Abstract
The specific recognition of carbohydrates by lectins plays a major role in many cellular processes. Galectin-1 belongs to a family of 15 structurally related β-galactoside binding proteins that are able to control a variety of cellular events, including cell cycle regulation, adhesion, proliferation, and apoptosis. The three-dimensional structure of galectin-1 has been solved by x-ray crystallography in the free form and in complex with various carbohydrate ligands. In this work, we used a combination of two-dimensional NMR titration experiments and molecular-dynamics simulations with explicit solvent to study the mode of interaction between human galectin-1 and five galactose-containing ligands. Isothermal titration calorimetry measurements were performed to determine their affinities for galectin-1. The contribution of the different hexopyranose units in the protein-carbohydrate interaction was given particular consideration. Although the galactose moiety of each oligosaccharide is necessary for binding, it is not sufficient by itself. The nature of both the reducing sugar in the disaccharide and the interglycosidic linkage play essential roles in the binding to human galectin-1.
Introduction
Carbohydrates are involved in a wide range of cellular processes, including cell recognition, cell adhesion, and cell differentiation, and are considered as the third class of information-encoding biological macromolecules (1). Their biological activities are usually mediated by their interactions with proteins at the surface of cells (2). Mammalian glycans rely on a group of 10 common monosaccharide units, resulting in a large number of possible hexasaccharides, of which 2000 structures of N-linked glycans are known. The complexity of the glycome is increased by the mode of attachment to glycoproteins and by further chemical modification, such as methylation, sulfation, acetylation, and phosphorylation. This diversity poses a real challenge to investigators attempting to study the role of glycosylation in biological phenomena (3). In addition, the flexibility of oligo- and polysaccharides allows them to adapt to a variety of interactions in biological recognition processes (4). Therefore, characterizing the dynamic properties of carbohydrates in complex with their binding partners is a crucial step toward gaining a better understanding of carbohydrate recognition.
Galectins constitute a class of carbohydrate-binding proteins (lectins) that share both a conserved sequence in the carbohydrate recognition domain (CRD) and an affinity for β-galactosides. They are found in all taxa of the living world. The galectin family is composed of 15 members subdivided into three categories: 1), the prototype (galectin-1, 2, 5, 7, 10, 11, 13, 14, and 15); 2), the tandem repeat characterized by the presence of two distinct CRDs joined by a linker peptide (galectin-4, 6, 8, 9, and 12); and 3), the chimera type, represented by the only member, galectin-3 (5).
All galectin ligands share a β-galactose moiety at the nonreducing end (6–8). However, biochemical studies have demonstrated that there is a higher level of complexity involved for carbohydrate recognition by the galectins than just the primary requirement of the galactose sugar. The oligosaccharide interaction can be very specific, as various galectins interact with a different range of galactose oligosaccharide ligands (2,9). Studies indicating that galectins are implicated in cancer development (10–12) have opened the way for the development of new therapeutic strategies to combat cancer (13–16). Gaining a detailed description and understanding of the protein-carbohydrate interaction is a key step in the development of new drugs.
The structure of galectin-1 has been characterized by x-ray crystallography (17) as a dimer containing two identical monomers related by a twofold symmetry axis. The folding of the homodimer involves a β-sandwich that consists of two antiparallel β-sheets as found in most carbohydrate-binding modules (18). The same β-sheets are in continuity in the dimer and make two mega β-sheets stabilizing the dimer. The dimer interface is formed by the N- and C-terminal of each monomer. The structure of galectin-1 has been solved in complex with various oligosaccharides, revealing a highly conserved core-binding site for lactose, N-acetyllactosamine, and biantennary saccharides of N-acetyllactosamine with features in agreement with specificity studies (19,20). The carbohydrate-binding sites (CBS) are located at the top of each monomer on the same face of the dimer. Each CBS is formed by a cavity that is approximately long enough to hold a linear tetrasaccharide (Fig. 1). One can schematically describe the galectin CBS as having five subsites (subsites A–E), which is supported by its specificity for small saccharides (9,21). In this model, subsite C corresponds to the actual β-galactoside-binding site of the galectins, and subsite D contributes to the second part of the conserved core disaccharide-binding site (Fig. 1). The shape of the binding site varies among galectins, suggesting that the same extensions would not be bound equally by the different galectins. Moreover, there are also variations in binding preference on the reducing side of the terminal galactose residue corresponding to subsites D–E in Fig. 1 (21). The structurally less defined subsite E can accommodate possible interactions with moieties linked at the reducing end of the saccharide in subsite D.
Figure 1.
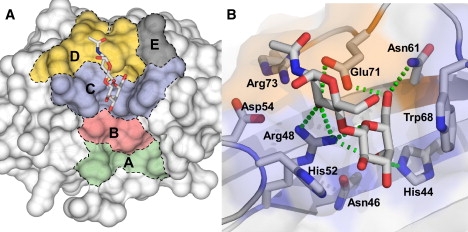
Surface representation of the CBS of human galectin-1. (A) The binding site can be divided into five subsites (subsites A–E) that can interact with the oligosaccharide ligands (21). The subsites are defined as follows: subsite A: Tyr-119, Ala-121, Ala-122, and Asp-123 (green); subsite B: Val-31, Leu-32, Asn-33, Leu-34, and Gly-35 (pink); subsite C: His-44, Phe-45, Asn-46, Arg-48, His-52, and Trp-68 (blue); subsite D: Gly-53, Asp-54, Val-59, Asn-61, Glu-71, and Arg-73 (orange); and subsite E: Gly-69 and Thr-70 (gray). An N-acetyllactosamine oligosaccharide ligand is shown inside the CBS to illustrate the binding to subsites C and D of the disaccharide in x-ray structures of galectin-1. (B) Detail of the interaction between the carbohydrate ligand and the protein. Residues from subsites C and D are highlighted. Hydrogen bonds are shown as dashed green lines. (PDB structure 1W6P from Lopez-Lucendo et al. (17).)
The carbohydrate-galectin interaction is characterized by a stacking interaction between the sugar ring and an aromatic residue in the protein-binding site, and by a series of specific hydrogen bonds between polar groups of the sugars and protein backbone and side-chain atoms (22–24). In the case of galectin-1, the main interaction involves a hydrogen bond between His-44 and Arg-48 with O4 of galactose residue. Aromatic residue Trp-68 is conserved within the galectin family and is responsible for positioning the carbohydrate ligand inside the cavity through a stacking interaction (17). The presence of an aromatic amino acid residue (Trp, Tyr, or Phe) near the nonpolar face of galactose is a common feature of galactose-binding lectins (24).
In a previous study, molecular dynamics (MD) simulations of bovine galectin-1 in complex with several N-acetyllactosamine-containing ligands were performed (25). The MD results indicated that bovine galectin-1 can accommodate different oligosaccharidic substituents at the nonreducing terminus of the oligosaccharide extending into the remainder of a characteristic surface groove (subsite B). Here, we used MD simulations in explicit water with a combination of the Charmm-22 (26) and GLYCAM06 (27) parameter sets to investigate the binding of galactose monosaccharide and four galactose-containing disaccharides to human galectin-1. The disaccharides differed in terms of the nature of the reducing unit (Glc, GlcNAc, or GalNAc) and interglycosidic linkage between the two sugars (β1→4/β1→3). We analyzed the MD trajectories in the light of NMR and isothermal titration calorimetry (ITC) data by monitoring structural and energetic properties such as atom and residue fluctuations, hydrogen bonds, the stability of the ligands within the cavity, and the interaction energy between the carbohydrates and galectin-1 residues. The analysis of the different trajectories revealed that MD simulations could be used to discriminate between high-affinity ligands (such as lactose, N-acetyllactosamine, and lacto-N-biose) and low-affinity ligands (such as galactose and galacto-N-biose), and to determine the contribution of the different hexopyranose units of the oligosaccharide to the binding of human galectin-1.
Materials and Methods
Galectin-1 expression and purification
A construct including a 6-His sequence added at the C-terminal extremity of galectin-1 was obtained. His6-galectin-1 was overexpressed in Escherichia coli M15. Protein production was induced using 1 mM isopropyl-β-D-thiogalactopyranoside for 4 h after growing to OD600nm 0.6. Recombinant galectin-1 was purified using a cobalt affinity column (TALON metal affinity). The protein was eluted with the use of imidazole buffer (150 mM). Galectin-1 was dialyzed overnight in the appropriate buffer for NMR or ITC experiments. 15N-labeling of the protein was achieved with the use of 15N-ammonium chloride added to M9 media.
Chemical compounds
D-galactose was obtained from Prolabo, D-lactose, and N-acetyl-D-lactosamine were obtained from Sigma (www.sigmaaldrich.com, Saint Quentin Fallavier, France), and galacto-N-biose (N-acetyl-neogalactosamine) and lacto-N-biose (N-acetyl-neolactosamine) were purchased from Dextra-Labs (www.dextra-labs.co.uk, Readding, UK).
NMR experiments
NMR experiments were performed on a 500 MHz Avance III Bruker spectrometer equipped with a QXI probe. The recent full assignment of human galectin-1 was used to map residues affected by the addition of the carbohydrate ligands (28). NMR titrations of the complex formation were performed by recording 15N-1H heteronuclear single quantum coherence (HSQC) using a 0.1 mM 15N-labeled galectin-1 sample in 10 mM phosphate buffer at pH 6.0. The interaction was mapped by comparing chemical shifts of bound and unbound protein. The spectral widths were 2000 Hz for 1H and 2027 Hz for 15N; 1024 data points in t2 and 64 transients for each of 128 t1 points were used.
ITC analysis
The isothermal titration microcalorimetry experiments were performed using a 2277 Thermal Activity Monitor calorimeter (Thermometric, Sweden) at 298 K with a 100 μL Hamilton injection syringe. The microcalorimeter was dynamically calibrated at the beginning of each experiment. Data acquisition and analyses were carried out using DIGITAM 4.1 software (Thermometric, Sweden). In a typical titration experiment, small aliquots (3–4 μL) of concentrated sugar dissolved in phosphate buffer (60 mM, pH 7.6) were injected into the calorimeter reaction vessel (900 μL) containing galectin-1 (200 μM) dissolved in the same buffer. Except for galactose (200 mM), the concentrated solutions for the other sugars were 20 mM. The enthalpy of interaction between galectin-1 and sugar was corrected by measuring the enthalpy change after injection of the sugar solution into buffer solution using identical procedures and experimental conditions. The heat released by galectin-1 dilution is negligible.
MD simulations
Programs used
All MD simulations were performed with the CHARMM package version 33b1 and force-field CHARMM version 22 (29,30) using a Dell precision 380 or a Linux cluster composed of eight biprocessors with hyperthreading technology. VMD was used for visualization and analyses.
Initial x-ray structures
Several x-ray structures were characterized for the unbound state of galectin-1. The C2S mutant was used because of the availability of crystallographic data of the protein, as well as many protein complexes with different ligands. We used a structure previously solved at a resolution of 1.65 Å (PDB ID: 1W6N) by Lopez-Lucendo et al. (17). As initial structures to study protein-sugar interactions, we also used the structures of galectin-1 bound to galactose (1W6M), lactose (1W6O), and N-acetyllactosamine (1W6P) characterized by the same authors. Carbohydrates exist as a mixture of α and β anomers in solution; however, in silico the anomer is fixed. We used the anomer found in the x-ray structure; therefore, the β anomer for galactose and lactose and the α anomer for N-acetyllactosamine were used in all simulations. In the case of lacto-N-biose (Gal-β-1,3-GlcNAc) and galacto-N-biose (Gal-β-1,3-GalNAc), for which no three-dimensional (3D) data were available, we generated the initial complexes by superimposing the nonreducing terminal galactose moieties in the 1W6P complex (N-acetyllactosamine), and thus used the same anomer as for N-acetyllactosamine (α). Galectin-1 structures were solved as a dimer (two molecules were present in the asymmetric unit), and only coordinates corresponding to chain A were used in the MD simulations. Nonprotein derivatives such as β-mercapto-ethanol and SO4 ions were not taken into account. S-hydroxy-cysteine was manually modified to cysteine.
Protonation of histidine and orientation of the side chains of Asn, Gln, and His residues were checked manually and using the WHAT IF web interface (http://swift.cmbi.kun.nl/). Residues His-44 and His-52 were assigned to HSP and HSD types, respectively. Crystallographic water molecules within 3 Å of the protein monomer were included in the initial model. All hydrogen atoms were included explicitly in the calculation. Coordinates of missing hydrogen atoms were added using the hbuild algorithm in CHARMM.
The galectin-1 complexes were solvated using a 63 × 63 × 63Å3 cubic water box of previously equilibrated TIP3 molecules, leading to a total average of ∼7200 water molecules (depending on the complex). Finally, sodium ions were added to ensure a neutral system.
Energy minimization
Unfavorable contacts were removed by 3000 steps of energy minimization with conjugate gradient. Harmonic constraints were applied and decreased gradually from 100 to 0 kcal/mole · Å2 to allow smooth minimization. In the first 1000 cycles, protein atoms were not allowed to move. We used the switch function for the van der Waals energy and electrostatic interactions with cuton, cutoff, and cutnb values of 9, 11, and 13 Å, respectively.
MD simulations
MD simulations were performed using Charmm-22 (26) parameters for proteins, and the GLYCAM06 parameters (27) for carbohydrates. Carbohydrate topology and parameter files were obtained in CHARMM format using the GLYCAM-Web carbohydrate builder (http://www.glycam.com/). Using this interface, it was possible to generate files for CHARMM MD simulations that were based on the GLYCAM force field.
The Shake algorithm was applied to all hydrogen-containing bonds (31), and a 1 fs integration step was used. The simulation used the periodic boundary condition approximation.
A typical trajectory was prepared as follows: The minimized initial structures were heated gradually to 300 K (30 ps), equilibrated (75 ps), and finally subjected to free MD (≈2 ns). Snapshots of the coordinates were saved every 500 steps (0.5 ps), leading to ∼4000 instantaneous conformations for each trajectory. At least two independent trajectories were generated for each complex. A typical 2 ns simulation required ∼3 weeks CPU time on a 3.5 GHz processor.
MD analyses
Trajectories were analyzed using a combination of CHARMM, in-house-written Perl scripts, and the VMD package. Overall root mean-square deviation (RMSD) variations were computed with CHARMM after superimposition of the CA atoms of galectin-1. Hydrogen bonds were identified using CHARMM scripts and a 3.0 Å cutoff. The interaction energies correspond to CHARMM energies (i.e., van der Waals and electrostatic interactions). The orientation of galactose in the CBS was measured as follows: 1), the orientation at the end of the equilibration step was used as reference; 2), all of the structures generated during the trajectory were superimposed onto the CA atoms of galectin-1; and 3), the angle between the vector formed by galactose atoms C1–C4 in the current structure and the reference was determined. The dihedral Φ/Ψ torsion angles were defined as (H1′-C1′-O-C3/C1′-O-C3-H3 and H1′-C1′-O-C4/C1′-O-C4-H4) for 1→3 and 1→4 interglycosidic linkages, respectively. The size of the binding site was estimated by measuring the distance between the center of geometries of CA atoms belonging to residues Asn-50, His-52, and Ala-55 on the one hand, and residues Ser-62, Gly-65, and Trp-68 on the other hand. Structural figures (Fig. 1; see Fig. S8 in the Supporting Material) were generated with PyMOL software (http://pymol.sourceforge.net/).
Results and Discussion
MD simulations
To investigate the characteristics of the binding of galectin-1 with various carbohydrates, we performed MD analyses of free galectin-1 and various lectin-carbohydrate complexes (Fig. S1 and Fig. S2).
MD free galectin-1
Galectin-1 (1W6N) was characterized as a dimer (17). However, since the two binding sites exhibit nearly identical structural properties, in the simulation studies we were able to focus on the protein-carbohydrate interaction of a single subunit. All MD simulations were then performed on a galectin-1 monomer solvated in a water box with periodic conditions. MD simulations of unbound galectin-1 monomer were performed first. The system was heated to 300 K and equilibrated before a free MD production run. The structure of the binding site at the end of the equilibration is shown in Fig. S3. At least two independent trajectories of 2 ns each were generated. The same procedure was used for the different simulations with galectin-1 bound to the various oligosaccharides. The results of the two independent trajectories were combined together to yield better statistics (32). The binding site is fairly well defined in the free protein and experiences only minor movements upon complexation.
Monitoring of the overall RMSD fluctuation of CA atoms during the trajectory showed that the galectin-1 3D structure was stable throughout the whole trajectory (Fig. S4). Analysis of the overall residue fluctuation during the time course of the simulation revealed that galectin-1 is a rigid structure (Fig. 2 A) because its overall fold is maintained by a network of hydrogen bonds within the two main β sheets. Apart from the N- and C-termini, the highest flexibility was observed within the two loops surrounding the CBS (residues 51–53 on one side and residues 64–66 on the other side). These loops surrounding subsites C and D (Fig. 1) contain glycine residues, which could account for their higher flexibility.
Figure 2.
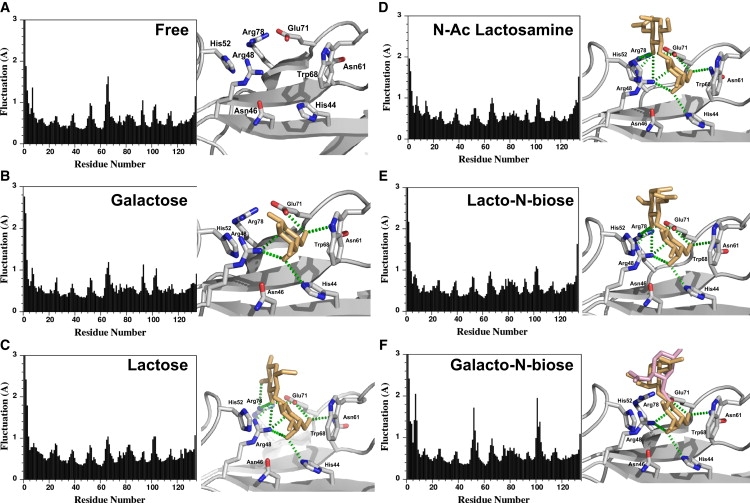
Overall fluctuation of CA atoms (left) and average structure (right) of galectin-1 for the different trajectories: (A) free galectin-1, (B) galectin-1 bound to galactose, (C) lactose, (D) N-acetyllactosamine, (E) lacto-N-biose, and (F) galacto-N-biose. Residues in close contact with the oligosaccharide ligand are shown as sticks and labeled. The carbohydrate ligand is shown as orange sticks. In the case of galacto-N-biose, two average conformations corresponding to two different populations of ligands during the trajectory are indicated as orange and pink sticks, respectively. Hydrogen bonds are indicated as green dashed lines.
Galectin-1 was crystallized in complex with different galactoside-containing oligosaccharides, namely, galactose, lactose, and N-acetyllactosamine (1W6M, 1W6O, and 1W6P, respectively). These x-ray structures were used as initial conformations to study protein-carbohydrate interactions by MD.
MD of galectin-1 with galactose
Because human galectin-1 belongs to the galactose-binding protein family, we first studied its interaction with galactose alone (1W6M). MD simulations were performed as described for unbound galectin-1. The overall RMSD fluctuation of galectin-1 CA atoms during the trajectory indicated that the overall structure was maintained during the time course of the simulations (Fig. S4). The average residue fluctuation was calculated and showed no major differences compared to free galectin-1 (Fig. 2 B). The interaction between galactose and the CBS was analyzed over the full trajectories to evaluate the stability of the carbohydrate in the binding site. Hydrogen bonds with galactose O4 atom (His-44 and Arg-48), O5 (Arg-48), and O6 (Asn-61 and Glu-71) were observed throughout the simulations (Fig. 3 A). However, the hydrogen bond between galactose O4 and His-44 was only observed 60% of the time, suggesting that the interaction with galactose was not highly stable. As an illustration, the orientation of galactose in the CBS was monitored, and the results showed that the monosaccharide was not stabilized in the CBS—it moved away from the initial binding conformation and was able to rotate freely due to the lack of stabilizing interactions (Fig. S5). Finally, the interaction energy between galactose and galectin-1 was computed with CHARMM, giving an average energy of −37 ± 9 kcal.mol−1 with a large fluctuation during the timescale of the simulation (Fig. S6).
Figure 3.

Hydrogen-bond network between galectin-1 and galactose (A), lactose (B), N-acetyllactosamine (C), lacto-N-biose (D), and galacto-N-biose during the trajectories.
MD simulations indicate that although the galactose is essential for oligosaccharide binding to human galectin-1, it cannot bind efficiently by itself. Therefore, we considered the interaction with some galactose-containing disaccharides to study the contribution of the hexopyranose unit on the reducing end of the galactose. For this, we used lactose (Galβ1-4Glc), N-acetyllactosamine (Galβ1-4GlcNAc), and galacto-N-biose (Galβ1-3GalNAc), which differ in terms of the nature of the reducing sugar unit (Fig. S1 and Fig. S2).
Importance of the reducing sugar unit
MD of galectin-1 with lactose, N-acetyllactosamine
The 3D structures solved by x-ray diffraction were used as initial conformations of the complex (1W6O and 1W6P for lactose and N-acetyllactosamine complexes, respectively). The simulations were carried out as described above. In both cases, the overall RMSD variations showed the stability of the structure during the trajectory (Fig. S4). The loops surrounding the active site were stabilized during the simulations by the addition of a second sugar to the galactose moiety, as indicated by the average residue fluctuation, especially for loop Ala-51–Asp-54 (Fig. 2, C and D, for lactose and N-acetyllactosamine, respectively).
Hydrogen bonds with OH groups at positions 4 and 6, and O5 of the galactose moiety were maintained in the interaction with both lactose and N-acetyllactosamine (Fig. 3, B and C). However, these interactions were more stable in the case of lactose and N-acetyllactosamine derivatives than with galactose alone (Table S1). Additional stable hydrogen bonds between Asn-61 and galactose 06 were observed with the two disaccharides studied. Furthermore, the reducing sugar (glucose or N-acetyl-glucosamine) was involved in hydrogen bonds with Arg-48 (subsite C), Glu-71, and Arg-73 (subsite D) at positions O3 and O4. Lactose and N-acetyllactosamine were stabilized in the CBS during the simulations and constant cavity size was maintained during the whole trajectory (Fig. S5). Contrary to what was observed with the galactose monosaccharide, the two disaccharides were well maintained in the CBS during the time course of the simulations. The trajectories displayed stable protein-carbohydrate complexes. The stability of the disaccharide was assessed by monitoring Ф and Ψ torsion angles of the interglycosidic linkage (Fig. 4, magenta and red). These dihedral angles were stable throughout the simulations with values of ∼60° and 20° for Ф and Ψ, respectively, which correspond to low-energy conformations of the disaccharides as reported previously (33). Interaction energies between galectin-1 and the two disaccharides were computed with CHARMM (Fig. S6) and are in the same range for lactose (−79 ± 3 kJ.mol−1) and N-acetyllactosamine (−75 ± 3 kJ.mol−1).
Figure 4.
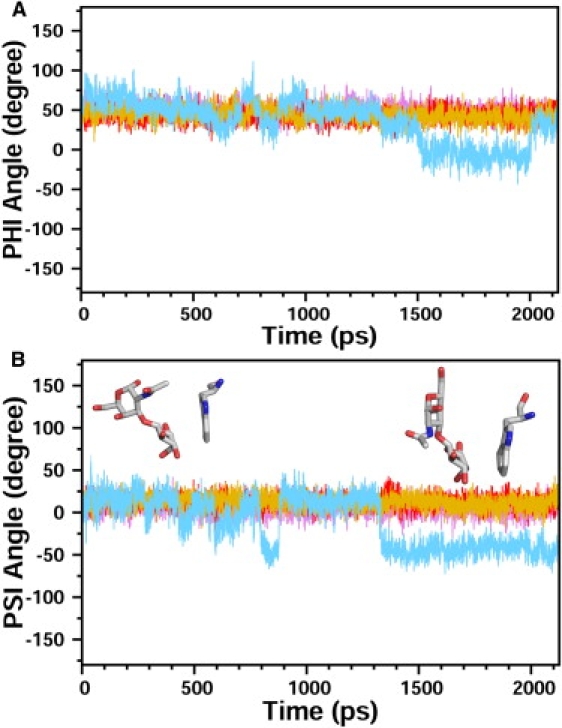
Stability of the different disaccharide sugars in the CBS. Variation of the Φ (A) and Ψ (B) torsion angles for the interglycosidic bond during the time course of the trajectory (lactose, magenta; N-acetyllactosamine, red; lacto-N-biose, gold; and galacto-N-biose, light blue). Three out of four disaccharides are stable during the whole trajectory, with Φ and Ψ angles of ∼60° and 20°, respectively. In contrast, a larger variation of these torsion angles is observed with galacto-N-biose, and different conformations can be observed. As an illustration, two orientations of galacto-N-biose relative to the conserved tryptophan residue are shown in stick representation.
MD with galacto-N-biose
In the case of galacto-N-biose (N-acetyl-neogalactosamine), no structural information about the complex was known. A complex was generated by fixing the galactose ring of galacto-N-biose in the position defined by the x-ray structure of the galectin-1/N-acetyllactosamine complex (1W6P), and a minimized binding conformation was found. The different trajectories were performed and analyzed as described above. The overall RMSD variation was similar to that of the previously studied saccharides (Fig. S4). Overall, galacto-N-biose behaved like galactose monosaccharide, as it was not stabilized in the binding site. The overall fluctuation showed a high flexibility of the loops surrounding the CBS, especially near residue His-52 (Fig. 2). At least two different populations of conformations could be observed during the different simulations (Fig. 2). Most hydrogen bonds were not preserved (Fig. 3 D and Table S1), especially for the N-acetyl-galactose reducing moiety, which mainly interacted with water at the end of the simulation, rather than with galectin-1 residues. As a consequence, this disaccharide was not strongly bound to galectin-1 and was able to move inside the binding site. Analysis of the Φ and Ψ dihedral angles showed a larger fluctuation of these torsion angles and revealed that the disaccharide could adopt alternate conformations due to the lack of stabilization of the sugar in the CBS (Fig. 4). The average interaction energy of −35 ± 5 kJ.mol−1 found between galectin-1 and galacto-N-biose indicates a weak binding (Fig. S6).
All hydrogen bonds between galectin-1 and the reducing glucose moiety observed in the presence of lactose and N-acetyllactosamine are lost in the interaction with galacto-N-biose (Table S1). The nature of the sugar on the reducing end of the galactose is important because glucose derivatives (Glc and GlcNAc) enhance binding by increasing the number of stable hydrogen bonds between the ligand and the protein.
Importance of the interglycosidic linkage
MD with lacto-N-biose
We performed MD simulations starting with lacto-N-biose (neolactosamine), which only differs from N-acetyllactosamine by the nature of the interglycosidic linkage (β1–3 vs. β1–4). Because no data were available for the structure of the complex, the ligand was docked in the CBS by superimposing the galactose moiety in the galectin-1/N-acetyllactosamine complex (1W6P). The newly formed protein-carbohydrate complex was then gradually minimized and subjected to MD as described for other galectin-sugar complexes. The different trajectories were analyzed as mentioned above and showed that lacto-N-biose behaved like lactose and N-acetyllactosamine. Although we started with a complex obtained by docking lacto-N-biose in the binding site, rather than with an x-ray structure, the disaccharide was bound tightly in the CBS in a similar manner to N-acetyllactosamine. The overall fluctuation showed that residues within loop Ala-51–Asp-54 were stabilized by the interaction with the disaccharide (Fig. 2). The stabilization was mainly due to the network of hydrogen bonds between galectin-1 and lacto-N-biose (Fig. 3 E and Table S1). The profile of hydrogen bonds observed was similar to that seen with lactose and N-acetyllactosamine. It is interesting to note, however, that the different interglycosidic linkage leads to a completely different orientation of the reducing GlcNAc sugar moiety between lacto-N-biose and N-acetyllactosamine (34). The hydrogen-bonding patterns showed that the bonds between residues Glu-71 and Arg-73 of human galectin-1 and the O3 atom of the GlcNAc unit of N-acetyllactosamine were lost in the case of the β(1–3) linkage; instead, the hydroxyl group at position O4 formed hydrogen bonds with the protein (Table S1). The conformational properties of lacto-N-biose around the interglycosidic bond were similar to those of lactose and N-acetyllactosamine, as indicated by Φ and Ψ torsion angles (Fig. 4). The ensemble of major conformations observed correspond to a low-energy population in solution (35). In addition, the size of the binding site was maintained almost constant during the entire simulation (Fig. S5). The interaction energy between galectin-1 and lacto-N-biose was in the same range as for the N-acetyllactosamine (−81 ± 2 kJ.mol−1; Fig. S6). Lacto-N-biose only differs from galacto-N-biose by the configuration of the OH group in position 4 of the reducing sugar (Fig. S1 and Fig. S2). This hydroxyl group forms hydrogen bonds with Arg-48 and Arg-73 in the complex with lacto-N-biose, whereas no stable interaction was detected with galacto-N-biose.
It is worth pointing out that the type of disaccharide (β1–3 or β1–4 linked) in subsite C and D determines the direction of the glycosidic bond leading into subsite E, and may thereby play a role in determining specificity, even in cases where the two disaccharide types bind with similar affinity by themselves. To address this point, the position of the hemiacetal OH group at the reducing end of the carbohydrate was monitored for the different carbohydrates (Fig. S8). The different simulations were performed with the anomer found in the x-ray complex when available. Since these anomers are fixed in silico, the anomer that was not used in the calculation was simply generated by exchanging the CH and COH groups of the reducing carbon atom during the analysis. The three oligosaccharide compounds that exhibit a higher affinity show a more compact clustering of the reducing terminal OH group, indicating a better defined orientation in the active site. In contrast, galacto-N-biose exhibits a more disperse distribution, which could be attributed to the configuration of the 4-hydroxyl group of the reducing sugar compared to its position in lacto-N-biose (Fig. S1 and Fig. S2). The anomeric configuration of the reducing sugar determines the orientation of a third sugar moiety. In the case of lactose and N-acetyllactosamine, extension of the disaccharide on the reducing end with β- or α-linked sugars would lead to main interactions with subsites D/E and C/D, respectively, whereas in the case of lacto-N-biose, the opposite would be observed.
To confirm the results obtained from the MD simulations, we used a combination of NMR and ITC experimental methods.
NMR experiments
NMR is a useful technique for mapping the interaction site of a ligand on a protein. In the case of galectin-1, a titration of the complex formation with various carbohydrates gives one the opportunity to observe the binding site in solution. The NMR assignment of human galectin-1 was reported very recently (28). A similar pattern was observed for the [1H-15N] HSQC NMR spectra recorded on 6His-Gal-1, and therefore the reported assignment was used to map galectin-1 residues involved in the binding of the various carbohydrates.
NMR titrations of galectin-1 complexes with the various oligosaccharides were carried out using HSQC experiments in the absence and presence of galactose, lactose, N-acetyllactosamine, lacto-N-biose, and galacto-N-biose (Fig. 5). Two different types of effects induced by the five carbohydrates were observed: galactose and galacto-N-biose had no effect on galectin-1 NMR spectra up to 10 excesses, whereas the presence of lactose, N-acetyllactosamine, and lacto-N-biose induced significant variations in the NMR spectra. Residues located in subsite C of galectin-1 (His-44, His-52, and Trp-68) were affected in the presence of the three carbohydrates, with various chemical shift variations indicating a different effect for the three molecules. Residues located in the proximity of subsite D (Gly-53, Val-59, and Ser-62) and in loops surrounding the binding site (Asn-39, Asn-40, and Gly-66) were also affected by the presence of the ligands. These chemical shift perturbations were attributed to residues in proximity of the carbohydrate in the x-ray structures and therefore corresponded to an equivalent mode of binding. However, additional chemical shift variations were observed corresponding to residues close to subsite B (Leu-34 and Gly-35) that were not in direct contact with the carbohydrate in the crystallographic structures. These data could be indicative of indirect long-distance effects or of an alternate mode of binding of the disaccharides in the CBS involving subsites B. It has been suggested that a linear tetrasaccharide composed of two lactose or N-acetyllactosamine subunits can be accommodated in the binding-site groove (21,36). One subunit could interact with subsites C and D, as seen in the x-ray structures, and the other subunit could interact with additional subsites A and B, as suggested by our NMR data. These conclusions are supported by the recent structural characterization of human galectin-9 in the presence of poly-N-acetyllactosamine, which revealed two putative modes of binding (37). The comparison of the observed chemical shift variations for the different carbohydrates suggests a higher affinity for N-acetyllactosamine than for lactose, and a poor affinity for galacto-N-biose and galactose.
Figure 5.
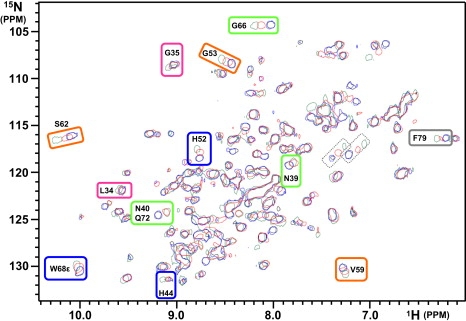
1H/15N HSQC spectra of galectin-1 (black) in the presence of 10 excess of galactose (blue), lactose (red), and N-acetyllactosamine (green). 6-His-galectin-1 was at 0.1 mM concentration in 10 mM sodium phosphate buffer, pH 6.0. Residues showing significant variations are indicated based on the recent assignment reported by Nesmelova et al. (32). The color code for subsites A–E is the same as in Fig. 1. Peaks circled by dotted lines were not assigned; they can correspond to Asn side chains near the binding site (Asn-46 and Asn-61).
ITC
The binding of the different sugars with galectin-1 was investigated using ITC. The results of a typical experiment for the binding of lactose are shown in Fig. 6. The complex formation produced an exothermic heat of binding (Fig. 6, top), providing a typical titration curve (Fig. 6, bottom). The exothermic heat of interaction decreased regularly with successive injections until saturation was achieved. In agreement with NMR experiments for galactose and galacto-N-biose, the binding affinities were too low to determine the thermodynamic parameters (Table 1). Lactose, N-acetyllactosamine, and lacto-N-biose form a 2:1 complex with galectin-1 dimer (one oligosaccharide per monomer). Affinity constants of 4896, 17067, and 6077 M−1 for lactose, N-acetyllactosamine, and lacto-N-biose, respectively, are in good accordance with the values reported previously in binding studies of different carbohydrates with bovine heart galectin-1, murine recombinant galectin-3, and human recombinant galectin-7 (38). The interaction of galectin-1 with saccharide ligands is enthalpically driven and is counterbalanced by an unfavorable entropic contribution. Such an enthalpy-entropy compensation phenomenon agrees well with results obtained for lactose and N-acetyllactosamine in wild-type hgalectin-1 and the C2S and R111H mutants (17).
Figure 6.

ITC experiment for galectin-1/lactose complex formation: heat of binding (top) and binding isotherm (bottom). The titration was obtained at 298 K for galectin-1 at 200 μM and 4 μL injections of 20 mM lactose in 60 mM phosphate buffer, pH 7.6.
Table 1.
Thermodynamic parameters obtained from ITC titrations
| Ligand (mM) | Ka (M−1) | ΔH° (kJ.mol−1) | ΔG° (kJ.mol−1) | TΔS° (kJ.mol−1) | n | |
|---|---|---|---|---|---|---|
| Gal | 200 | <1000 | −11.8 ± 0.7 | – | – | 2.0 ± 0.1 |
| Lac | 20 | 4896 ± 439 | −33.9 ± 3.3 | −21.0 ± 0.3 | −12.9 ± 3.0 | 2.2 ± 0.7 |
| LacNAc | 20 | 17067 ± 2002 | −26.3 ± 0.8 | −24.1 ± 0.3 | −2.2 ± 0.5 | 1.9 ± 0.2 |
| NeoLac | 20 | 6077 ± 452 | −21.2 ± 1.4 | −21.6 ± 0.2 | 0.4 ± 1.2 | 1.8 ± 0.1 |
| NeoGal | 20 | <1000 | nd | – | – | nd |
Thermodynamic parameters of the binding determined from microcalorimetry experiments for galactose (Gal), lactose (Lac), N-acetyllactosamine (LacNAc), lacto-N-biose (NeoLac), and galacto-N-biose (NeoGal). Values are the mean ± SE of three independent experiments.
Conclusions
In this work, we used NMR and computational methods to study galectin-1 binding to various oligosaccharide ligands. The results of the MD simulations obtained with the different saccharides allow for an interpretation of the binding affinities measured by ITC and NMR experiments, and indicate that it is possible to use MD simulations to discriminate between ligands of high and low affinities, as was shown previously for bovine galectin-1 (25). However, the interaction energies computed are only qualitative. To precisely calculate the binding energies, one would have to take into account the solvation energies and entropy using methods such as generalized Born (39) and normal-mode analyses, which was beyond the scope of this study. In addition, ITC is a macroscopic method, and the existence of alternate subsites of binding, as shown by x-ray for galectin-9 (37) and NMR (this work), is a major limitation to correlating MD simulation parameters and experimental data. MD simulations using alternate binding modes as starting conformation, such as those observed with galectin-9, may be useful to determine the contribution of each subsite to the affinity of the carbohydrate ligands.
Our results show that the galactose monosaccharide itself binds poorly to galectin-1, and a second monosaccharide moiety (at least) on the reducing end is necessary to provide a higher-affinity ligand. The interglycosidic linkage does not appear to be essential, as both N-acetyllactosamine (1–4) and lacto-N-biose (1–3) can interact with galectin-1. The different linkages lead to different orientations of the sugar in the CBS. However, the network of hydrogen bonds can adapt to optimize the interaction due to the flexibility of the saccharides. On the other hand, the nature of the reducing sugar is crucial; among the four disaccharides analyzed, only those containing a glucopyranose ring were able to bind efficiently to galectin-1. The conformation of the galacto-N-biose in the CBS led to a different orientation of the reducing sugar that could not be stabilized by the network of hydrogen bond. Finally, although no structural data were available for the complex between galectin-1 and lacto-N-biose, we were able to reproduce the efficient binding of this disaccharide. We obtained our initial structure by superimposing the nonreducing galactose moiety, which enabled us to study the galactose-containing oligosaccharides without prior knowledge of the 3D structure of the complex. Our approach allowed us to define some key interactions between galectin-1 and carbohydrate ligands that could serve as a starting point for designing new drug protocols. For example, the configuration of hydroxyl group at position 4 of the reducing sugar plays a crucial role in the stabilization of the ligand in the binding site.
Computational modeling of the sugar-lectin interaction showed that binding sites accommodate more than one carbohydrate subunit, i.e., they bind dimers and oligomers as well as monomers. In the case of linear carbohydrate chains, the free energy of binding often increases quickly for the first few carbohydrate subunits, but then the further addition of sugar subunits leads to only small increments in binding energy (40). The N-acetyllactosamine (Galβ1-4GlcNAc) and lacto-N-biose (Galβ1-3GlcNAc) disaccharide moieties studied here are commonly found epitopes in N-glycans; however, the nature of the subsequent sugars in the glycan sequence is probably more associated with the specificity of the ligand than with its affinity. MD simulation is a promising approach to address this question because it can be used to study the interaction between different members of the galectin family and various oligosaccharides.
Supporting Material
One table and nine figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01507-0.
Supporting Material
Acknowledgments
We thank Bernard Chetrit for his assistance in managing the Linux PC cluster, and Olivier Bornet for recording the heteronuclear NMR experiments. We also thank Dr. James Brown (Wellcome Trust Centre, Oxford, UK) for reading the manuscript and making valuable suggestions.
This study was supported by a grant from the French National Research Agency.
Footnotes
Christophe Meynier and Mikael Feracci contributed equally to this work.
References
- 1.Aoki-Kinoshita K.F. An introduction to bioinformatics for glycomics research. PLOS Comput. Biol. 2008;4:e1000075. doi: 10.1371/journal.pcbi.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 3.Pilobello K.T., Mahal L.K. Deciphering the glycocode: the complexity and analytical challenge of glycomics. Curr. Opin. Chem. Biol. 2007;11:300–305. doi: 10.1016/j.cbpa.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 4.DeMarco M.L., Woods R.J. Structural glycobiology: a game of snakes and ladders. Glycobiology. 2008;18:426–440. doi: 10.1093/glycob/cwn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirabayashi J., Kasai K. The family of metazoan metal-independent β-galactoside-binding lectins: structure, function and molecular evolution. Glycobiology. 1993;3:297–304. doi: 10.1093/glycob/3.4.297. [DOI] [PubMed] [Google Scholar]
- 6.Yang R.Y., Rabinovich G.A., Liu F.T. Galectins: structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008;10:e17. doi: 10.1017/S1462399408000719. [DOI] [PubMed] [Google Scholar]
- 7.Elola M.T., Wolfenstein-Todel C., Troncoso M.F., Vasta G.R., Rabinovich G.A. Galectins: matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell. Mol. Life Sci. 2007;64:1679–1700. doi: 10.1007/s00018-007-7044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camby I., Le Mercier M., Lefranc F., Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 9.Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim. Biophys. Acta. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- 10.Lahm H., Andre S., Hoeflich A., Kaltner H., Siebert H.C. Tumor galectinology: insights into the complex network of a family of endogenous lectins. Glycoconj. J. 2004;20:227–238. doi: 10.1023/B:GLYC.0000025817.24297.17. [DOI] [PubMed] [Google Scholar]
- 11.van den Brule F., Califice S., Castronovo V. Expression of galectins in cancer: a critical review. Glycoconj. J. 2004;19:537–542. doi: 10.1023/B:GLYC.0000014083.48508.6a. [DOI] [PubMed] [Google Scholar]
- 12.Salatino M., Croci D.O., Bianco G.A., Ilarregui J.M., Toscano M.A. Galectin-1 as a potential therapeutic target in autoimmune disorders and cancer. Expert Opin. Biol. Ther. 2008;8:45–57. doi: 10.1517/14712598.8.1.45. [DOI] [PubMed] [Google Scholar]
- 13.Wada J., Makino H. Galectins, galactoside-binding mammalian lectins: clinical application of multi-functional proteins. Acta Med. Okayama. 2001;55:11–17. doi: 10.18926/AMO/32029. [DOI] [PubMed] [Google Scholar]
- 14.Kato T., Ren C.H., Wada M., Kawanami T. Galectin-1 as a potential therapeutic agent for amyotrophic lateral sclerosis. Curr. Drug Targets. 2005;6:407–418. doi: 10.2174/1389450054021846. [DOI] [PubMed] [Google Scholar]
- 15.Ingrassia L., Camby I., Lefranc F., Mathieu V., Nshimyumukiza P. Anti-galectin compounds as potential anti-cancer drugs. Curr. Med. Chem. 2006;13:3513–3527. doi: 10.2174/092986706779026219. [DOI] [PubMed] [Google Scholar]
- 16.Sharon N. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim. Biophys. Acta. 2006;1760:527–537. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Lucendo M.F., Solis D., Andre S., Hirabayashi J., Kasai K. Growth-regulatory human galectin-1: crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J. Mol. Biol. 2004;343:957–970. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne Y., Bolgiano B., Liao D.I., Strecker G., Cantau P. Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat. Struct. Biol. 1994;1:863–870. doi: 10.1038/nsb1294-863. [DOI] [PubMed] [Google Scholar]
- 20.Seetharaman J., Kanigsberg A., Slaaby R., Leffler H., Barondes S.H. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution. J. Biol. Chem. 1998;273:13047–13052. doi: 10.1074/jbc.273.21.13047. [DOI] [PubMed] [Google Scholar]
- 21.Leffler H., Carlsson S., Hedlund M., Qian Y., Poirier F. Introduction to galectins. Glycoconj. J. 2004;19:433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 22.Ambrosi M., Cameron N.R., Davis B.G. Lectins: tools for the molecular understanding of the glycocode. Org. Biomol. Chem. 2005;3:1593–1608. doi: 10.1039/b414350g. [DOI] [PubMed] [Google Scholar]
- 23.Weis W.I., Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 24.Sujatha M.S., Balaji P.V. Identification of common structural features of binding sites in galactose-specific proteins. Proteins. 2004;55:44–65. doi: 10.1002/prot.10612. [DOI] [PubMed] [Google Scholar]
- 25.Ford M.G., Weimar T., Kohli T., Woods R.J. Molecular dynamics simulations of galectin-1-oligosaccharide complexes reveal the molecular basis for ligand diversity. Proteins. 2003;53:229–240. doi: 10.1002/prot.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell A.D., Bashford D., Bellott M., Dunbrack J.R.L., Evanseck J.D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesmelova I.V., Pang M., Baum L.G., Mayo K.H. 1H, 13C and 15N backbone and side-chain chemical shift assignments for the 29 kDa human galectin-1 dimer. Biomol. NMR Assign. 2008;2:203–205. doi: 10.1007/s12104-008-9121-9. [DOI] [PubMed] [Google Scholar]
- 29.Brooks B.R., Bruccoleri R.E., Olafson D.J., States D.J., Swaminathan S. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 30.MacKerel A.D., Brooks C.L., Nilsson L., Roux B., Won Y. CHARMM: The energy function and its parameterization with an overview of the program. In: Schleyer P.V.R., Allinger N.L., Clark T., Gasteiger J., Kollman P.A., Schaefer H.F., Schreiner P.R.E., editors. Encyclopedia of Computational Chemistry. John Wiley & Sons; Chichester, UK: 1998. pp. 271–277. [Google Scholar]
- 31.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–336. [Google Scholar]
- 32.Caves L.S., Evanseck J.D., Karplus M. Locally accessible conformations of proteins: multiple molecular dynamics simulations of crambin. Protein Sci. 1998;7:649–666. doi: 10.1002/pro.5560070314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Aparicio V., Sollogoub M., Bleriot Y., Colliou V., Andre S. The conformation of the C-glycosyl analogue of N-acetyl-lactosamine in the free state and bound to a toxic plant agglutinin and human adhesion/growth-regulatory galectin-1. Carbohydr. Res. 2007;342:1918–1928. doi: 10.1016/j.carres.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Lemieux R.U., Bock K., Delbaere L.T.J., Koto S., Rao V.S. The conformations of oligosaccharides related to the ABH and Lewis human blood group determinants. Can. J. Chem. 1980;56:631–653. [Google Scholar]
- 35.Vidal P., Vauzeilles B., Bleriot Y., Sollogoub M., Sinay P. Conformational behaviour of glycomimetics: NMR and molecular modelling studies of the C-glycoside analogue of the disaccharide methyl β-D-galactopyranosyl-(1→3)-β-D-glucopyranoside. Carbohydr. Res. 2007;342:1910–1917. doi: 10.1016/j.carres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Stowell S.R., Dias-Baruffi M., Penttila L., Renkonen O., Nyame A.K. Human galectin-1 recognition of poly-N-acetyllactosamine and chimeric polysaccharides. Glycobiology. 2004;14:157–167. doi: 10.1093/glycob/cwh018. [DOI] [PubMed] [Google Scholar]
- 37.Nagae M., Nishi N., Murata T., Usui T., Nakamura T. Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain. Glycobiology. 2009;19:112–117. doi: 10.1093/glycob/cwn121. [DOI] [PubMed] [Google Scholar]
- 38.Brewer C.F. Thermodynamic binding studies of galectin-1, -3 and -7. Glycoconj. J. 2004;19:459–465. doi: 10.1023/B:GLYC.0000014075.62724.d0. [DOI] [PubMed] [Google Scholar]
- 39.Dominy B.N., Brooks C.L. Methodology for protein-ligand binding studies: application to a model for drug resistance, the HIV/FIV protease system. Proteins. 1999;36:318–331. doi: 10.1002/(sici)1097-0134(19990815)36:3<318::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 40.Neumann D., Lehr C.M., Lenhof H.P., Kohlbacher O. Computational modeling of the sugar-lectin interaction. Adv. Drug Deliv. Rev. 2004;56:437–457. doi: 10.1016/j.addr.2003.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


