trait value combinations were observed (Fig. S1). These three properties are highly correlated: AC is negatively correlated with GC (−0.91) and GT value (−0.46), whereas GC is positively correlated with the GT value (0.50).
To avoid false association, population structure was evaluated with Admixture Model using simple sequence repeat (SSR) marker data. The resulting subpopulation membership percentage (Q-values) agreed with the subspecies classification. The majority of indica varieties have high AC, low GC, and low GT values, while the majority of japonica varieties show low AC, high GC, and high GT values (Fig. S2).
Nucleotide Analysis of SSRGs.
Eighteen SSRGs were selected as candidate genes in this study (Table S1). The entire genomic regions of these genes were sequenced for 16 representative core varieties (Fig. S1). Nucleotide analysis showed that the diversities of the coding sequences were much lower than those of whole genes in all 18 SSRGs, and the diversities of the nonsynonymous substitution were lower than the synonymous except for GBSSII (Table S2). This result suggested that these SSRGs had likely undergone artificial selection during domestication and the linkage disequilibrium (LD) analysis provided further evidence (Fig. S3). Because most SSRGs showed a stable LD decay, we chose the most diverse fragments of each gene and sequenced these fragments in the remaining varieties. The polymorphic sites were filtered out (Table S3) for the association analysis.
Genes Affecting Rice ECQs.
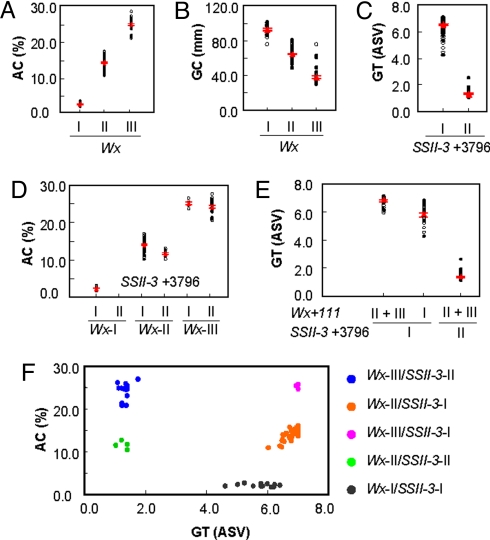
The effect of SSRGs was tested against each ECQ property (i.e., AC, GC, or GT) across the panel using a mixed model method (25). Association analysis with individual SSRGs revealed that two genes, Wx and SSII-3, mainly control AC. Wx sites have much smaller P-values (3 × 10−15 and 4 × 10−24) than SSII-3 (2 × 10−5), suggesting that Wx is likely the major gene affecting AC. Further analysis showed strong evidence of haplotype effects (P value 1 × 10−51): Wx-I, Wx-II, and Wx-III. Among these, Wx-I bears a loss-of-function mutation that results in glutinous rice varieties with extremely low AC, Wx-II shows a leaky phenotype that leads to a medium level of AC, and Wx-III functions as the wild type allele with high AC (Fig. 2A). To eliminate the effect of this major gene, a second model containing the Wx haplotype was used to search for evidence of minor genes (see Methods). The model detected five genes that affect AC additively with Wx: AGPlar, PUL, SSI, SSII-3, and SSIII-2 (Table 1). Based on the association sites, each of the minor genes could also be classified into two haplotypes: haplotype I and haplotype II (Table S4). For each associated gene, the estimate AC of haplotype I is significantly higher than haplotype II under the control of the Wx haplotype (Table 1).
Fig. 2.
Effects of Wx and SSII-3 on grain ECQs in rice. (A) Wx functions as a major gene controlling AC. (B) Wx functions as a major gene controlling GC. (C) SSII-3 functions as a major gene controlling GT. (D) SSII-3 also functions additively with Wx to control grain AC. (E) Wx works with SSII-3 in controlling grain GT. (F) The haplotype combination of Wx and SSII-3. The red triangles and lines are mean ± SE, and the trait values of each variety are represented as solid (2005) or open (2006) circles. Numbers behind genes refer to the association sites and the Roman numerals above the lines stand for haplotypes.
Table 1.
Highly associated SNPs
| Trait | Model | SNP | Estimate difference (HapII–HapI) | F value | P value |
|---|---|---|---|---|---|
| AC | Q + K | Wx-1160 | 16.3484 | 104.09 | 3 × 10−15 |
| Wx+111 | 16.5511 | 267.77 | 4 × 10−24 | ||
| QK+Wx | AGPlar-1633 | −0.8694 | 5.31 | 3 × 10−2 | |
| PUL+855 | −0.9351 | 4.15 | 5 × 10−2 | ||
| SSI+3216 | −0.8892 | 5.13 | 3 × 10−2 | ||
| SSII-3+3796 | −1.2193 | 12.20 | 2 × 10−3 | ||
| SSIII-2-1078 | −0.9584 | 7.82 | 8 × 10−3 | ||
| GC | Q + K | Wx-1160 | 36.6758 | 66.65 | 7 × 10−11 |
| Wx+111 | 39.2079 | 150.40 | 1 × 10−18 | ||
| QK+Wx | AGPiso-511 | 6.6442 | 7.24 | 9 × 10−3 | |
| SBE3+3577 | 6.0568 | 6.98 | 1 × 10−2 | ||
| ISA-1326 | 17.6971 | 8.73 | 5 × 10−3 | ||
| GT | Q + K | SSII-3+3796 | −5.1413 | 1423.32 | 5 × 10−44 |
| QK+SSII-3 | Wx+111 | −1.0150* | 132.72 | 2 × 10−17 | |
| SBE3+3577 | −0.3821 | 5.76 | 2 × 10−2 | ||
| ISA-1499 | −0.5977 | 5.90 | 2 × 10−2 | ||
| SSIV-2+437 | −0.9660 | 8.59 | 5 × 10−3 |
*The estimate difference between haplotype I and II + III.
For the properties of GC, Wx was identified as the major gene with a halpotype P value of 2 × 10−19 (Table S5). Varieties with Wx-I show high GC values, those with Wx-II show medium GC values, and those with Wx-III have low GC values (Fig. 2B). As with AC, three minor genes, AGPiso, SBE3, and ISA, affect GC once the model accounted for the Wx haplotype (Table 1). Each of the minor genes was classified into two haplotypes: haplotype I and haplotype II (Table S4). The estimate GC value of haplotype I for each additive gene is significantly lower than haplotype II, while the Wx haplotype is controlled (Table 1).
For the properties of GT, SSII-3 has highly significant P value (5 × 10−44), suggesting that SSII-3 plays a major role in regulating GT. SSII-3 has two allelic states: SSII-3-I and SSII-3-II. Varieties with SSII-3-I have higher GT values than those with SSII-3-II (Fig. 2C). Again, with the model controlling for SSII-3, a further search identified Wx, SBE3, ISA, and SSIV-2 as minor genes that affect GT additively (Table 1). The estimate GT value of haplotype I of Wx is significantly lower than haplotypes II+III, while the SSII-3 haplotype is controlled (Table 1). Each of the other minor genes was also classified into two haplotypes: haplotype I and haplotype II (Table S4). The estimate GT value of haplotype I for each additive gene is significantly higher than haplotype II, while the SSII-3 haplotype is defined (Table 1).
Interestingly, Wx and SSII-3 are diversified and cooperated in affecting AC and GT. Besides acting as a major gene controlling GT, SSII-3 also affected AC as a minor gene. Consistent with its effect on GT, SSII-3-I contributed to higher AC under the same Wx background, whereas SSII-3-II led to lower AC (Fig. 2D). This means that varieties with the SSII-3-I allele would show higher AC and GT values, and varieties with the SSII-3-II allele would have lower AC and GT values. On the other hand, Wx also cooperated with SSII-3 to affect GT. Wx-I not only led to lower AC but also decreased GT value under the SSII-3-I background, whereas Wx-II and Wx-III increased GT value (Fig. 2E). Therefore, we concluded that the effect of either Wx or SSII-3 on AC and GT values falls into a consistent pattern.
To understand why AC and GT values are negatively correlated, we checked the haplotype combination of Wx and SSII-3 in the panel and found a natural grouping pattern of each haplotype combination when AC and GT values were plotted (Fig. 2F). The number of varieties within each haplotype combination was different. With 35 varieties belonging to Wx-II/SSII-3-I (middle AC and high GT values) and 15 varieties belonging to Wx-III/SSII-3-II (high AC and low GT values), 71% of the germplasms were accounted for. At the same time, varieties with Wx-III/SSII-3-I haplotype had high AC and high GT values, and those with Wx-II/SSII-3-II had low AC and low GT values. When correlation was conducted collectively across the panel, the correlation coefficient became −0.46 (P value, 7 × 10−5).
Network Controlling Grain ECQs.
These results strongly suggest that SSRGs, which determine the rice grain ECQs by affecting AC, GC, and GT, form a fine network (Fig. 3) with the following features. First, Wx and SSII-3 are central in determining grain ECQs by affecting all three properties: AC, GC, and GT. Wx functions as the sole major gene for both AC and GC but as a minor gene affecting GT, whereas SSII-3 is the sole major gene controlling GT but as a minor gene affecting AC and GC. Second, two genes affect two properties simultaneously: both ISA and SBE3 affect GC and GT. Third, several minor genes are specific for each property: SSIII-2, AGPlar, PUL, and SSI for AC, AGPiso for GC, and SSIV-2 for GT. Fourth, the correlations among AC, GC, and GT were caused by the joint action of these associated genes and unequal haplotype combination. Summarizing the associated genes into the starch biosynthesis pathway, we can see that different SSRGs affect different characters at the different starch biosynthesis stage (Fig. S4). For example, at the step from Glc1P to ADPG, AC is mainly affected by AGPlar, whereas GC is mainly affected by AGPiso.
Fig. 3.
Summary of genes controlling rice grain ECQs. The three large ovals represent AC (green), GC (blue), and GT (pink), and the role of each gene is proportionally represented by a small oval. Numbers in the overlapped region indicate the correlation coefficients between the two properties. UHC: Unequal haplotype combination.
Verification by Transgenic Tests.
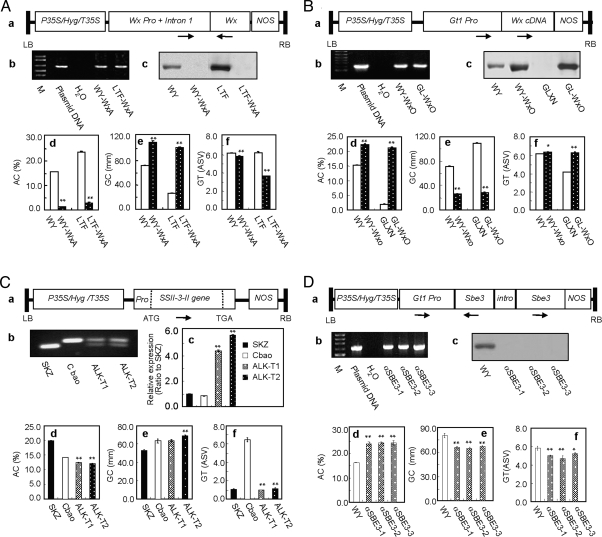
To validate the effects of major and minor genes identified with the association analysis, we carried out transgenic studies by introducing Wx and SSII-3, the two major genes, and SBE3, one minor gene, into different background rice cultivars. To determine how the properties were modified when Wx-II or Wx-III were altered to Wx-I (a loss-of-function mutation), an antisense Wx RNA under control of the rice Wx promoter (Fig. 4A, a) was introduced into a Wx-II variety, Wuyunjing 7 (WY, medium AC, medium GC value, and high GT value) and a Wx-III variety, Longtefu B (LTF, high AC, very low GC value, and high GT value). We randomly chose two representatives, WY-WxA and LTF-WxA, from 58 stable transgenic rice lines (Fig. 4A, b). Protein analyses showed that grains of the two transgenic lines produced nearly undetectable Wx protein (Fig. 4A, c), indicating that the Wx expression was successfully inhibited in transgenic plants. Compared with untransformed controls, AC was decreased significantly (Fig. 4A, d), and the GC values were increased remarkably for the homozygous transgenic lines (Fig. 4A, e). As expected, the grain GT value levels of the two transgenic lines were also decreased (Fig. 4A, f).
Fig. 4.
Transgenic verification. (A) Analysis of antisense Wx transgenic lines. (a) The construct of the antisense Wx gene. (b) Identification of transgenic lines. Levels of Wx (c), AC (d), GC (e), and GT (f) in the transgenic lines. (B) Analysis of Wx-III overexpression transgenic lines. (a) The construct of overexpressing of Wx-III. (b) Identification of transgenic lines. Levels of Wx (c), AC (d), GC (e), and GT (f) in the transgenic lines. (C) Expression of SSII-3-II in the SSII-3-I background. (a) The construct containing Shuangkezao SSII-3-II. (b) Identification of transgenic lines. (c) Transcriptional levels of SSII-3 in transgenic lines revealed by qPCR. Values of AC (d), GC (e), and GT (f) in the transgenic lines. (D) Transgenic analysis of the repression of minor gene SBE3. (a) The RNAi construct of SBE3. (b) Identification of transgenic lines. (c) Western analyses of transgenic lines. Significant changes of AC (d), GC (e), or GT (f) in the transgenic lines. The error bar for each value represents the mean ± SE, * and ** indicate the least significant difference at 0.05 or 0.01 probability level, respectively.
To further confirm the functions of Wx, we also generated homozygous transgenic lines that overexpress Wx under the control of the rice glutelin (Gt1) promoter that specifically expresses in the endosperm (Fig. 4B, a) by transforming WY and Guanglingxiangnuo (GLXN), a Wx-I variety with extremely low AC, high GC value, and medium GT value (Fig. 4 B, d–f). We randomly chose two representatives, WY-WxO and GL-WxO, from 75 stable transgenic lines (Fig. 4B, b). The Wx protein levels in these transgenic lines were increased significantly (Fig. 4B, c), indicating that the transgenes were strongly expressed. As expected, the AC of the homozygous transgenic grains was increased dramatically compared with the nontransgenic grains (Fig. 4B, d). Consistent with the previous experiment, the GC value of the transgenic grains was decreased remarkably (Fig. 4B, e). As in the antisense Wx RNA experiment, the GT of the transgenic grains was also affected (Fig. 4B, f).
To confirm the effect of SSII-3 on ECQs, we isolated the entire SSII-3 gene including its fully functional promoter region from Shuangkezao (SKZ) (Fig. 4C, a). SKZ is a SSII-3-II haplotype variety whose grains have an extremely low GT value, high AC, and a medium GC value. We introduced the SKZ SSII-3 gene into Cbao, a SSII-3-I haplotype variety with a high GT value, medium AC, and a medium GC value. We chose ALK-T1 and ALK-T2 randomly as representatives of the five transgenic lines (Fig. 4C, b); the transgene expression levels were significantly increased (Fig. 4C, c). Comparing with the untransformed SKZ, the grain ECQs of the homozygous transgenic lines showed a dramatic decrease in the GT value (Fig. 4C, d). In the grains of the two transgenic lines, the AC levels were significantly decreased (Fig. 4C, e). But the GC values were increased significantly in one transgenic line and slightly in the other (Fig. 4C, f).
To test the effect of minor genes on grain ECQs, we performed transgenic studies by repressing the expression of SBE3 in the variety WY by RNA interference (RNAi) under the control of the Gt1 promoter (Fig. 4D, a). Among the 57 stable homozygous transgenic lines, three representative SBE3 RNAi lines, αSBE3–1 to αSBE3–3, were randomly chosen and compared with untransformed controls (Fig. 4D, b). Protein blotting analyses showed that the expression of SBE3 was entirely inhibited in all three SBE3 RNAi lines (Fig. 4D, c, loading control, see Fig. S5). Compared with untransformed controls, the AC levels were substantially increased in the homozygous transgenic lines, whereas the GC and GT values were significantly decreased (Fig. 4D, d–f).
In addition to exploring the effects of the transgenic approach on grain ECQs, we also carefully investigated and compared traits of agronomic importance between transgenic lines and their wild type controls. In a normal field trial of Wx transgenic rice lines (Table S6), the data showed minimal effect on the major agronomic traits of transgenic lines when expression of the major gene Wx was inhibited or enhanced. Similar results were found between transgenic rice lines with other transgenes and their untransformed wild type.
Discussion
Elucidation of SSRGs Functions in Regulating Rice ECQs with a Diverse Background.
Over the past decades, properties of AC, GC, and GT of rice grains have been extensively studied, but no consensus has been reached. For example, AC has been reported to be governed only by Wx (3, 5, 26, 27), by several gene loci (28, 29), or even by some unidentified non-Wx genes (2). The discrepancies in different reports were due to the limitation of the QTLs mapping approach, which depends on haplotype configurations of both parents in a specific study (30, 31). Through association analysis, our study provides a clearer picture of how the allelic diversities at SSRGs collectively regulate rice ECQs through the starch biosynthetic network. Our study provides strong evidences that Wx not only affects AC, as reported in refs. 10 and 32, but also regulates GC as a major gene and GT as a minor one. Furthermore, our results also clearly demonstrated that SSII-3 plays an essential role not only in controlling GT, but also AC and GC. In addition, we also showed that some other SSRGs affect additively AC, GC, and GT as minor genes and all theses associated genes form a fine complex network controlling ECQs of rice grains. Moreover, besides the minor genes that function in regulating these grain properties, different haplotype combinations of major genes are present in rice germplasms. The correlations between these ECQ properties, as demonstrated in the current study, add another layer of complexity to the process of unraveling genetic architecture of the ECQs.
Grain quality has long been considered in rice production. Starch comprises ≈90% of milled rice, and its structure determines grain quality properties. The genes associated with grain quality properties would have been subjected to artificial selection during breeding and production. Our results indicated that the diversities of the coding sequences were much lower than those of the whole genes and the diversities of the nonsynonymous substitution were significantly lower than the synonymous in the associated genes. In addition, the associated genes were from different classes of enzymes. We propose that the SSRGs affect the starch structure at different starch synthesis steps during starch development and further affect the grain quality.
In this study, we showed that the negative correlation between AC and GT values was a result of unequal trait values for different haplotype combinations of Wx and SSII-3 and unequal frequencies of these haplotypes in rice germplasm. Because Wx and SSII-3 are closely located on chromosome 6, it is possible that these two genes may have undergone coselection during domestication. This is consistent with our previous work on the marker-assisted selection, in which it was found that under the same SSII-3 background, when Wx-III was substituted for Wx-II, not only were AC and GC changed, but the GT value was also down-regulated (33).
Association Analysis and Genetic Transformation Provide Critical Information to Rice Breeding.
Improving the grain quality of hybrid rice requires long term effort from many breeding programs. Genetic approaches to breeding rice varieties with desired grain ECQs, either by genetic engineering or molecular marker-assisted selection, will definitely improve the efficiency of rice ECQs breeding. Our results indicate that the selection for any single gene in rice breeding would be insufficient because modification of one gene may lead to changes in all three properties of ECQs. Rather, future network-based multitrait selection would more efficiently improve ECQ properties. Moreover, we demonstrated that it is possible to breed varieties with desired grain ECQs by genetic modification without adversely affecting general agronomic performance. Our genetic characterization of widely used breeding germplasms provides essential information for future marker-assisted selection.
In summary, we demonstrated in this study that a combination of association analysis and genetic transformation can be exploited to understand and manipulate economic traits of relevance to consumers. Findings described herein should also facilitate investigation of other complex agronomic traits in rice and are applicable to other important crops.
Methods
Measurement of Grain ECQs.
Mature rice grains were milled after being harvested, air dried, and stored at room temperature for 3 months. Their ECQs were measured according to methods reported in ref. 5. GT was evaluated as the alkali spreading value (ASV). Grain ECQs were independently analyzed in two successive years with three repeats each year.
Identification of SSRGs.
DNA sequences of rice SSRGs were searched and retrieved from all available DNA databases in 2005 with Blastn and analyzed with CLUSTAL W (34). A total of 50 full-length cDNA sequences in rice were identified, representing 18 SSRGs from AGP, SS, SBE, and DBE gene families (Table S1).
Statistic Analysis.
Population structure was evaluated by the program STRUCTURE (35) with 49 SSR markers distributed on 12 chromosomes in rice (SI Experimental Procedures) using admixture model. The relative kinship (K) matrix was calculated with the software package SPAGeDi (36). Diversities of genomic DNA and CDS sequences were analyzed with TASSEL (37) and DnaSP (38), respectively. The LD was evaluated with TASSEL (37). Association analysis followed the unified mixed model reported in ref. 25, using SAS 9.0 (39). Models with both one-single-gene and two-single-gene effects and interaction terms were applied.
Additional Experimental Procedures.
A detailed description of plant materials, DNA preparation and sequencing (primer sequences are shown in Table S7), SSR primers used for population structure evaluations, and protein blotting analysis can be found in the SI Experimental Procedures.
Supplementary Material
Acknowledgments.
We thank Dr. Guiai Jiao at China National Rice Research Institute for measuring rice grain ECQs and Guojun Dong for assistance in growing plant materials. This work was supported by the Ministry of Science and Technology of China Grants 2006AA10A1 and 2005CB12, United States Department of Agriculture Cooperative State Research, Education and Extension Service (2006-35300-17155), and the National Natural Science Foundation of China Grant 30530470.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ150815–GQ152092).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912396106/DCSupplemental.
References
- 1.Bao JS, Corke H, Sun M. Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in nonwaxy rice (Oryza sativa L.) Theor Appl Genet. 2006;113:1185–1196. doi: 10.1007/s00122-006-0394-z. [DOI] [PubMed] [Google Scholar]
- 2.Li J, et al. QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome. 2004;47:697–704. doi: 10.1139/g04-029. [DOI] [PubMed] [Google Scholar]
- 3.Zhou PH, Tan YF, He YQ, Xu CG, Zhang Q. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor Appl Genet. 2003;106:326–331. doi: 10.1007/s00122-002-1023-0. [DOI] [PubMed] [Google Scholar]
- 4.Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet. 2002;104:1–8. doi: 10.1007/s001220200000. [DOI] [PubMed] [Google Scholar]
- 5.Tan YF, et al. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor Appl Genet. 1999;99:642–648. doi: 10.1007/s001220051279. [DOI] [PubMed] [Google Scholar]
- 6.Reddy KR, Ali SZ, Bhattacharya KR. The fine structure of rice-starch amylopectin and its relation to the texture of cooked rice. Carbohydr Polym. 1993;22:267–275. [Google Scholar]
- 7.Little R, Hilder G, Dawson E. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 1958;35:111–126. [Google Scholar]
- 8.Cagampang GB, Perez CM, Juliano BO. A gel consistency test for eating quality of rice. J Sci Food Agric. 1973;24:1589–1594. doi: 10.1002/jsfa.2740241214. [DOI] [PubMed] [Google Scholar]
- 9.Juliano B. Rice chemistry and technology. Saint Paul: American Association of Cereal Chemists; 1985. Criteria and test for rice grain quality; pp. 443–513. [Google Scholar]
- 10.Wang ZY, et al. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995;7:613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, et al. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China C Life Sci. 2003;46:661–668. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- 12.Hannah LC, James M. The complexities of starch biosynthesis in cereal endosperms. Curr Opin Biotechnol. 2008;19:160–165. doi: 10.1016/j.copbio.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- 14.James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Curr Opin Plant Biol. 2003;6:215–222. doi: 10.1016/s1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 15.Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ball S, et al. From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- 17.Dinges JR, Colleoni C, Myers AM, James MG. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 2001;125:1406–1418. doi: 10.1104/pp.125.3.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulton DC, et al. Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato. J Biol Chem. 2002;277:10834–10841. doi: 10.1074/jbc.M111579200. [DOI] [PubMed] [Google Scholar]
- 19.Van de Wal M, et al. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem. 1998;273:22232–22240. doi: 10.1074/jbc.273.35.22232. [DOI] [PubMed] [Google Scholar]
- 20.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 22.Thornsberry JM, et al. Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 23.Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ESt. Genetic diversity and selection in the maize starch pathway. Proc Natl Acad Sci USA. 2002;99:12959–12962. doi: 10.1073/pnas.202476999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson LM, et al. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell. 2004;16:2719–2733. doi: 10.1105/tpc.104.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 26.Septiningsih EM, Trijatmiko KR, Moeljopawiro S, McCouch SR. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet. 2003;107:1433–1441. doi: 10.1007/s00122-003-1376-z. [DOI] [PubMed] [Google Scholar]
- 27.Mikami I, et al. Allelic diversification at the Wx locus in landraces of Asian rice. Theor Appl Genet. 2008;116:979–989. doi: 10.1007/s00122-008-0729-z. [DOI] [PubMed] [Google Scholar]
- 28.He P, et al. Genetic analysis of rice grain quality. Theor Appl Genet. 1999;98:502–508. [Google Scholar]
- 29.Aluko G, et al. QTL mapping of grain quality traits from the interspecific cross Oryza sativa x O. glaberrima. Theor Appl Genet. 2004;109:630–639. doi: 10.1007/s00122-004-1668-y. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitt SR, Buckler ESI. Using natural allelic diversity to evaluate gene function. In: Grotewold E, editor. Plant functional genomics: Methods and protocols. Vol. 236. Totowa, New Jersey: Humana Press; 2003. pp. 123–139. [DOI] [PubMed] [Google Scholar]
- 32.Olsen KM, et al. Selection under domestication: Evidence for a sweep in the rice Waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, et al. Molecular marker-assisted selection for improved cooking and eating quality of two elite parents of hybrid rice. Crop Sci. 2006;46:2354–2360. [Google Scholar]
- 34.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy OJ, Vekemans X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- 37.Bradbury PJ, et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 38.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 39.SAS Institute. Cary, North Carolina: SAS Institute; 1999. SAS/STAT User's Guide Version 8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.