Abstract
Isothermal titration calorimetry has been applied to characterize the thermodynamics of ligand binding to wild-type lactose permease (LacY) and a mutant (C154G) that strongly favors an inward facing conformation. The affinity of wild-type or mutant LacY for ligand and the change in free energy (ΔG) upon binding are similar. However, with the wild type, the change in free energy upon binding is due primarily to an increase in the entropic free energy component (TΔS), whereas in marked contrast, an increase in enthalpy (ΔH) is responsible for ΔG in the mutant. Thus, wild-type LacY behaves as if there are multiple ligand-bound conformational states, whereas the mutant is severely restricted. The findings also indicate that the structure of the mutant represents a conformational intermediate in the overall transport cycle.
The lactose permease of Escherichia coli (LacY), a member of the Major Facilitator Superfamily of membrane transport proteins, couples the stoichiometric translocation of a galactoside and an H+ (reviewed in Refs. 1, 2). As such, LacY utilizes free energy stored in an electrochemical H+ gradient (ΔμH+) to drive accumulation of galactosidic sugars against a concentration gradient. Conversely, in the absence of ΔμH+, LacY utilizes free energy released from downhill translocation of galactosides to drive uphill translocation of H+ with generation of ΔμH+, the polarity of which depends on the direction of the sugar gradient. Composed of 417 amino acid residues, ~70% of which are hydrophobic, LacY has been solubilized, purified, and reconstituted into proteoliposomes in a fully functional state. Because galactoside gradients by themselves generate a ΔμH+ of either polarity, it has been postulated that the primary driving force for turnover is binding and dissociation of sugar on either side of the membrane.
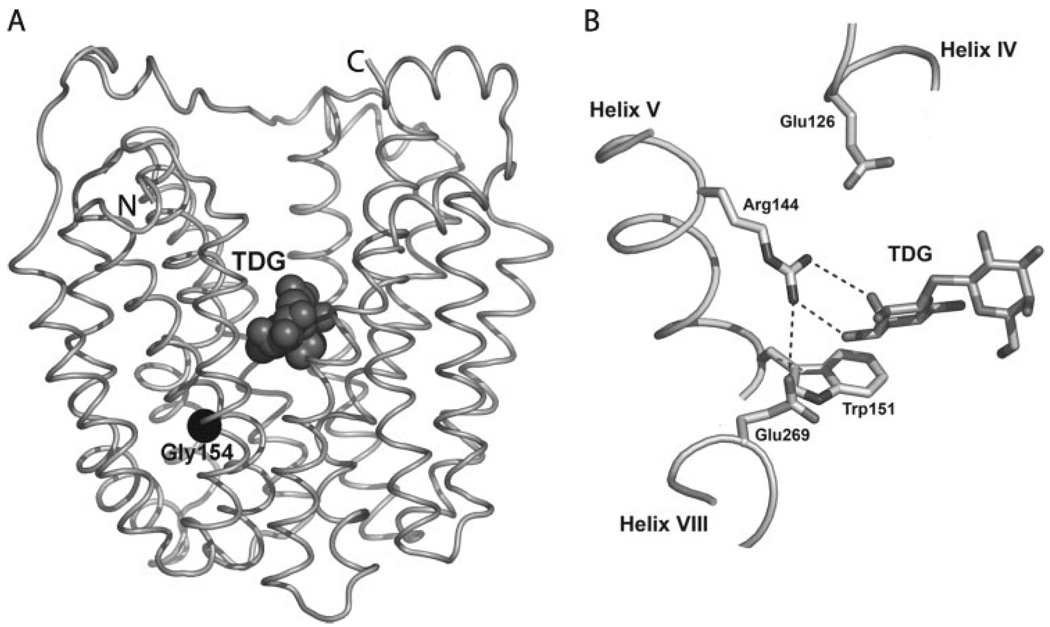
An x-ray structure of mutant C154G, which binds ligand as well as wild-type LacY but catalyzes very little transport and is compromised conformationally (3–6), has been solved in an inward facing conformation with bound ligand (Fig. 1) (7). Notably, wild-type LacY has the same global fold (2, 8). LacY contains 12 transmembrane helices organized in two pseudo-symmetrical α-helical bundles. The N- and C-terminal 6-helix domains form a large internal cavity open to the cytoplasm. A single sugar-binding site is observed at the apex of the cavity near the approximate middle of the molecule. The structure confirms many previous findings obtained from site-directed biochemical and biophysical studies (1, 2).
FIGURE 1.
A, backbone representation of C154G LacY crystal structure with bound TDG (Protein Data Bank accession code 1PV7; Ref. 7). LacY is viewed parallel to the membrane. Twelve transmembrane helices are organized in two pseudo-symmetrical α-helical bundles. The N- and C-terminal 6-helix domains form a large internal cavity open to the cytoplasm. TDG, represented by dark gray spheres, is observed at the apex of the cavity near the approximate middle of the molecule. B, aminoacyl side chains involved in galactopyranoside binding and/or proton translocation (Glu269 is involved in both).
Only a handful of side chains are essential with respect to the symport mechanism. Arg144 (helix V) forms a bi-dentate H-bond with the O4 and O3 atoms of the galactopyranosyl ring (7), confirming the critical role of this residue in sugar binding and recognition (2). Glu126 (helix IV), another important residue for binding, is in close proximity to Arg144 and may interact with the O4, O5, or O6 atoms of the galactopyranosyl ring via water molecules. An aromatic residue at position 151 (helix V), preferably Trp, is irreplaceable for sugar binding (9), stacking hydrophobically with the galactopyranosyl ring (10, 11). Glu269 (helix VIII) in the C-terminal domain, which is involved in both sugar binding and H+ translocation and interacts with the O3 atom of the galactopyranosyl ring, forms a salt bridge with Arg144 and is in close proximity to Trp151 (7, 11–13). His322 (helix X), Glu325 (helix X), and Arg302 (helix IX), as well as Tyr236 (helix VII), are likely involved directly in H+ translocation (2, 14). His322 may be the immediate H+ donor to Glu325, and Arg302 may interact with Glu325 to drive deprotonation (14, 15).
In the ligand-free state, the essential residues for substrate binding and specificity (Arg144, Glu126, and Glu269) are not in the correct configuration to bind substrate, and the following sequence of events has been suggested based upon structural changes (13). Sugar initially recognizes Trp151 through nonspecific hydrophobic stacking between the galactopyranosyl and indole rings (9, 10). This interaction orients the galactopyranosyl moiety for recognition by Arg144, Glu126, and Glu269 (i.e. induced fit) and disrupts the salt bridge between Arg144 and Glu126, and a bi-dentate H-bond is formed with Arg144 through the O4 and O3 groups on the galactopyranosyl ring. Subsequently, protonated Glu269 moves out of a relatively hydrophobic environment, deprotonates, and forms a salt bridge with Arg144 and an H-bond with sugar to complete ligand binding, which triggers the global conformational change that allows accessibility of the binding site to the other side of the membrane.
Here, we report a thermodynamic characterization of sugar binding with wild-type LacY and the C154G mutant by using isothermal titration calorimetry (ITC),2 a technique that allows measurement of changes in free energy (ΔG), enthalpy (ΔH), entropic free energy component (TΔS), and heat capacity (ΔCp) (16). β-Galactopyranosyl 1-thio-β-d-galactopyranoside (TDG) or p-nitrophenyl α-d-galactopyranoside (NPG) bind to wild-type LacY, and the entropic free energy component is the major component that contributes to ΔG. In striking contrast, enthalpy alone is responsible for ΔG in the mutant. Thus, multiple ligand-bound conformational states are present in the wild-type protein, whereas the mutant is severely restricted with respect to conformational changes.
EXPERIMENTAL PROCEDURES
Materials
E. coli XL-1 Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZDM15 Tn10 (Tetr)]) was obtained from Stratagene (La Jolla, CA). TDG and NPG were purchased from Sigma. Isopropyl 1-thio-β-d-galactopyranoside and n-dodecyl β-d-maltopyranoside (DDM) were purchased from Calbiochem. TALON Superflow Metal Affinity Resin was from BD Biosciences, and Vivaspin 20 concentrators (30-kDa cutoff) were purchased from Vivascience (Sartorius AG, Goettingen, Germany). Micro BCA protein assay kits were from Pierce. All other materials were of reagent grade and obtained from commercial sources.
Protein Expression and Purification
Plasmid pT7–5 encoding wild-type or C154G LacY with a His6 tag at the C terminus was used essentially as described (5) with minor modifications. Briefly, wild-type and C154G LacY were expressed in E. coli XL-1 Blue after induction with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside. Membranes were prepared, washed with 5.0 m urea, and solubilized in 2% DDM. LacY was then purified by TALON Superflow metal affinity chromatography. The column was washed with 15 mm imidazole to elute weakly bound contaminants, and LacY was eluted with 150 mm imidazole, dialyzed against 50 mm NaPi (pH 7.5)/0.01% DDM, concentrated with a Vivaspin 20 concentrator (30-kDa cutoff), frozen in liquid nitrogen, and stored at −80 °C. The Micro BCA method was used to measure protein concentration. All LacY preparations were at least 90–95% pure as judged by silver staining after sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
ITC
Heat flow resulting from binding of given ligands and associated conformational changes was measured by using a high sensitivity VP-ITC instrument (Microcal, LLC, Northampton, MA). Titration calorimetry experiments were performed as follows. Purified LacY proteins were dialyzed extensively against the target buffer (either 50 mm NaPi, (pH 7.5)/0.01% DDM or 50 mm Tris-HCl (pH 7.5)/0.01% DDM), and the sugars were dissolved in the last dialysate in order to minimize heats of dilution upon injection into the protein solution. Prior to use, solutions were degassed under vacuum to eliminate air bubbles. Each test ligand (0.25–2.0 mm) was injected into the calorimeter cell (volume 1.43 ml containing 50 – 100 µm protein) at 5-min intervals. The titration cell was stirred continuously at 300 rpm. To measure heats of dilution, ligands (at the same concentration used for test titrations) were titrated against reaction buffer without protein. Phenyl α-d-glucopyranoside (0.5 mm), which does not bind to LacY (17), was used as a negative control and gives no heat change after the addition to wild-type or C154G LacY. N-ethylmaleimide (5 mm)-inactivated LacY, which does not bind ligand, also exhibits no heat change. Each measurement was repeated a minimum of three times, and the data did not vary by more than 10%.
The heat produced from each injection was determined by integrating the heat flow tracings. Heats of dilution in the presence of complex obtained after saturation were used to correct for heat of dilution. The heat of binding for each injection was obtained by integrating the area under the peak using ORIGIN (version 7.0; Microcal LLC). The heat evolved (Q) on addition of ligand is represented by the equation (18, 19) Q = V0ΔH[M]t Ka[L]/(1 + Ka[L]), where V0 is the volume of the cell, ΔH is the enthalpy of binding per mole of ligand, [M]t is the total macromolecule concentration including bound and free fractions, Ka is the association constant, and [L] is the free ligand concentration. ΔH and Ka were determined directly from the isotherm. ΔG(change in free energy) and TΔS (change in the entropic free energy component) were computed from the following equations, ΔG = − RT lnKa and ΔG = ΔH − TΔS, where R is the gas constant and T is the absolute temperature in degrees Kelvin.
Binding constants (Ka) of wild-type LacY or mutant C154G for TDG or NPG were calculated from experimental data by using the differential heat mode equation or integral heat mode (20). The integral heat mode calculation resulted in lower Ka values with only 5–10% lower estimates for the ΔG calculations. For consistency, only the differential heat mode calculation program built into the Microcal Origin software package was used.
Calorimetric titrations with NPG were carried out at various temperatures ranging from 6 to 25 °C, and the change in heat capacity (ΔCp) was determined from the equation ΔCp = ΔH/dT (21). The values of ΔCp were obtained from the slopes of a linear regression analysis of ΔH versus T. All other titrations were carried out at 20 °C.
To measure changes in protonation upon binding, apparent binding enthalpies were measured in NaPi (ΔHapp(Pi)), or Tris (ΔHapp-(Tris)) at pH 7.5. The two buffers have very different enthalpies of protonation. The number of protons involved was calculated from Equation 1 (19)
| (Eq.1) |
where ΔHiTris (the enthalpy of ionization for Tris) is 11.3 kcal·mol−1, ΔHiPi (the enthalpy of ionization for Pi) is 1.13 kcal·mol−1 (22, 23), and nH+ is the number of protons taken up or released.
RESULTS
Ligand Binding Affinity
The heat associated with binding of lactose analogs TDG and NPG was measured by ITC (Fig. 2). Binding constants (Ka) of wild-type LacY or mutant C154G for TDG or NPG were calculated from experimental data and converted into dissociation constants (Kd = 1/Ka). Values of Kd are 83 µm (Ka = 1.2 × 104 m−1) for TDG binding to wild-type LacY and 33 µm (Ka = 3.0 × 104m−1) for C154G LacY at 20 °C (293 K; Table 1). NPG, a ligand with higher affinity (24, 25), binds to wild-type or C154G LacY with a Kd of 13 µm(Ka = 7.8 × 104m−1) or 5 µm(Ka = 2.1 × 105 m−1), respectively (Table 2), ~7-fold better affinity than TDG with either protein (Table 1). Relative to the wild type, C154G LacY has 2–3 times better affinity for both ligands, as shown previously (4–6), a difference that is hardly significant energetically.
FIGURE 2. ITC of ligand binding to wild-type LacY and the C154G mutant.
Experiments were conducted at 20 °C as described under “Experimental Procedures.” A series of injections of 10 or 5 µl of 2mm TDG (A and B, respectively) or 10 µl of 0.25mm NPG (C and D) were made into 1.43 ml of 100 µm wild-type LacY (A and C) or 50 µm C154G LacY (B and D) in 50 mm NaPi (pH 7.5)/0.01% DDM. Raw data are shown in the upper panels, and the area under each peak corresponds to the heat change (absorbed as in panel A or released as in panels B–D) upon injection of a single aliquot of TDG or NPG. The lower panels show plots of total energy exchanged (kcal·mol−1 of injectant) as a function of the molar ratio of ligand to protein (i.e. the integrated data obtained from the raw data after subtraction of the heat of dilution). The solid lines in the bottom panels represent the best-fit curves to the data, using the one-site model from Microcal Origin. The thermodynamic parameters derived are listed in Table 1 and Table 2.
TABLE 1. Thermodynamic parameters for TDG binding.
Experiments were carried out in NaPi, pH 7.5, at 20 °C (293 K) as described under“Experimental Procedures.”
| LacY | Ka | Kda | ΔH | ΔGb | TΔSc | ΔS |
|---|---|---|---|---|---|---|
| M −1 | µM | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | cal·mol−1·K−1 | |
| Wild type | 1.2 × 104 | 83 | 0.3 | −5.4 | 5.7 | 19.5 |
| C154G | 3.0 × 104 | 33 | −5.6 | −6.0 | 0.4 | 1.3 |
Kd = 1/Ka.
ΔG was calculated by the equation ΔG = −RT lnKa.
TΔS was calculated by the equation TΔS = ΔH − ΔG.
TABLE 2. Thermodynamic parameters for NPG binding.
Experiments were carried out in NaPi, pH 7.5, as described under “Experimental Procedures.”
| LacY | T | Ka | Kda | ΔH | ΔGb | TΔSc | ΔS | ΔCpd |
|---|---|---|---|---|---|---|---|---|
| K | M−1 | µM | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | cal·mol−1·K−1 | cal·mol−1·K−1 | |
| Wild type | 279 | 8.4 × 104 | 12 | −0.3 | −6.3 | 6.0 | 21.6 | −156.5 |
| 280 | 7.5 × 104 | 13 | −0.3 | −6.3 | 6.0 | 21.2 | ||
| 288 | 1.0 × 105 | 10 | −1.2 | −6.6 | 5.4 | 18.9 | ||
| 293 | 7.8 × 104 | 13 | −2.2 | −6.6 | 4.4 | 15.0 | ||
| 298 | 7.6 × 104 | 13 | −3.3 | −6.7 | 3.4 | 11.5 | ||
| C154G | 279 | 4.6 × 105 | 2 | −11.6 | −7.2 | −4.4 | −15.8 | −124.5 |
| 288 | 3.0 × 105 | 3 | −13.4 | −7.2 | −6.2 | −21.4 | ||
| 293 | 2.1 × 105 | 5 | −13.5 | −7.1 | −6.4 | −21.8 | ||
| 298 | 1.7 × 105 | 6 | −14.1 | −7.1 | −7.0 | −23.5 |
Kd = 1/Ka.
ΔG was calculated by the equation ΔG = −RT lnKa.
TΔS was calculated by the equation TΔS = ΔH − ΔG.
ΔCp = ΔH/dT was determined from the slope of the linear fit in Fig. 4.
Thermodynamic Parameters for TDG Binding
Thermodynamic parameters, the enthalpy change (ΔH), the free energy change (ΔG), and the entropic free energy component change (TΔS) accompanying binding of TDG were calculated from experimental data. Values of ΔG are similar for TDG binding to wild-type LacY (−5.4 kcal·mol−1) and to C154G LacY (−6.0 kcal·mol−1) (Table 1). However, the contributions from ΔH and TΔS are markedly different. For wild-type LacY, a change in the entropic free energy component (TΔS = 5.7 kcal·mol−1) makes the sole contribution to ΔG, whereas a change in enthalpy (ΔH = −5.6 kcal·mol−1) is the major contributor to the interaction between C154G LacY and TDG with an almost negligible change in the entropic free energy component (TΔS = 0.4 kcal·mol−1) (Fig. 2, A and B, and Table 1). Furthermore, ligand binding with the C154G mutant is exothermic, and the magnitude of ΔH is much larger (−5.6 kcal·mol−1) than for wild-type LacY (0.3 kcal·mol−1). Thus, the interaction between TDG and C154G LacY is maintained by more or stronger interactions relative to the wild-type LacY.
Thermodynamic Parameters for NPG Binding
At 20 °C (293 K), the interactions of NPG with both wild-type and C154G LacY are exothermic (Figs. 2, C and D). Binding enthalpy is weakly negative for wild-type LacY (ΔH = −2.2 kcal·mol−1), whereas the entropic free energy component is positive and large in magnitude (TΔS = 4.4 kcal·mol−1) (Table 2). Thus, binding between NPG and wild-type LacY is characterized by both entropic and enthalpic changes, but the entropic change is clearly the dominant component. In contrast, binding of NPG to C154G LacY is characterized by a large negative enthalpy change (ΔH = −13.5 kcal·mol−1), which is compensated by a large but negative change in the entropic free energy component (TΔS = −6.4 kcal·mol−1) (Table 2), indicating a substantial decrease in conformational flexibility upon ligand binding.
Over the range of temperatures tested (6–25 °C or 279–298 K), the thermodynamic parameters exhibit similar characteristics (Table 2). TΔS is the major contributor to the interaction of NPG with wild-type LacY, whereas ΔH contributes exclusively to NPG binding with C154G LacY. For all of these interactions, ΔH contributes to ΔG(Fig. 3, Table 2). However, in comparison with NPG binding to wild-type LacY, ΔH of the interaction between NPG and C154G LacY remains negative and large (Table 2, Fig. 3). Concomitantly, the entropic free energy component, TΔS, decreases due to tightening of the system, but ΔG changes little (Fig. 3, Table 2), indicating enthalpy/entropy compensation in the mutant (26).
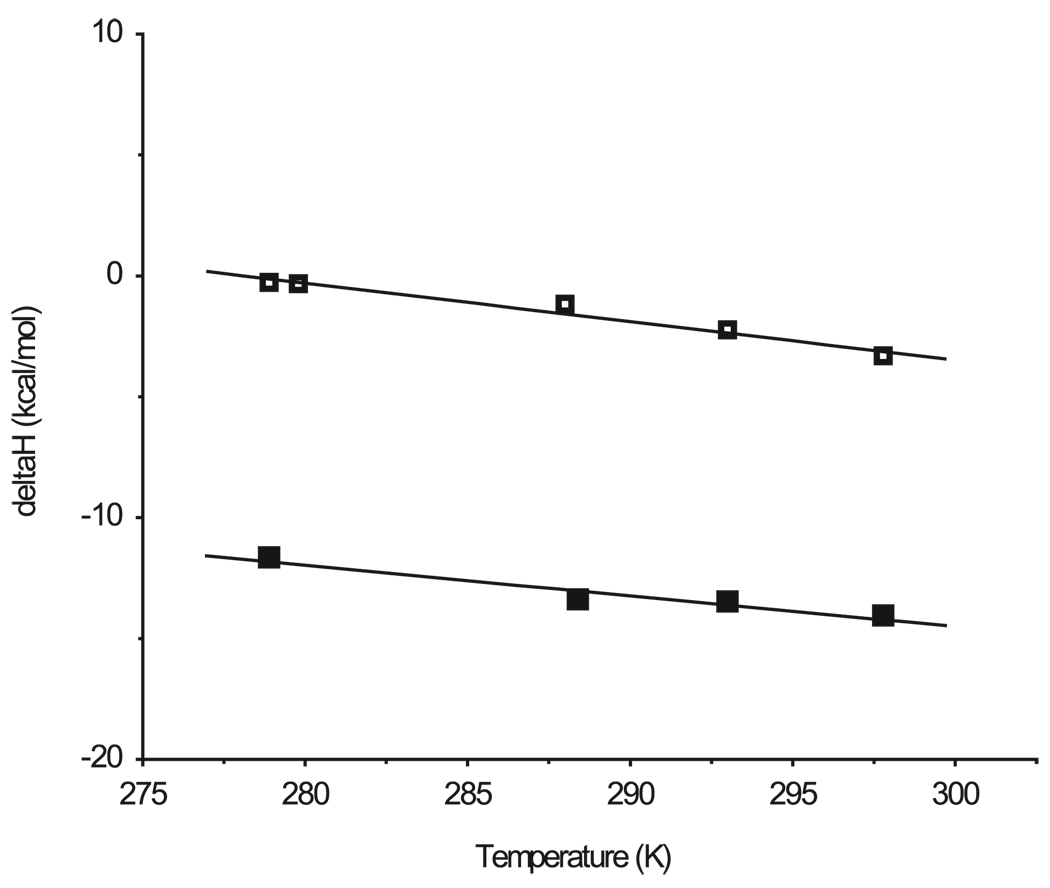
FIGURE 3. Temperature dependence of thermodynamic parameters for NPG binding to wild-type (A) or C154G LacY (B).
ΔH (■),−TΔS (▲), and ΔG (●) for NPG binding are plotted against temperature. ΔH was obtained directly from ITC (Table 2), and ΔG and −TΔS were calculated from equations ΔG = −RTlnKa and ΔG = ΔH − TΔS, respectively.
From the change of ΔH with temperature (Fig. 4), the change in heat capacity (ΔCp) is calculated to be −156.5 and −124.5 cal·mol−1·K−1 for NPG interaction with wild-type and C154G LacY, respectively (Table 2). Both exhibit a similar negative ΔCp, suggesting that the binding sites in both proteins have a similar interface (7, 13) and the decrease in the entropic free energy component for NPG binding to C154G LacY is due to decreased conformational flexibility.
FIGURE 4. Enthalpy of NPG binding as a function of temperature.
ITC experiments were carried out as described under “Experimental Procedures” at the temperatures given. The values for ΔCp (Table 2) were obtained from the slopes of linear regression analyses of ΔH as a function of temperature. Wild type, □ C154G, ■.
NPG Binding and Protonation of LacY
All available evidence (see Ref. 2) indicates that protonation of LacY precedes ligand binding. Therefore, the thermodynamics of NPG binding was studied in phosphate (Pi) versus Tris. These two buffers have very different enthalpies of protonation (ΔHi Pi = 1.13 kcal·mol−1 and ΔHi Tris = 11.3 kcal·mol−1) (22) and can be used to determine whether ligand binding induces a change in the protonation state of LacY. NPG binding to wild-type LacY in Tris yields a ΔHapp(Tris) of −1.7 kcal·mol−1, whereas the same reaction in NaPi exhibits a ΔHapp(Pi) of −2.2 kcal·mol−1. NPG binding to C154G LacY yields a ΔHapp(Tris) of −13.9 kcal·mol−1 and a ΔHapp(Pi) of −13.5 kcal·mol−1 (Table 3). The number of protons (nH+) involved was calculated (19, 27) as 0.05 for NPG binding to wild-type LacY and −0.04 for NPG binding to C154G LacY. Clearly, these values approximate zero, indicating no protonation change at this step of the mechanism.
TABLE 3. Thermodynamic parameters for NPG binding in NaPi or Tris.
Experiments were carried out in NaPi or Tris, pH 7.5, as indicated, at 20 °C as described under “Experimental Procedures.”
| LacY | Buffer | ΔHapp | ΔGa | TΔSb | ΔS | nH+c |
|---|---|---|---|---|---|---|
| kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | cal·mol−1·K−1 | |||
| Wild type | Pi | −2.2 | −6.6 | 4.4 | 15.0 | 0.05 |
| Tris | −1.7 | −6.8 | 5.1 | 17.4 | ||
| C154G | Pi | −13.5 | −7.1 | −6.4 | −21.8 | −0.04 |
| Tris | −13.9 | −6.6 | −7.3 | −25.0 |
ΔG was calculated from the equation ΔG = −RT lnKa.
TΔS were calculated from the equation TΔS = ΔH − ΔG.
nH+ = [ΔHapp(Pi) − ΔHapp(Tris)]/[HiPi − ΔHiTris].
DISCUSSION
Wild-type LacY is a highly dynamic membrane transport protein in which ligand binding induces widespread conformational changes (1, 2, 13). In contrast, C154G LacY binds galactosidic sugars with relatively high affinity but is conformationally constrained and catalyzes very little translocation across the membrane (3–6). In addition, C154G LacY exhibits higher thermostability and has less tendency to aggregate than the wild type (5), which led to its crystallization and x-ray structure determination (7). Because the C154G mutation in helix V abuts Gly24 in helix I where the two helices cross at the approximate middle of the membrane and replacement of Gly24 with Cys rescues activity (6), it was suggested that tighter packing between helices V and I may be responsible for the properties of the C154G mutant. As shown previously (4, 5), the C154G mutation results in a small increase in affinity for TDG or NPG relative to wild-type LacY at all temperatures tested (Table 1 and Table 2). The effect may result from the tighter interactions between ligand and specific residues in the binding site. By allowing tighter packing between helices I and V, the C154G mutation may directly inhibit conformational flexibility near the binding site and indirectly alter positioning of residues that make direct contact with the sugar (e.g. Glu269, Trp151, Arg144, Glu126, Met23, and/or Asp237/Lys358) (6, 7, 13). In this study, the thermodynamics of binding between wild-type or C154G LacY and two galactosidic ligands are analyzed calorimetrically.
Remarkably, although the change in free energy of binding (ΔG) is similar for both proteins as expected, in wild-type LacY the change is due primarily to an increase in the entropic free energy component (TΔS), whereas in the C154G mutant an increase in enthalpy (ΔH) markedly predominates. Thus, ΔH for TDG binding is shifted from a small positive value in wild-type LacY to a relatively large negative value in C154G LacY (Table 1). The large negative value observed with the mutant suggests tighter packing upon ligand binding, which extends the conclusion that the C154G mutation causes decreased conformational flexibility (5, 6) and that the mutant favors a particular conformation(s). Thus, a specific conformer of C154G LacY is selected from the dynamic ensemble of conformations upon ligand binding.
Wild-type LacY is in a highly dynamic state (14, 28), and widespread conformational changes accompany ligand binding (2, 13). Therefore, the number of conformers for ligand-bound wild type is very likely greater than that for the unliganded protein, resulting in an increase in the entropic free energy component upon ligand binding. However, TΔS is reversed from positive in wild-type LacY to negative in the C154G mutant (Fig. 3 and Table 2). A negative change in the entropic free energy component is observed with H-bond formation, a decrease in the number of isoenergetic conformations, or a decrease in soft internal vibrational modes (29–31). Therefore, the ligand-bound mutant exhibits significantly increased conformational constraint(s). In other words, C154G LacY is compromised by the entropic free energy component, which restricts the number of conformers that can be occupied (32). With this restriction, the mutant is able to undergo the induced-fit phenomenon associated with ligand binding (13) but can hardly overcome the energy barrier to achieve the outward facing conformation and catalyzes translocation extremely poorly (Fig. 5).
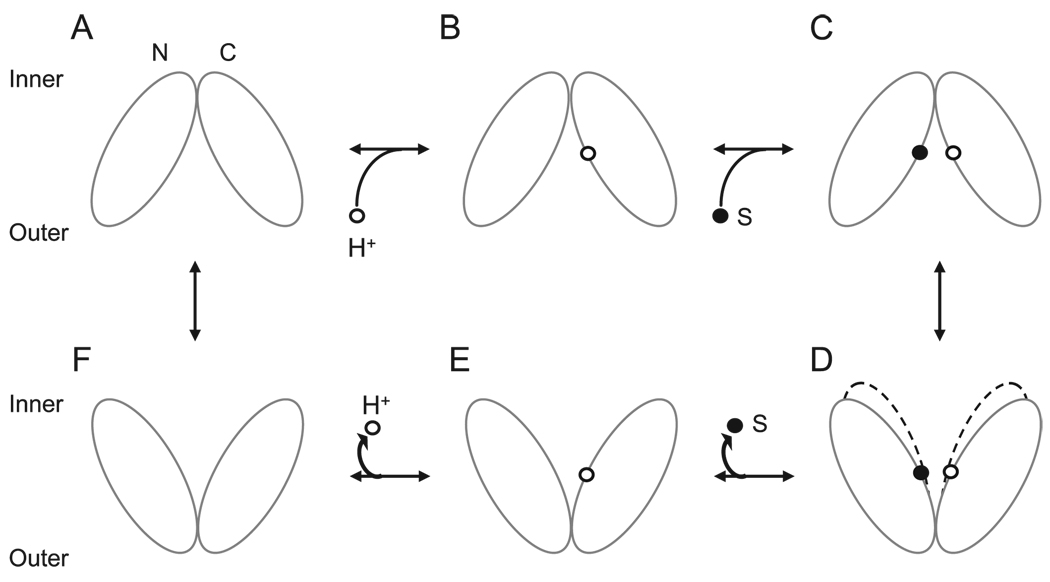
FIGURE 5. Transport cycle of wild-type and C154G LacY.
Deprotonated wild-type LacY in outward facing conformation (A) is unstable, accepts a proton (B), and binds substrate (C) on the outer face. Substrate binding induces a global conformational change, resulting in the inward facing conformation (D). Substrate is released (E), and the proton is released (F) to the inner face. The protein returns to the outward facing conformation (A). C154G LacY in the inward facing conformation (D) binds sugar from the inner face but, as indicated by the broken lines, is conformationally constrained with respect to the transition to the outward facing conformer. N- and C-terminal domains are shown as white ovals; bound proton and sugar are represented as small white and black circles, respectively.
Finally, as observed by using Pi or Tris, buffers with very different enthalpies of protonation, no significant change in protonation state of either wild-type or C154G LacY is observed upon ligand binding. The observation is consistent with a number of lines of evidence (2) indicating that protonation of LacY occurs prior to ligand binding and that the apparent pKa for TDG binding is between pH 9 and 10.3
Acknowledgments
We thank Junichi Sugihara for help with fermentation and Lan Guan, Shushi Nagamori, and Natalia Ermolova for advice and discussion. We also thank Lan Guan for critically reading the manuscript. All the ITC experiments were performed on the VP-ITC instrument in Prof. J. Fraser Stoddart’s laboratory, and we thank Dr. Diego Benitez and Dr. Taichi Ikeda for help with the ITC experiments.
Footnotes
This work was supported by National Institutes of Health Grant DK51131 (to H. R. K.), DK069463, GM073210 and GM07929. And also NSF grant 0450970.
The abbreviations used are: ITC, isothermal titration calorimetry; LacY, lactose permease; TDG, β-d-galactopyranosyl 1-thio-β-d-galactopyranoside; NPG, p-nitrophenyl α-d-galactopyranoside; DDM, n-dodecyl β-d-maltopyranoside.
I. N. Smirnova, V. N. Kasho, and H. R. Kaback, unpublished observations.
REFERENCES
- 1.Kaback HR. C. R. Biol. 2005;328:557–567. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Guan L, Kaback HR. Annu. Rev. Biophys. Biomol. Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menick DR, Sarkar HK, Poonian MS, Kaback HR. Biochem. Biophys. Res. Commun. 1985;132:162–170. doi: 10.1016/0006-291x(85)91002-2. [DOI] [PubMed] [Google Scholar]
- 4.van Iwaarden PR, Driessen AJ, Lolkema JS, Kaback HR, Konings WN. Biochemistry. 1993;32:5419–5424. doi: 10.1021/bi00071a017. [DOI] [PubMed] [Google Scholar]
- 5.Smirnova IN, Kaback HR. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 6.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 7.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 8.Guan L, Smirnova IN, Verner G, Nagamoni S, Kaback HR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1723–1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan L, Hu Y, Kaback HR. Biochemistry. 2003;42:1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez-Ibar JL, Guan L, Weinglass AB, Verner G, Gordillo R, Kaback HR. J. Biol. Chem. 2004;279:49214–49221. doi: 10.1074/jbc.M407408200. [DOI] [PubMed] [Google Scholar]
- 12.Weinglass A, Whitelegge JP, Faull KF, Kaback HR. J. Biol. Chem. 2004;279:41858–41865. doi: 10.1074/jbc.M407555200. [DOI] [PubMed] [Google Scholar]
- 13.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. EMBO J. 2006 doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaback HR, Sahin-Toth M, Weinglass AB. Nat. Rev. Mol. Cell. Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 15.Sahin-Tóth M, Kaback HR. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundle DR, Sigurskjold BW. Methods Enzymol. 1994;247:288–305. doi: 10.1016/s0076-6879(94)47022-7. [DOI] [PubMed] [Google Scholar]
- 17.Sahin-Tóth M, Akhoon KM, Runner J, Kaback HR. Biochemistry. 2000;39:5097–5103. doi: 10.1021/bi0000263. [DOI] [PubMed] [Google Scholar]
- 18.Indyk L, Fisher HF. Methods Enzymol. 1998;295:350–364. doi: 10.1016/s0076-6879(98)95048-0. [DOI] [PubMed] [Google Scholar]
- 19.Pierce MM, Raman CS, Nall BT. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 20.Sigurskjold BW, Altman E, Bundle DR. Eur. J. Biochem. 1991;197:239–246. doi: 10.1111/j.1432-1033.1991.tb15904.x. [DOI] [PubMed] [Google Scholar]
- 21.Jelesarov I, Bosshard HR. J. Mol. Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Beres L, Sturtevant JM. Biochemistry. 1971;10:2120–2126. doi: 10.1021/bi00787a025. [DOI] [PubMed] [Google Scholar]
- 23.Christensen JJ, Hansen LD, Izatt RM. Handbook of Proton Ionization Heats and Related Thermodynamic Quantities. New York: John Wiley; 1976. pp. 148–159. [Google Scholar]
- 24.Rudnick G, Schuldiner S, Kaback HR. Biochemistry. 1976;15:5126–5131. doi: 10.1021/bi00668a028. [DOI] [PubMed] [Google Scholar]
- 25.Sahin-Tóth M, Lawrence MC, Nishio T, Kaback HR. Biochemistry. 2001;43:13015–13019. doi: 10.1021/bi011233l. [DOI] [PubMed] [Google Scholar]
- 26.Dunitz JD. Chem. Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 27.Raman CS, Allen MJ, Nall BT. Biochemistry. 1995;34:5831–5838. doi: 10.1021/bi00017a015. [DOI] [PubMed] [Google Scholar]
- 28.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturtevant JM. Proc. Natl. Acad. Sci. U. S. A. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janin J. Structure. 1997;5:473–479. doi: 10.1016/s0969-2126(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Pawley NH, Nicholson LK. J. Mol. Biol. 2001;313:873–887. doi: 10.1006/jmbi.2001.5083. [DOI] [PubMed] [Google Scholar]
- 32.Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, van der Merwe PA. Immunity. 1999;10:357–365. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]